Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(8):1518-1526. doi:10.7150/jca.22691 This issue Cite
Research Paper
Expression of Minichromosome Maintenance Proteins (MCM) and Cancer Prognosis: A meta-analysis
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China.
Received 2017-9-4; Accepted 2018-1-19; Published 2018-4-6
Abstract
Minichromosome maintenance proteins (MCM) played a critical role in replication and cell cycle progression. However, their prognostic roles in cancer remain controversial. Therefore, we performed a meta-analysis to investigate the prognostic value of MCMs in cancers. Totally 31 eligible articles with 7653 cancer patients were included in this meta-analysis. We evaluated the relationship between MCMs expression and overall survival (OS) in various cancer patients by using pooled hazard ratios (HRs) and risk ratios (RRs) with 95% confidence intervals (CIs). The meta-analysis showed that carriers with high expression of MCM5 and MCM7 were significantly associated with short OS for pooled HR (HR=1.04, 95% CI=1.01-1.08, P=0.020, HR=1.78, 95% CI=1.04-3.02, P=0.035, respectively). For pooled RR, individuals with increased MCM2 and MCM7 expression were significantly correlated with poor OS (RR=2.30, 95% CI=1.14-4.63, P=0.019; RR=3.52, 95% CI=2.01-6.18, P<0.001, respectively). The findings suggest that high expression of MCM2, MCM5 and MCM7 might serve as predictive biomarkers for poor prognosis in cancers.
Keywords: MCM, meta-analysis, prognosis
Introduction
Based on GLOBOCAN estimates, approximately 14.1 million new cancer cases and 8.2 million deaths occurred in 2012 worldwide [1]. Although diagnosis and therapy for cancer has made great progress, the prognosis of most cancers was still poor on account of recurrence, metastasis and chemoradio-resistance [2, 3]. Clinical characteristics, such as stage, grade and histologic type, etc. are the most crucial prognostic factors to predict clinical outcomes [4]. However, individuals with the same classical parameters often end up with various outcomes [5]. In recent years, molecular classification beyond to stage and grade is just unfolding for providing accurate prediction of clinical outcome based on the expression of cancer-related biomarkers, by which treatment could be adjusted according to molecular status[6].
The MCMs are ubiquitously expressed proteins, including MCM1-10. Among these proteins, MCM2-7 form a hexamer called the MCM complex which are all AAA+ ATPases and share partial homologous sequences[7]. MCM complex plays an important role in the initiation of DNA replication. In G1-phase, MCM2-7 replicative helicase binds around double-stranded DNA (dsDNA) in the form of inactive head-to-head dimer. In S-phase, the active MCM2-7 double hexamer (MCM DH) conducts bidirectional DNA synthesis at eukaryotic origins[8]. In addition, the MCM complex contributes to replication elongation, cohesion, condensation, transcription and recombination of DNA molecule[9]. Each member of MCM complex may plays a distinct or similar role in the regulation of cell behavior. Previous evidence showed that MCM complex subunits have been implicated in cell proliferation, invasion and metastasis [10-12].
Controversial results have been reported among a variety of cancers. A number of studies demonstrated that overexpression of MCMs were found to be significantly correlated to a poor outcome in cancers, such as gastric cancer(GC) [13, 14], non-benign epithelial ovarian tumor [15], breast cancer [16] , glioma [17], non-pure-(BAC)bronchioloalveolar carcinoma [18], gallbladder [19], osteosarcoma[20] and muscle-invasive urothelial cancer [21]. High MCMs expression was correlation with high TNM stage, lymph node metastasis and recurrence in above cancers, which suggested increased expression of MCM2 might be associated with increased malignancy of these cancers. However, other studies for ER-positive breast cancer [22], small lung adenocarcinomas [23] and colorectal cancer (CRC) [24] demonstrated the potential role of MCMs overexpression in predicting better prognosis. In ER-positive breast cancer study, Ali et al. pointed out that high MCM2 expression was correlated with human epidermal growth-factor receptor 2 (HER-2)-positive, and HER-2-positive was known as a good biomarker for prognosis of breast cancer [22]. In addition, some researches indicated that MCMs expression was not related with survival of non-small cell lung cancer (NSCLC)[25] and hepatocellular carcinoma (HCC)[26]. The effects of MCMs expression in prognosis of cancers have been investigated but the results have not yet reached a consensus. Up to now, no meta-analysis has investigated the prognosis of various cancers in relation to MCMs expression. To explore whether expression of MCMs was correlated with prognosis of overall cancer and specific cancer subtypes, we performed this meta-analysis.
In our study, 31 studies with 7653 patients were included. Our results indicated that positive or high expression of MCM2, MCM5 and MCM7 may predict worse prognosis of cancers. Our results may be helpful to provide clinical evidence for effective treatment of cancer patients.
Materials and Methods
Search Strategy
Literature search was performed in the electronic platforms of PubMed, Web of Science, Cochrane Library and Embase. The last search date was October 10, 2017. The search strategy was used as: 'MCM2/CDCL1/BM28', 'MCM3/P102/RLF', 'MCM4/CDC21', 'MCM5/P1Cdc46', 'MCM6/Mis5', 'MCM7/CDC47' and 'cancer/carcinoma/neoplasm/neoplasia/tumor/tumour'. Article language was limited to English and Chinese. The references of all relevant articles were manually reviewed to find potentially relevant articles. To evaluate the quality of studies, we used the Newcastle-Ottawa Scale (NOS). We assigned the studies of high quality a scored ≥ 6 stars. The results are shown in Table 1. Two investigators assessed the eligibility of the studies independently and reached agreement by discussion.
Inclusion and Exclusion Criteria
Inclusion criteria: (1) studies concerning the survival outcomes of cancer patients with high/positive MCMs expression versus low/negative MCMs expression; (2) studies with complete information for assessment of hazard ratios (HRs) or risk ratios (RRs) and their 95% confidence intervals (CIs) for overall survival (OS); (3) original articles in English. Exclusion criteria: (1) study without sufficient data; (2) letters, editorials, case reports, reviews, comments or meeting abstracts.
Data Extraction
Two authors (Kaihua Gou and Xue Feng) extracted the data of included studies. The following extracted information was recorded: first author's name, year of publication, number of patient, ethnicity, tumor stage, detection method, cut-off value, analytical method, HRs or RRs with their 95 % CIs for OS and study quality. If the above-mentioned data was not available, items were regarded as 'not reported'.
Statistical Analyses
STATA (Version 11.0; StataCorp, College Station, TX) was used to conduct statistical analysis. Pooled HRs or RRs and their 95% CIs were calculated to measure the impact of MCMs expression on the survival of patients. I2 test and Q test were performed to assess heterogeneity among the studies (P<0.10 indicates significant heterogeneity between studies). A fixed-effect model was used to calculate the pooled HRs or RRs when heterogeneity between studies was not significant. Otherwise, a random-effect model was applied. Sensitivity analysis was carried out to explore heterogeneity when significant heterogeneity was manifested. Subgroup analyses were conducted to explore the effects of source of controls and cancer types. In addition, Egger's test and Begg's test were performed to assess publication bias among included studies. P value<0.05 was considered as statistically significant. We followed the PRISMA statement (S1) to design and report our meta-analysis.
Characteristics of eligible studies in this meta-analysis.
| Author | Year | Cancer type | Ethnicity | Number | Cutoff | TNM | U/M | Expression | Detection method | Study quality |
|---|---|---|---|---|---|---|---|---|---|---|
| MCM2 | ||||||||||
| Cheng, D. D. | 2017 | Osteosarcoma | Chinese | 129 | 2 scores | NR | M | Protein | IHC | 6 |
| Liu, Z. | 2016 | GBC | Chinese | 1060 | 25% | I-IV | M | Protein | IHC | 7 |
| Ali, H. R. | 2012 | Breast cancer | British | 1064 | 3 scores | NR | U/M | Protein | TMA & IHC | 7 |
| Yang, C. | 2012 | Gastric cancer | Chinese | 264 | 3 scores | I-IV | M | Protein | TMA & IHC | 7 |
| Zhao, D. B. | 2011 | CRC | Chinese | 306 | 51.20% | I-III | M | Protein | TMA & IHC | 6 |
| Wojnar, A. | 2011 | Breast cancer | Polish | 117 | 25% | I-IV | U | Protein | IHC | 9 |
| Fujioka, S. | 2009 | Lung ACs | Japanese | 100 | 14.60% | I | M | Protein | IHC | 7 |
| Loddo, M. | 2009 | Breast cancer | British | 182 | 30% | NR | U | Protein | IHC | 7 |
| Kayes, O. J. | 2009 | Penile carcinoma | British | 84 | 4% | I-IV | M | Protein | IHC | 6 |
| Tokuyasu, N. | 2008 | Gastric cancer | Japanese | 43 | 44% | NR | M | Protein | IHC | 8 |
| Gakiopoulou, H. | 2007 | Ovarian AC | Greek | 128 | 20% | I-IV | M | Protein | IHC | 6 |
| Yang, J. | 2006 | NSCLC | American | 128 | 25% | I-IIIA | U | Protein | IHC & WB | 6 |
| Korkolopoulou, P. | 2005 | UC | Greek | 65 | 18% | II-IV | M | Protein | IHC | 6 |
| Gonzalez, M. A. | 2004 | Breast cancer | British | 164 | 50% | NR | U | Protein | TMA & IHC | 7 |
| Hashimoto, K. | 2004 | Lung AC | Japanese | 122 | 40% | I-III | U | Protein | IHC | 7 |
| Kato, H. | 2003 | OSCC | Japanese | 93 | 62.70% | I-IV | M | Protein | IHC | 7 |
| MCM5 | ||||||||||
| Yu, S. Y. | 2014 | Oral SCC | Chinese | 97 | 60% | I-IV | M | Protein | IHC | 6 |
| Giaginis, C. | 2011 | Gastric cancer | Greek | 66 | NR | I-IV | M | Protein | IHC | 7 |
| Gakiopoulou, H. | 2007 | Ovarian cancer | Greek | 128 | 25% | I-IV | M | Protein | IHC | 6 |
| MCM7 | ||||||||||
| Almadori, G. | 2017 | Laryngeal SCC | Italian | 61 | 50% | I-IV | M | Protein | IHC | 6 |
| Karavias, D. | 2016 | HCC | Greek | 111 | 5 scores | I-IV | U | Protein | IHC | 8 |
| Deraco, M. | 2015 | DMPM | Italian | 170 | NR | NR | U | Protein | IHC & TMA | 6 |
| Zhong, X. | 2015 | OSCC | Chinese | 139 | 50% | I-IV | M | Protein | IHC & TMA | 7 |
| Hua, C. | 2014 | Glioma | Chinese | 59 | NR | II-IV | M | Protein & RNA | WB & qPCR | 6 |
| Zhong, X. | 2014 | NSCLC | Chinese | 270 | 50% | I-IV | M | Protein | IHC | 8 |
| Liu, Y. Z. | 2012 | NSCLC | Chinese | 494 | 50% & 10% | I-IV | M | Protein | IHC & ICC | 6 |
| Zhou, Y. M. | 2012 | HCC | Chinese | 87 | 30% | I-IV | M | Protein | IHC | 7 |
| Tolonen, T. T. | 2011 | Prostate cancer | Finnish | 292 | 20% | I-IV | U | Protein | IHC | 6 |
| Haruki, T. | 2011 | Lung AC | Japanese | 100 | 15.60% | NR | M | Protein | IHC | 7 |
| Hamamoto, Y. | 2010 | STS | Japanese | 109 | 17.40% | I-IV | M | Protein | IHC | 6 |
| Fujioka, S. | 2009 | Lung AC | Japanese | 100 | 20.20% | NR | M | Protein | IHC | 7 |
| Nishihara, K. | 2009 | CRC | Japanese | 191 | 58% | I-IV | U | Protein | IHC | 7 |
IHC: immunohistochemistry; TMA: tissue microarray; qPCR: quantitative PCR; WB: western blot; HR: hazard ration; H: high expression; L: low expression; P: positive expression; N: negative expression; TNM: tumor-node-metastasis; M: multivariate analysis; U: univariate analysis; NR: not reported; NSCLC: non-small cell lung cancer; SCC: squamous cell cancer; AC: adenocarcinoma; GBC: gallbladder cancer; CRC: colorectal cancer; Laryngeal SCC: laryngeal squamous cell carcinoma; HCC: hepatocellular carcinoma; DMPM: Diffuse Malignant Peritoneal Mesothelioma; OSCC: oesophageal squamous cell carcinoma; STS: soft tissue sarcomas.
Results
Study Characteristics
In total, 2813 potentially non-duplicated studies were obtained after the initial database searches. After excluding 2546 articles based on title/abstract review, 267 articles were retrieved. Then, another 217 studies were excluded after review of the full texts, including 97 articles of insufficient data, 61 articles of meeting abstract, 23 reviews, 2 article without full text, 1 non-English article and 2 articles quality ≤ 5stars. Finally, 31eligible studies which fulfilled all inclusion criteria were selected in this meta-analysis [13-24, 26-44]. The study selection procedure is outlined in Figure 1. The principal characteristics of the included studies are summarized in Table 1. The overall sample-size added up to 7653 participants. Of the 31 studies, the populations of 17 studies were Asian [13, 14, 17-20, 23, 24, 28-30, 32, 34, 37-40, 44], and the remaining 13 studies were Caucasian [15, 16, 21, 22, 26, 27, 31, 33, 35, 36, 41, 42]. Nine studies investigated the association of MCM2 expression with OS for HR [14-16, 20, 22, 24, 31, 35, 41, 42] and 8 for RR [13, 17-19, 21, 23, 29, 36] separately; 3 articles investigated the association of MCM5 with OS for HR [15, 27, 37]; 6 studies in association with MCM7 were included respectively for HR [26, 30, 38-40, 43] and 7 for RR[17, 23, 28, 33, 41, 44, 45]. The types of cancers included HCC, OSCC, NSCLC and so on. Data concerning different cancers and ethnicity were considered as separate studies in the subgroup analysis.
Association of MCMs with OS
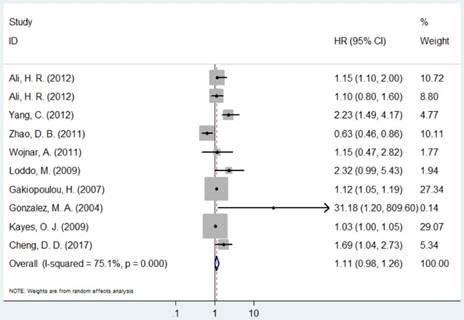
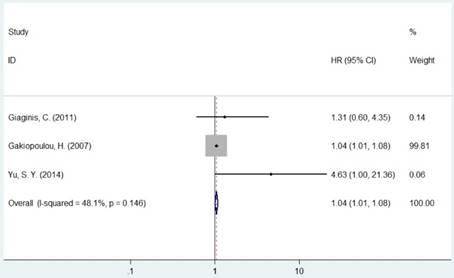
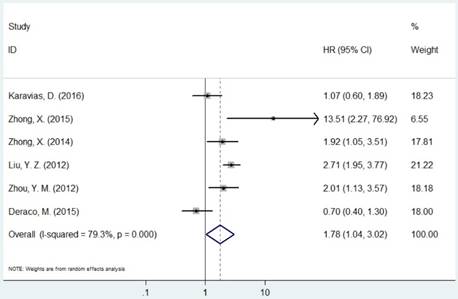
The pooled HR was presented in Table 2. Carriers with increased MCM2 expression were not associated with worse OS compared with decreased expression (HR=1.11, 95% CI=0.98-1.26, P=0.094, Figure 2). However, individuals with high expression of MCM5 and MCM7 were correlated with worse OS compared with low expression (HR=1.04, 95% CI=1.01-1.08, P=0.020; HR=1.78, 95% CI=1.04-3.02, P=0.035, respectively, Figures 4 and 5). As for ethnicity, patients with increased expression of MCM7 were significantly associated with shorter OS in Asian (HR=2.49, 95%CI=1.93-3.21, P<0.001). In regard to cancer types, the pooled HR of MCM7 high/positive expression was 2.51 in lung cancer (Table 2).
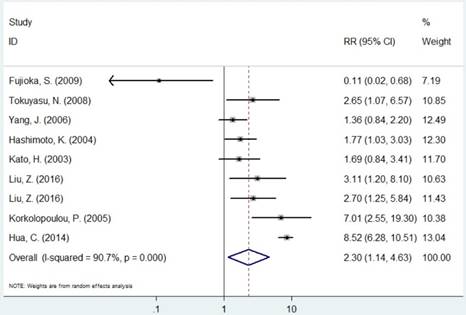
The pooled RR of included studies are presented in Table 3. Patients with increased expression of MCM2 and MCM7 were significantly associated with shorter OS (RR=2.30, 95% CI=1.14-4.63, P=0.019; RR=3.52, 95% CI=2.01-6.18, P<0.001, respectively, Figures 3 and 6). In the subgroup analysis of cancer type, increased expression of MCM2 was related to poor OS of digestive system cancer (RR=2.36, 95% CI=1.57-3.55, P<0.001) but no significant association was found for lung cancer (RR=1.01, 95% CI=0.42-2.41, P=0.992). In the comparison of OS between low/negative and high/positive MCM7 expression, low/negative was significantly associated with a better OS in Asian and Caucasian (HR=3.81, 95%CI=1.84-7.87, P<0.001; HR=2.51, 95%CI=1.57-4.00, P<0.001, respectively). In lung cancer, increased expression of MCM7 were significantly associated with shorter OS (HR=7.84, 95%CI=2.14-28.74, P=0.002).
Meta-analysis results of the association between MCMs expression and overall survival for pooled HR.
| Categories | Group/subgroup | Data set number | HR(95%CI) | P value | Model | Phet | I² (%) | |
|---|---|---|---|---|---|---|---|---|
| MCM2 | Overall | 10 | 1.11(0.98-1.26) | 0.0941 | R | <0.001 | 75.0% | |
| Digestive system cancer | 2 | 1.17(0.34-4.02) | 0.809 | R | <0.001 | 94.1% | ||
| Breast cancer | 5 | 1.20(0.97-1.48) | 0.095 | F | 0.166 | 38.2% | ||
| Asian | 3 | 1.31(0.571-3.00) | 0.525 | R | <0.001 | 91.4% | ||
| Caucasian | 7 | 1.09(1.00-1.19) | 0.061 | R | 0.030 | 57.1% | ||
| MCM5 | Overall | 3 | 1.04(1.01-1.08) | 0.020 | F | 0.146 | 48.1% | |
| MCM7 | Overall | 6 | 1.78(1.04-3.02) | 0.035 | R | <0.001 | 79.3% | |
| Digestive system cancer | 3 | 2.17(0.86-5.49) | 0.101 | R | 0.017 | 75.5% | ||
| Lung cancer | 2 | 2.51(1.88-3.45) | <0.001 | F | 0.323 | 0.0% | ||
| Asian | 5 | 2.49(1.93-3.21) | <0.001 | F | 0.169 | 40.5% | ||
R: random effect model; F: fixed effect model.
Flow diagram of studies selection procedure

Forest plot of HR for the association between MCM2 and OS (H/P vs. L/N).
Forest plot of RR for the association between MCM2 and OS (H/P vs. L/N).
Forest plot of HR for the association between MCM5 and OS (H vs. L).
Forest plot of HR for the association between MCM7 and OS (H/P vs. L/N).
Heterogeneity Test, Sensitivity Analysis, and Publication Bias
For MCMs, significant heterogeneity was observed except for MCM5, which could not be completely explained by design or subgroup analysis. Because the numbers of included studies for MCM5 was <5, we did not perform sensitivity analyses. The results of the sensitivity analysis for MCM2 and MCM7 showed that the exclusion of each single study did not change the statistical significance except MCM7 for HR.
Forest plot of RR for the association between MCM7 and OS (H/P vs. L/N).
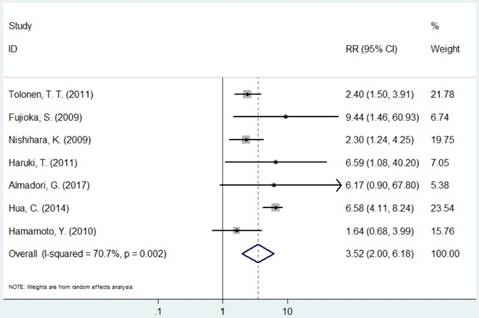
Meta-analysis results of the association between MCMs expression and overall survival for pooled RR.
| Categories | Group/subgroup | Data set number | RR(95%CI) | P value | Model | Phet | I² (%) |
|---|---|---|---|---|---|---|---|
| MCM2 | Overall | 9 | 2.30(1.14-4.63) | 0.019 | R | <0.001 | 90.7% |
| Lung cancer | 3 | 1.01(0.42-2.41) | 0.992 | R | 0.013 | 77.1% | |
| Digestive system cancer | 4 | 2.36(1.57-3.55) | <0.001 | F | 0.711 | 0.0% | |
| Asian | 7 | 2.15(0.96-4.82) | 0.064 | R | <0.001 | 90.5% | |
| Caucasian | 2 | 2.90(0.58-14.40 | 0.193 | R | 0.004 | 87.8% | |
| MCM7 | Overall | 7 | 3.50(2.01-6.18) | <0.001 | R | 0.002 | 70.7% |
| Asian | 5 | 3.81(1.84-7.87) | <0.001 | R | 0.005 | 73.1% | |
| Caucasian | 2 | 2.51(1.57-4.00) | <0.001 | F | 0.403 | 0.0% | |
| Lung cancer | 2 | 7.84(2.14-28.74) | 0.02 | F | 0.786 | 0.0% |
Publication bias.
| z value | P value | t value | P value | |
|---|---|---|---|---|
| HR | ||||
| MCM2 | 0.720 | 0.474 | 1.600 | 0.148 |
| MCM5 | 1.040 | 0.296 | 1.650 | 0.347 |
| MCM7 | 1.36 | 0.175 | 1.01 | 0.344 |
| RR | ||||
| MCM2 | 0.730 | 0.466 | -2.430 | 0.046 |
| MCM7 | 0.30 | 0.764 | -0.34 | 0.751 |
We performed the Begg's and Egger's tests to identify potential publication bias. The detailed results for publication bias test were summarized in Table 4. No significant publication bias was found in this meta-analysis.
Discussion
Since the prognosis significance of MCM family proteins in cancers is controversial, a quantitative meta-analysis is employed in our study. As far as we are concerned, this is the first meta-analysis to evaluate the correlation between expression levels of MCMs and survival of cancer. By analyzing the data extracted from 31 full-text publications, we revealed that high expression of MCM2, MCM5 and MCM7 might be associated with poor OS.
The pooled RR results showed that high MCM2 expression was associated with patients' poor OS. A number of researches have indicated the role of MCM2 in cancer development. Liu et al. reported that positive MCM2 expression was significantly correlation with high TNM stage, large tumor size, lymph node metastasis and invasion in squamous cell (SC)/adenosquamous carcinoma (ASC) and adenocarcinoma (AC) of the gallbladder [19]. Similarly, Giaginis et al. found that MCM2 expression was significantly correlation with the tumors grade, vascular invasion and Dukes' stage in CRC [46]. In addition, MCM2 expression was found to be independent predictors of recurrence in bladder cancer [47]. Mutation of TP53 is associated with a poorer prognosis and this abnormality is common in tumors with high expression of MCM2 and MCM7 [48]. These findings suggested that high MCM2 expression in cancers tends to indicate higher biological malignant aggressiveness, which are consistent with our results.
The present study pointed out that the OS of patients with high expression of MCM5 was significantly shorter than that of patients with low expression. For MCM5, significant correlation was found between the higher MCM5 expression and OSCCs with larger tumor size, higher clinical stage, higher histological grade, lymph node metastasis and deeper invasion depth [37]. Additionally, the expression levels of MCM5 were also found to be increased in advancing tumor stage of epithelial ovarian adenocarcinoma [15] and muscle-invasive urothelial cancer [21]. Similarly, it has been reported that MCM5 silencing reduced cell proliferation in human anaplastic thyroid cancer-derived cell lines [49]. In human melanocyte cell line, Sox10 inhibited proliferation by down-regulating the expression of MCM5. [50]. Estrogen receptor beta increased cell proliferation and invasion by up-regulating expression of MCM5 in bladder cancer cell lines [51]. Therefore, overexpression of MCM5 might be linked with increased proliferative rate of cancer cells. These results, at least in part, explained the neoplasms with higher level expression of MCM5 own more aggressive biological behaviors. Effective therapeutic target is very essential for the clinical treatment of cancers. Our results would provide useful information about the potential of MCM5 as a therapeutic target.
In the present study, we have found that high MCM7 expression was correlated with poor OS both in HR and RR. We suggest possibly following explanations of why MCM7 expression affected OS. The levels of MCM7 protein expression was higher in Grade II than in Grade I in meningioma [52]. Guan et al. found MCM7 expression was elevated with increased tumor grade in papillary urothelial neoplasia [53]. Feng et al. revealed that MCM7 were associated with the lymph nodes metastasis and the clinical stage in OSCC [54]. However, Ishibashi et al. conducted a study on the correlation between MCM7 expression and clinicopathological characteristics of CRC which was no statistical significance [55]. In vitro, low MCM7 expression significantly inhibited cell proliferation, colony formation and migration in esophageal carcinoma cell lines [56]. Similarly, Qu et al. indicated that MCM7 downregulation reduced proliferation by suppressing the expression of extracellular regulated kinase 2 (ERK2), ERK3, ERK4 and ERK7 which were proteins of MAPK signaling pathway in HepG2 cell line [57]. Cell proliferation, worse clinical tumor stage, positive lymph nodes metastasis and recurrence were all unfavorable cancer parameters. The relations of MCM7 expression with these factors could support our finding of its potential as a prognostic biomarker. As for different populations, the current findings suggested that MCM7 expression might be a useful predictor for prognosis in Asian patients but not in Caucasian patients. The results of different ethnic background should be confirmed by future studies.
Kwok, H. F et al. suggested that MCM2-7 gene may be closely co-regulated by common transcription factors (AML-1a, GATA-1, SRY) in breast cancer [58]. Similar to above result, our study indicated that the pooled HR or RR of MCM2, MCM5 and MCM7 were all >1, although MCM2 pooled HR do not reach statistical significance. Therefore, we point out that MCMs expression may be associated with the prognosis of cancers as a complex. However, contrary to our result, a conclusion that the presence of MCM2 protein disturbs the assembly of MCM4, MCM6, and MCM7 proteins to suppress the DNA helicase activity was draw from study on Hela cells [59]. In that case, high level expression of MCM2 would predict better prognosis. The discrepancy between above publication and our conclusion may be due to the condition that MCM2 protein modification lead to the function change of MCM4, 6 and 7 complex beside the amount of MCM2 protein.
Several limitations should be acknowledged in this meta-analysis. First, the sample size was not sufficiently large for MCM5. Second, all the studies included in the meta-analysis were published in English and Chinese, therefore publication bias might present in our study although the bias test did not show it. Third, the heterogeneity could not be totally eliminated by subgroup analysis and sensitivity analysis. The detecting methods of MCMs expression, cut-off value, source of antibodies, dilution ratios and surgical operation were different, which may cause heterogeneity between the included studies. Finally, the different survival analysis methods might affect the accuracy of outcome, although the most of the studies conducted multivariate analysis in Cox proportional hazards model.
Conclusions
In summary, this meta-analysis found that high expression of MCM2, MCM5 and MCM7 were related with worse survival for cancer patients. However, before MCMs expression are routinely used in patient management, large-scale and well-designed studies on different ethnicities are still needed to validate the results of our meta-analysis.
Acknowledgements
This study is supported by grants from Public Welfare Foundation of Liaoning Province (No. 2015005002) and the National Science and Technology Support Program (2015BAI13B07).
Author Contributions
KG. designed the research; KG JL HL XF. conducted the studies; KG JL. analyzed the data and prepared the manuscript; CX YY. guided the experiments and edited the paper. All of the authors read and approved the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87-108
2. Ferrandina G, Lauriola L, Distefano MG, Zannoni GF, Gessi M, Legge F, Maggiano N, Mancuso S, Capelli A, Scambia G. et al. Increased cyclooxygenase-2 expression is associated with chemotherapy resistance and poor survival in cervical cancer patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20(4):973-981
3. Inoue K, Slaton JW, Karashima T, Yoshikawa C, Shuin T, Sweeney P, Millikan R, Dinney CP. The prognostic value of angiogenesis factor expression for predicting recurrence and metastasis of bladder cancer after neoadjuvant chemotherapy and radical cystectomy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6(12):4866-4873
4. Margulis V, Shariat SF, Matin SF, Kamat AM, Zigeuner R, Kikuchi E, Lotan Y, Weizer A, Raman JD, Wood CG. Outcomes of radical nephroureterectomy: a series from the Upper Tract Urothelial Carcinoma Collaboration. Cancer. 2009;115(6):1224-1233
5. Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168(6):2395-2400
6. Engel RH, Kaklamani VG. HER2-positive breast cancer: current and future treatment strategies. Drugs. 2007;67(9):1329-1341
7. Costa A, Renault L, Swuec P, Petojevic T, Pesavento J, Ilves I, MacLellan-Gibson K, Fleck R, Botchan M, Berger J. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife. 2014;3:e03273
8. Hyrien O. How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000Research. 2016:5
9. Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiology and molecular biology reviews: MMBR. 2004;68(1):109-131
10. Lau KM, Chan QK, Pang JC, Li KK, Yeung WW, Chung NY, Lui PC, Tam YS, Li HM, Zhou L. et al. Minichromosome maintenance proteins 2, 3 and 7 in medulloblastoma: overexpression and involvement in regulation of cell migration and invasion. Oncogene. 2010;29(40):5475-5489
11. DaFonseca CJ, Shu F, Zhang JJ. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3034-3039
12. Zheng T, Chen M, Han S, Zhang L, Bai Y, Fang X, Ding SZ, Yang Y. Plasma minichromosome maintenance complex component 6 is a novel biomarker for hepatocellular carcinoma patients. Hepatology research: the official journal of the Japan Society of Hepatology. 2014;44(13):1347-1356
13. Tokuyasu N, Shomori K, Nishihara K, Kawaguchi H, Fujioka S, Yamaga K, Ikeguchi M, Ito H. Minichromosome maintenance 2 (MCM2) immunoreactivity in stage III human gastric carcinoma: clinicopathological significance. Gastric Cancer. 2008;11(1):37-46
14. Yang C, Wen Y, Li H, Zhang D, Zhang N, Shi X, Jiang B, Ma X, Yang P, Tang H. et al. Overexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancer. Oncology reports. 2012;27(1):135-142
15. Gakiopoulou H, Korkolopoulou P, Levidou G, Thymara I, Saetta A, Piperi C, Givalos N, Vassilopoulos I, Ventouri K, Tsenga A. et al. Minichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: relationship with cell cycle regulators and prognostic implications. British journal of cancer. 2007;97(8):1124-1134
16. Gonzalez MA, Tachibana KE, Chin SF, Callagy G, Madine MA, Vowler SL, Pinder SE, Laskey RA, Coleman N. Geminin predicts adverse clinical outcome in breast cancer by reflecting cell-cycle progression. The Journal of pathology. 2004;204(2):121-130
17. Hua C, Zhao G, Li Y, Bie L. Minichromosome Maintenance (MCM) Family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer. 2014;14:526
18. Hashimoto K, Araki K, Osaki M, Nakamura H, Tomita K, Shimizu E, Ito H. MCM2 and Ki-67 expression in human lung adenocarcinoma: prognostic implications. Pathobiology. 2004;71(4):193-200
19. Liu Z, Yang Z, Jiang S, Zou Q, Yuan Y, Li J, Li D, Liang L, Chen M, Chen S. MCM2 and TIP30 are prognostic markers in squamous cell/adenosquamous carcinoma and adenocarcinoma of the gallbladder. Molecular medicine reports. 2016;14(5):4581-4592
20. Cheng DD, Zhang HZ, Yuan JQ, Li SJ, Yang QC, Fan CY. Minichromosome maintenance protein 2 and 3 promote osteosarcoma progression via DHX9 and predict poor patient prognosis. Oncotarget. 2017;8(16):26380-26393
21. Korkolopoulou P, Givalos N, Saetta A, Goudopoulou A, Gakiopoulou H, Thymara I, Thomas-Tsagli E, Patsouris E. Minichromosome maintenance proteins 2 and 5 expression in muscle-invasive urothelial cancer: a multivariate survival study including proliferation markers and cell cycle regulators. Human pathology. 2005;36(8):899-907
22. Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Aurora kinase A outperforms Ki67 as a prognostic marker in ER-positive breast cancer. British Journal of Cancer. 2012;106(11):1798-1806
23. Fujioka S, Shomori K, Nishihara K, Yamaga K, Nosaka K, Araki K, Haruki T, Taniguchi Y, Nakamura H, Ito H. Expression of minichromosome maintenance 7 (MCM7) in small lung adenocarcinomas (pT1): Prognostic implication. Lung Cancer. 2009;65(2):223-229
24. Zhao DB, Chandler I, Chen ZM, Pan HC, Popat S, Shao YF, Houlston RS. Mismatch repair, minichromosome maintenance complex component 2, cyclin A, and transforming growth factor beta receptor type II as prognostic factors for colorectal cancer: results of a 10-year prospective study using tissue microarray analysis. Chinese medical journal. 2011;124(4):483-490
25. Werynska B, Pula B, Muszczynska-Bernhard B, Piotrowska A, Jethon A, Podhorska-Okolow M, Dziegiel P, Jankowska R. Correlation between expression of metallothionein and expression of Ki-67 and MCM-2 proliferation markers in non-small cell lung cancer. Anticancer research. 2011;31(9):2833-2839
26. Karavias D, Maroulis I, Papadaki H, Gogos C, Kakkos S, Karavias D, Bravou V. Overexpression of CDT1 Is a Predictor of Poor Survival in Patients with Hepatocellular Carcinoma. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2016;20(3):568-579
27. Giaginis C, Giagini A, Tsourouflis G, Gatzidou E, Agapitos E, Kouraklis G, Theocharis S. MCM-2 and MCM-5 expression in gastric adenocarcinoma: clinical significance and comparison with Ki-67 proliferative marker. Digestive diseases and sciences. 2011;56(3):777-785
28. Haruki T, Shomori K, Hamamoto Y, Taniguchi Y, Nakamura H, Ito H. Geminin expression in small lung adenocarcinomas: implication of prognostic significance. Lung Cancer. 2011;71(3):356-362
29. Kato H, Miyazaki T, Fukai Y, Nakajima M, Sohda M, Takita J, Masuda N, Fukuchi M, Manda R, Ojima H. et al. A new proliferation marker, minichromosome maintenance protein 2, is associated with tumor aggressiveness in esophageal squamous cell carcinoma. Journal of surgical oncology. 2003;84(1):24-30
30. Liu YZ, Jiang YY, Hao JJ, Lu SS, Zhang TT, Shang L, Cao J, Song X, Wang BS, Cai Y. et al. Prognostic significance of MCM7 expression in the bronchial brushings of patients with non-small cell lung cancer (NSCLC). Lung Cancer. 2012;77(1):176-182
31. Loddo M, Kingsbury SR, Rashid M, Proctor I, Holt C, Young J, El-Sheikh S, Falzon M, Eward KL, Prevost T. et al. Cell-cycle-phase progression analysis identifies unique phenotypes of major prognostic and predictive significance in breast cancer. British journal of cancer. 2009;100(6):959-970
32. Nishihara K, Shomori K, Tamura T, Fujioka S, Ogawa T, Ito H. Immunohistochemical expression of geminin in colorectal cancer: Implication of prognostic significance. Oncology reports. 2009;21(5):1189-1195
33. Tolonen TT, Tammela TL, Kujala PM, Tuominen VJ, Isola JJ, Visakorpi T. Histopathological variables and biomarkers enhancer of zeste homologue 2, Ki-67 and minichromosome maintenance protein 7 as prognosticators in primarily endocrine-treated prostate cancer. BJU international. 2011;108(9):1430-1438
34. Toyokawa G, Masuda K, Daigo Y, Cho HS, Yoshimatsu M, Takawa M, Hayami S, Maejima K, Chino M, Field HI. et al. Minichromosome Maintenance Protein 7 is a potential therapeutic target in human cancer and a novel prognostic marker of non-small cell lung cancer. Molecular cancer. 2011;10:65
35. Wojnar A, Pula B, Piotrowska A, Jethon A, Kujawa K, Kobierzycki C, Rys J, Podhorska-Okolow M, Dziegiel P. Correlation of intensity of MT-I/II expression with Ki-67 and MCM-2 proteins in invasive ductal breast carcinoma. Anticancer research. 2011;31(9):3027-3033
36. Yang J, Ramnath N, Moysich KB, Asch HL, Swede H, Alrawi SJ, Huberman J, Geradts J, Brooks JS, Tan D. Prognostic significance of MCM2, Ki-67 and gelsolin in non-small cell lung cancer. BMC Cancer. 2006;6:203
37. Yu SY, Wang YP, Chang JY, Shen WR, Chen HM, Chiang CP. Increased expression of MCM5 is significantly associated with aggressive progression and poor prognosis of oral squamous cell carcinoma. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2014;43(5):344-349
38. Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, Wang E, Zhang L, Han Y. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. 2015;66(2):192-200
39. Zhong X, Guan X, Liu W, Zhang L. Aberrant expression of NEK2 and its clinical significance in non-small cell lung cancer. Oncology letters. 2014;8(4):1470-1476
40. Zhou YM, Zhang XF, Cao L, Li B, Sui CJ, Li YM, Yin ZF. MCM7 expression predicts post-operative prognosis for hepatocellular carcinoma. Liver international: official journal of the International Association for the Study of the Liver. 2012;32(10):1505-1509
41. Almadori G, Lauriola L, Coli A, Bussu F. Minichromosome maintenance protein 7 and geminin expression: Prognostic value in laryngeal squamous cell carcinoma in patients treated with radiotherapy and cetuximab. 2017, 39(4):684-693.
42. Kayes OJ, Loddo M, Patel N, Patel P, Minhas S, Ambler G, Freeman A, Wollenschlaeger A, Ralph DJ, Stoeber K. et al. DNA replication licensing factors and aneuploidy are linked to tumor cell cycle state and clinical outcome in penile carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(23):7335-7344
43. Deraco M, Cabras A, Baratti D, Kusamura S. Immunohistochemical Evaluation of Minichromosome Maintenance Protein 7 (MCM7), Topoisomerase IIalpha, and Ki-67 in Diffuse Malignant Peritoneal Mesothelioma Patients Using Tissue Microarray. Annals of surgical oncology. 2015;22(13):4344-4351
44. Hamamoto Y, Shomori K, Nosaka K, Haruki T, Teshima R, Ito H. Prognostic significance of Minichromosome maintenance protein 7 and Geminin expression in patients with 109 soft tissue sarcomas. Oncology Letters. 2010;1(4):703-709
45. Nishihara K, Shomori K, Fujioka S, Tokuyasu N, Inaba A, Osaki M, Ogawa T, Ito H. Minichromosome maintenance protein 7 in colorectal cancer: implication of prognostic significance. International journal of oncology. 2008;33(2):245-251
46. Giaginis C, Georgiadou M, Dimakopoulou K, Tsourouflis G, Gatzidou E, Kouraklis G, Theocharis S. Clinical significance of MCM-2 and MCM-5 expression in colon cancer: association with clinicopathological parameters and tumor proliferative capacity. Digestive diseases and sciences. 2009;54(2):282-291
47. Burger M, Denzinger S, Hartmann A, Wieland WF, Stoehr R, Obermann EC. Mcm2 predicts recurrence hazard in stage Ta/T1 bladder cancer more accurately than CK20, Ki67 and histological grade. British journal of cancer. 2007;96(11):1711-1715
48. Qiu WG, Polotskaia A, Xiao G, Di L, Zhao Y, Hu W, Philip J, Hendrickson RC, Bargonetti J. Identification, validation, and targeting of the mutant p53-PARP-MCM chromatin axis in triple negative breast cancer. NPJ breast cancer. 2017:3
49. Mio C, Lavarone E, Conzatti K, Baldan F, Toffoletto B, Puppin C, Filetti S, Durante C, Russo D, Orlacchio A. et al. MCM5 as a target of BET inhibitors in thyroid cancer cells. Endocrine-related cancer. 2016;23(4):335-347
50. Su Z, Zheng X, Zhang X, Wang Y, Zhu S, Lu F, Qu J, Hou L. Sox10 regulates skin melanocyte proliferation by activating the DNA replication licensing factor MCM5. Journal of dermatological science. 2017;85(3):216-225
51. Hsu I, Chuang KL, Slavin S, Da J, Lim WX, Pang ST, O'Brien JH, Yeh S. Suppression of ERbeta signaling via ERbeta knockout or antagonist protects against bladder cancer development. Carcinogenesis. 2014;35(3):651-661
52. Winther TL, Torp SH. MCM7 expression is a promising predictor of recurrence in patients surgically resected for meningiomas. Journal of neuro-oncology. 2017;131(3):575-583
53. Guan B, Wang X, Yang J, Zhou C, Meng Y. Minichromosome maintenance complex component 7 has an important role in the invasion of papillary urothelial neoplasia. Oncology letters. 2015;10(2):946-950
54. Feng CJ, Li HJ, Li JN, Lu YJ, Liao GQ. Expression of Mcm7 and Cdc6 in oral squamous cell carcinoma and precancerous lesions. Anticancer research. 2008;28(6a):3763-3769
55. Ishibashi Y, Kinugasa T, Akagi Y, Ohchi T, Gotanda Y, Tanaka N, Fujino S, Yuge K, Kibe S, Yoshida N. et al. Minichromosome maintenance protein 7 is a risk factor for recurrence in patients with Dukes C colorectal cancer. Anticancer research. 2014;34(8):4569-4575
56. Qiu YT, Wang WJ, Zhang B, Mei LL, Shi ZZ. MCM7 amplification and overexpression promote cell proliferation, colony formation and migration in esophageal squamous cell carcinoma by activating the AKT1/mTOR signaling pathway. Oncology reports. 2017;37(6):3590-3596
57. Qu K, Wang Z, Fan H, Li J, Liu J, Li P, Liang Z, An H, Jiang Y, Lin Q. et al. MCM7 promotes cancer progression through cyclin D1-dependent signaling and serves as a prognostic marker for patients with hepatocellular carcinoma. Cell death & disease. 2017;8(2):e2603
58. Kwok HF, Zhang SD, McCrudden CM, Yuen HF, Ting KP, Wen Q, Khoo US, Chan KY. Prognostic significance of minichromosome maintenance proteins in breast cancer. American journal of cancer research. 2015;5(1):52-71
59. Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272(39):24508-24513
Author contact
![]() Corresponding authors: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, ShenyangCity, 110001, Liaoning Province, China. E-mail address: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening, Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: yuanyuanedu.cn
Corresponding authors: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, ShenyangCity, 110001, Liaoning Province, China. E-mail address: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening, Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact