3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(9):1548-1559. doi:10.7150/jca.24566 This issue Cite
Review
The Emerging Role of Circular RNAs in Hepatocellular Carcinoma
1. Institute of Life Sciences, Jiangsu University, Zhenjiang 212013, Jiangsu, China
2. Department of Pharmacy, College of Life Sciences, China Jiliang University, Hangzhou 310018, Zhejiang, China
3. School of Pharmacy, Jiangsu University, Zhenjiang 212013, Jiangsu, China
Received 2017-12-26; Accepted 2018-2-12; Published 2018-4-12
Abstract
Hepatocellular carcinoma (HCC) ranks the third leading cause of cancer death in the world and has a notably low survival rate. Circular RNAs (circRNAs) are newly classed non-coding RNA (ncRNA) members that are capable of regulating gene expression at transcription or post-transcription levels. Recent studies demonstrate that some circRNAs are differentially expressed in HCC, and the deregulation of these circRNAs is associated with the clinical pathological and prognostic significance. They also play essential roles in HCC progression, and contribute to cell proliferation, migration, invasion and metastasis by targeting different microRNAs (miRNAs) and protein-coding genes. In this review, we concentrate on recent progress of some important circRNAs in HCC, with an emphasis on their deregulation, functions and regulatory mechanisms, and discuss their potential utility as diagnostic and/or prognostic biomarkers or therapeutic targets for HCC.
Keywords: circRNA, HCC, liver cancer, miRNA sponge, biomarker, proliferation
Introduction
Hepatocellular carcinoma (HCC), the most common malignant liver cancer, ranks the third leading cause of cancer death in the world and has a notably low survival rate [1]. Although advances in clinical and experimental settings in HCC, the survival rate of HCC patients remains quite low after five years [2]. Majority of HCC patients are only diagnosed at advanced stage before any curative therapy could be applied; for those patients after surgical resection and liver transplantation, relapse and metastasis are frequently happened; whereas for those patients who are not eligible for surgery or with metastasis, chemotherapy and radiotherapy do not contribute significantly to cure HCC, while these therapies have disadvantage of significant side effects. To improve survival outcome of HCC patients, comprehensive approaches are needed to find novel biomarkers for the early diagnosis of HCC, as well as novel therapeutic targets for the development of potential therapeutics against HCC.
Non-coding RNAs (ncRNAs) are functional transcripts that are not translated into proteins. Based on the transcript size, ncRNAs may be grouped into two major classes: small ncRNAs (18-200 nucleotides) and long ncRNAs (lncRNAs, exceeding 200 nucleotides). Small ncRNAs called microRNAs (miRNAs, generally 22-25 nucleotides) bind to miRNA response elements (MREs) on the 3'-untranslated regions (UTRs) of their target messenger RNAs (mRNAs) to promote mRNA degradation or repress protein translation; whereas lncRNAs interact with DNA, RNA or proteins, and regulate gene expression at multiple levels, including chromatin, transcription, post-transcription and translation [3-7]. To date, dozens of miRNAs and lncRNAs have been reported to be deregulated in HCC and contribute to HCC progression [8-11].
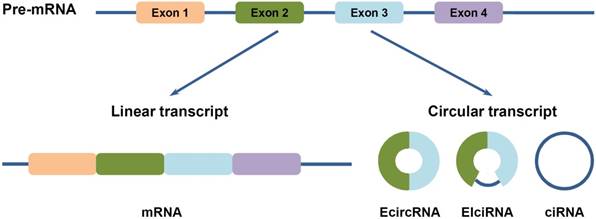
Three types of circular RNAs (circRNAs). CircRNAs are generated from backsplicing of exons, introns, or both. Accordingly, they are classified into three types: (1) EcircRNAs are composed of exons. They are predominatly cytoplasmic, may harbor miRNA response elements (MREs) and serve as miRNA sponges. (2) EIciRNAs are circularized with introns retained between the exons. (3) CiRNAs are composed of introns. Both of EIciRNAs and ciRNAs are abundant in the nucleus and may play functional roles on gene transcription and post-transcription.
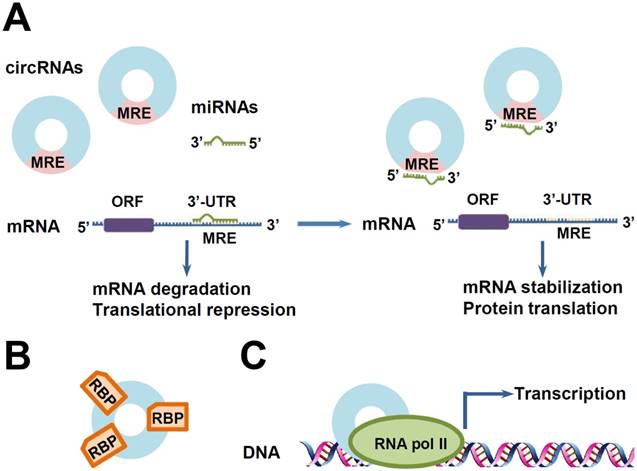
Some functions of circRNAs. (A) CircRNAs as miRNA sponges. MicroRNAs (miRNAs) promote mRNA degradation or repress protein translation by binding to MREs on the 3'-untranslated regions (UTRs) of their target mRNAs. CircRNAs containing shared MREs can competitively absorb miRNAs like sponges, resulting in the reduction of miRNAs. Then, the reduced miRNAs exhibit less inhibitory effects on their target genes, leading to the over-expression of these genes. (B) CircRNAs as RNA-binding protein (RBP) sponges. (C) Transcription regulation. CircRNAs can interact with RNA polymerase II (RNA Pol II) machinery and promote gene transcription.
Circular RNAs (circRNAs) are newly classed ncRNA members, which are generated from backsplicing of exons, introns, or both, to form circular exonic circRNAs (EcircRNAs), circular intronic RNAs (ciRNAs) and exon-intron circRNAs (EIciRNAs) (Figure 1) [12-14]. Unlike the better known linear RNAs, circRNAs form a covalently closed continuous loop structures with neither 5'-3' polarity nor a polyadenylated tail, which prevent them from degradation by RNA exonucleases or RNase R [12,15]. Hence, circRNAs are more stable than linear mRNAs, which make them more abundant than their canonical linear transcripts from the same genes [12]. Furthermore, they are highly conserved in different species, and also show tissue specific and developmental stage-specific features [12,13,16]. Moreover, circRNAs are capable of regulating gene expression at transcription or post-transcription levels: EcircRNAs, which make up of the majority of circRNAs, are predominantly cytoplasmic, and may harbor MREs and serve as miRNA sponges; whereas ciRNAs and EIciRNAs are abundant in the nucleus, and may regulate gene transcription and post-transcription (Figure 2) [12,13,16,17]. Besides, they have been found to exist in different extracellular body fluids, including saliva, blood and urine [18]. These suggest that circRNAs may be involved in different biological processes, such as cell proliferation, invasion and metastasis, apoptosis and autophagy, etc., and thus play important roles in HCC progression.
Recent studies demonstrate that circRNAs have essential roles in cancer progression, and contribute to cell proliferation, invasion and metastasis, and apoptosis of cancer by targeting different miRNAs and protein-coding genes [19-22]. For instance, circRNA_LARP4 can act as a miRNA sponge for miR-424, enhancing the expression of miR-424-target gene large tumor suppressor kinase 1 (LATS1), thus promoting proliferation and invasion of gastric cancer cells [19]. Circ-foxo3 can function by interacting with both miRNAs and proteins. By binding to and forming ternary complex with cyclin-dependent kinase (CDK) 2 and CDK inhibitor (CDKI) p21, circ-foxo3 arrested the function of CDK2 and inhibited cell cycle progression; whereas by sponging multiple miRNAs that regulate the production of Foxo3 mRNA, this circRNA promoted the expression of tumor suppressor Foxo3, leading to reduced cancer cell proliferation and survival [20,21]. Another circRNA, ci-mcm5, however, can serve as a positive regulator of Pol II machinery and enhance the expression of its parent gene minichromosome maintenance (MCM) 5, a protein which was associated with more advanced pathological stage and poorer prognosis of ovarian adenocarcinoma [14,22]. In HCC, several circRNAs have been found to be frequently deregulated, and the expression of some of these circRNAs is associated with the clinical pathological and prognostic significance. Moreover, some of them were found to regulate HCC progression by modulating cell proliferation, apoptosis, invasion and metastasis. Herein, we review recent advances of some important circRNAs in HCC, with an emphasis on their deregulation, functions and mechanisms of action, and discuss their potential significance as diagnostic and/or prognostic biomarkers and therapeutic targets for HCC (Table 1, Table 2) [23].
Down-regulated CircRNAs in HCC
CircMTO1
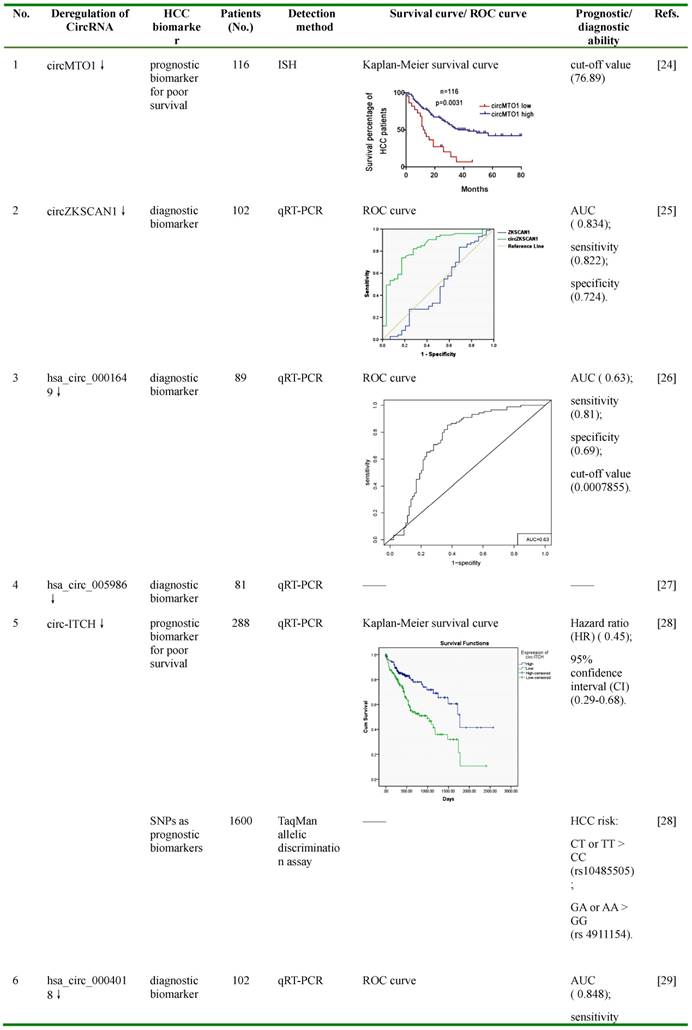
circMTO1, also known as hsa_circ_0007874 or hsa_circ_104135, was reported to be significantly down-regulated in HCC tissues compared with normal controls, and its lower expression was associated with poor prognosis of HCC patients with a cut-off value of 76.89 [24]. By bioinformatic prediction and RNA immunoprecipitation (RIP) assay using circMTO1 specific probe, miR-9, an up-regulated miRNA in HCC and an inducer of HCC cell proliferation and invasion, was found to be associated with circMTO1 [24,40-42]. Further fluorescence in situ hybridization (FISH) analysis showed that miR-9 was co-localized with circMTO1 in the cytoplasm, and this co-localization was reduced in HCC tissues compared with matched non-tumor tissues [24]. In HCC cell lines, silencing of circMTO1 significantly increased the expression of miR-9, promoted cell proliferation and invasion, and greatly reduced apoptosis; whereas over-expression of circMTO1 increased apoptosis [24]. Further studies suggested that the expression of p21, a target of miR-9 which inhibits cell cycle and proliferation, could be regulated by circMTO1 and miR-9 [24]. Silencing of circMTO1 or overexpression of miR-9 significantly reduced, while promotion of circMTO1 greatly increased, the expression of p21 [24].
Deregulated circRNAs in hepatocellular carcinoma (HCC) and their functions
| No. | CircRNA | Alias | Deregulation | Gene symbol | Genome position | Function | genes/ proteins affected | Refs. |
|---|---|---|---|---|---|---|---|---|
| 1 | circMTO1 | hsa_circ_0007874, hsa_circ_104135 | ↓ | MTO1 | chr6:74175931-74176329 | proliferation(-) ; invasion(-); apoptosis(+) | ┤miR-9, →p21 | [24] |
| 2 | circZKSCAN1 | hsa_circ_0001727 | ↓ | ZKSCAN1 | chr7:99621041-99621930 | proliferation(-); migration and invasion(-) | —— | [25] |
| 3 | hsa_circ_0001649 | hsa_circ_001599 | ↓ | SHPRH | chr6:146209155-146216113 | —— | ┤MMP9, MMP10, MMP13 | [26] |
| 4 | hsa_circ_005986 | —— | ↓ | PRDM2 | chr1:14057494-14068652 | proliferation (-) | ┤miR-129-5p, →Notch1 | [27] |
| 5 | circ-ITCH | hsa_circ_0001141, hsa_circ_001763 | ↓ | ITCH | chr20:33001547-33037285 | —— | —— | [28] |
| 6 | hsa_circ_0004018 | —— | ↓ | SMYD4 | chr17:1703150-1704318 | —— | —— | [29] |
| 7 | hsa_circ_0003570 | —— | ↓ | FAM53B | chr10:126370175-126384781 | —— | —— | [30] |
| 8 | circHIPK3 | hsa_circ_0000284 | ↑ | HIPK3 | chr11:33307958-33309057 | proliferation (+); migration (+) | ┤miR-124-3p, →IL6R, DLX2, AQP3 | [31,32] |
| 9 | hsa_circ_100338 | —— | ↑ | SNX27 | chr1: 151638888-151639119 | migration and invasion(+) | ┤miR-141-3p | [33] |
| 10 | hsa_circ_0005075 | —— | ↑ | EIF4G3 | chr1: 21377358-21415706 | —— | —— | [34] |
| 11 | hsa_circ_000839 | hsa_circ_0000497 | ↑ | SLAIN1 | chr13:78293666-78327493 | —— | —— | [35] |
| 12 | ciRS-7 | CDR1as, hsa_circ_0001946 | ↑or↓? | CDR1 | chrX:139865339-139866824 | proliferation(+) ; invasion(+); | ┤miR-7, →CCNE1, PIK3CD, EGFR | [36-38] |
| 13 | circARSP91 | hsa_circ_0085154 | —— | PABPC1 | Chr8:101721360-101721451 | proliferation(-) | —— | [39] |
"┤": inhibitory roles; "→": stimulatory roles.
circRNA: circular RNA; MMP: matrix metallopeptidase; Notch1: Notch homolog 1; IL6R: interleukin 6 receptor; DLX2: distal-less homeobox 2; AQP3: aquaporin 3; CCNE1: cyclin E1; PIK3CD: phosphoinositide 3-kinase catalytic subunit delta; EGFR: epidermal growth factor receptor; Refs: references.
Deregulated circRNAs in HCC as biomarkers
Figures in this table were reprinted by permission from Ref. 24-30,33,34, and 36.
ROC: receiver operating characteristic; AUC: area under ROC curve; ISH: In situ hybridization; qRT-PCR: quantitative reverse transcription PCR; SNP: single nucleotide polymorphism; censored: refers to survival cases at the end of follow-up; HR: hazard ratio; CI: confidence interval.
However, after silencing circMTO1, miR-9 inhibition significantly reduced the down-regulation of p21, and blocked the circMTO1 silencing-mediated promotion of proliferation and inhibition of apoptosis, of HCC cells [24]. In in vivo studies of HCC-bearing nude mouse model, intratumoral injection of cholesterol-conjugated circMTO1 siRNAs significantly promoted tumor growth and serum alpha-fetoprotein (AFP) levels [24]. Results from RT-PCR and immunohistochemistry staining demonstrated that, in tumor tissues after intratumoral circMTO1 knockdown, the expression of circMTO1 and p21 was greatly reduced, while the expression of proliferating cell nuclear antigen (PCNA) and matrix metalloproteinase (MMP) 2, two markers for cell proliferation and invasion, was significantly enhanced [24]. These data suggested that circMTO1 may suppress HCC progression by sponging miR-9 to increase the expression of miR-9 targeted gene p21. CircMTO1 could be used as a prognosis biomarker for poor survival of HCC patients, and as a potential therapeutic target for inhibiting HCC tumor growth.
CircZKSCAN1
CircZKSCAN1, also known as hsa_circ_0001727, was particularly abundant in human brain and liver, and was generated from the exons 2 and 3 of a zinc finger with KRAB and SCAN domains 1 (ZKSCAN1, also named ZNF139)[43]. In HCC tissues and cell lines, ZKSCAN1 gene is expressed in both linear (ZKSCAN1 mRNA) and circular (circZKSCAN1) forms of RNA, and both of these forms were found to be significantly lower compared with negative controls [25]. Unlike ZKSCAN1, the low expression of which was only reversely correlated to tumor size, the low expression of circZKSCAN1 was associated with multiple clinical parameters, including tumor numbers, cirrhosis, vascular invasion, microscopic vascular invasion, as well as the tumor grade [25]. Compared with the mRNA of ZKSCAN1, which showed an area under the receiver operating characteristic curve (AUC) of 0.474, the AUC of circZKSCAN1 was much higher (0.834), with the sensitivity and specificity of 0.822 and 0.724, respectively. Further in vitro studies by MTS, transwell migration and invasion assays revealed that over-expression of either ZKSCAN1 mRNA or circZKSCAN1 sharply reduced, whereas knocking down one of them greatly enhanced, the proliferation, migration and invasion of HCC cells [25]. The results from the in vivo experiments by subcutaneously injection of circZKSCAN1 over-expressing or knocking-down HCC cells in nude mice showed similar trends on tumor volumes and weights [25]. Cellular location analysis using FISH assay indicated that circZKSCAN1 was most likely located in the cytoplasm, which suggests that it may function as a competitive endogenous RNA (ceRNA) and participate in the regulatory network of the cell [25]. To further understand the underlying mechanisms by which circZKSCAN1 modulated cell proliferation, migration and invasion, RNA-seq was carried out to identify multiple differentially expressed genes in circZKSCAN1 knocking-down HCC cells. KEGG enrichment analysis suggested that these genes were more likely to be enriched in phosphoinositide-3-kinase (PI3K), migration, actin cytoskeleton, adhesion, and cytokine interation pathway [25]. Some of these were further validated by RT-PCR, including apoptosis-related genes (RAC2, EFNA3, caspase 3, BCL2), proliferation-related genes (TGFB1, ITGB4, CXCR4, survivin, CCND1), and invasion and metastasis-related genes (PDK1, MYB, CDH5, COL3A1) [25]. These results indicate that circZKSCAN1 may serve as a promising biomarker for HCC diagnosis, and may also be a potential target for HCC treatment.
Hsa_circ_0001649
Hsa_circ_0001649 was generated from the exons 26-29 of SNF2 histone linker PHD RING helicase (SHPRH), an E3 ubiquitin-protein ligase involved in DNA repair [44,45]. It was found to be down-regulated in several cancers, including HCC, gastric cancer and cholangiocarcinoma, and has shown its potential as a diagnostic marker and a tumor suppressor [26,46,47]. In HCC tissues, it was found to be significantly down-regulated compared with adjacent liver tissues, and the expression of this circRNA was correlated with tumor size and the occurrence of tumor embolus [26]. The AUC of hsa_circ_0001649 was 0.63, with the cut-off value, sensitivity, and specificity of 0.0007855, 0.81 and 0.69, respectively [26]. Further studies demonstrated that knockdown of hsa_circ_0001649 by siRNA in HCC cells greatly increased the expression of several MMPs, such as MMP9, MMP10, and MMP13, indicating the possible role of hsa_circ_0001649 in regulating the invasion and metastasis of HCC [26,48-50]. Bioinformatic analysis reveals that hsa_circ_0001649 have potential binding sites of several RNA-binding proteins (RBPs), including U2 auxiliary factor 65 kDa subunit (U2AF65), Eukaryotic initiation factor 4A-III (EIF4A3) and Regulator of nonsense transcripts 1 (UPF1), implying that it may function as a protein sponge or a transcription regulator [26]. These findings suggest that hsa_circ_0001649 may serve as a potential diagnostic biomarker of HCC, and might participate in the progression of HCC.
Hsa_circ_0005986
Another circRNA, hsa_circ_0005986, was also down-regulated in HCC tissues and HCC cell lines, and its lower expression was correlated with chronic hepatitis B family history, tumor diameters, microvascular invasion and Barcelona Clinic Liver Cancer (BCLC) stage [27]. Further studies demonstrated that knocking down of hsa_circ_0005986 increased miR-129-5p levels, leading to reduced expression of its target gene Notch homolog 1(NOTCH1), a receptor which plays an important role in HCC carcinogenesis and metastasis [27,51-54]. However, knocking down of miR-129-5p significantly promoted, while over-expression of this miRNA greatly inhibited, the expression of hsa_circ_0005986 and NOTCH1 [27]. These indicated that hsa_circ_0005986 may function as a ceRNA by sponging miR-129-5p and leading to the over-expression of its targeted gene Notch1. Functional studies revealed that knocking down of hsa_circ_0005986 in HCC cells accelerated G0/G1 to S phase transition, and thus promoted cell proliferation [27]. These indicated that hsa_circ_0005986 may be used as a potential diagnostic biomarker, and could also be an effective target for reducing HCC tumor growth.
Circ-ITCH
ITCH, an E3 ubiquitin protein ligase involved in ubiquitin-mediated protein degradation, has been reported to inhibit Wnt/β-catenin pathway, which plays important roles in HCC progression and associates with the susceptibility of chemotherapy of HCC cells [55-59]. Circ-ITCH, a circRNA spans exons 6-13 of gene ITCH, was recently demonstrated to be down-regulated and contribute to the progression of several cancers, including esophageal squamous cell carcinoma, lung and colorectal cancer [60-62]. By acting as miRNA sponges for miR-7, miR-17, and miR-214, circ-ITCH increased the expression of its parential gene ITCH, leading to suppression of Wnt/β-catenin signaling pathway and inhibition of cancer cell proliferation [60-61]. Recently, circ-ITCH was found to be significantly down-regulated in HCC tissues compared with matched adjacent tissues, while higher expression of circ-ITCH was correlated with favorable survival of HCC patients (hazard ratio (HR) =0.45; 95% confidence interval (CI) =0.29-0.68) [28]. Furthermore, two single nucleotide polymorphisms (SNPs) of circ-ITCH, including rs10485505 and rs4911154, were associated with HCC risk [28]. For rs10485505, genotype CT (odds ratio (OR) =1.18; 95% CI =1.03-1.35) and TT (OR =1.40; 95% CI =1.04-1.88) were greatly associated with increased HCC risk compared with the genotype CC; while for rs4911154, genotype GA (OR =1.27; 95% CI =1.10-1.46) and AA (OR =1.74; 95% CI =1.21-2.49) were markedly associated with increased HCC risk compared with the genotype GG [28]. These data suggested that circ-ITCH and its genetic variation may serve as biomarkers for HCC prognosis and susceptibility.
Hsa_circ_0004018
Hsa_circ_0004018 is transcribed from SMYD4, a potential tumor suppressor [63]. Hsa_circ_0004018 was reported to be decreased in HCC tissues and HCC cell lines compared with negative controls, and its expression was correlated with AFP level, tumor diameters, differentiation, barcelona clinic liver cancer (BCLC) stage and tumor-node-metastasis (TNM) stage [29]. Furthermore, hsa_cir_0004018 expression exerted HCC-stage-specific characteristics: it was gradually decreased from chronic hepatitis (F0-3), to liver cirrhosis (F4), and then to HCC tissues [29]. The AUC of hsa_circ_0004018 was 0.848 (95% CI =0.803-0.894), with the Youden index, sensitivity, and specificity of 0.531, 0.716 and 0.815, respectively. Bioinformatic analysis revealed that hsa_circ_0004018 might function as miRNA sponges for cancer-related miRNAs, such as hsa-miR-30e-5p and hsa-miR-626, thus participate in the carcinogenesis and metastasis of HCC [29]. These data indicated that hsa_cir_0004018 might be used as a potential diagnostic biomarker of HCC, and may also be involved in HCC development and progression.
Hsa_circ_0003570
Similar like hsa_circ_0004018, hsa_cir_0003570 also showed down-regulated expression in HCC tissues, and its down-regulation was correlated with serum AFP level, tumor diameters, differentiation, microvascular invasion, BCLC stage and TNM stage [30]. Its expression showed the same HCC-stage-specific characteristics with gradual decrease from chronic hepatitis, to liver cirrhosis, and to HCC tissues [30]. Hsa_cir_0003570 had poor performance for differentiating HCC from liver cirrhosis and chronic hepatitis, while had relatively good performance for differentiating liver cirrhosis from chronic hepatitis [30]. For the diagnosis of HCC, the AUC of hsa_circ_0003570 was 0.70, with the cut-off value, sensitivity, and specificity of 12.24, 0.449, and 0.868, respectively; whereas for the diagnosis of cirrhosis, the AUC of this circRNA was 0.778, with the cut-off value, sensitivity, and specificity of 11.28, 0.697, and 0.831, respectively. These data indicate that hsa_circ_0003570 might serve as a potential diagnostic biomarker of HCC.
Up-regulated CircRNAs in HCC
CircHIPK3
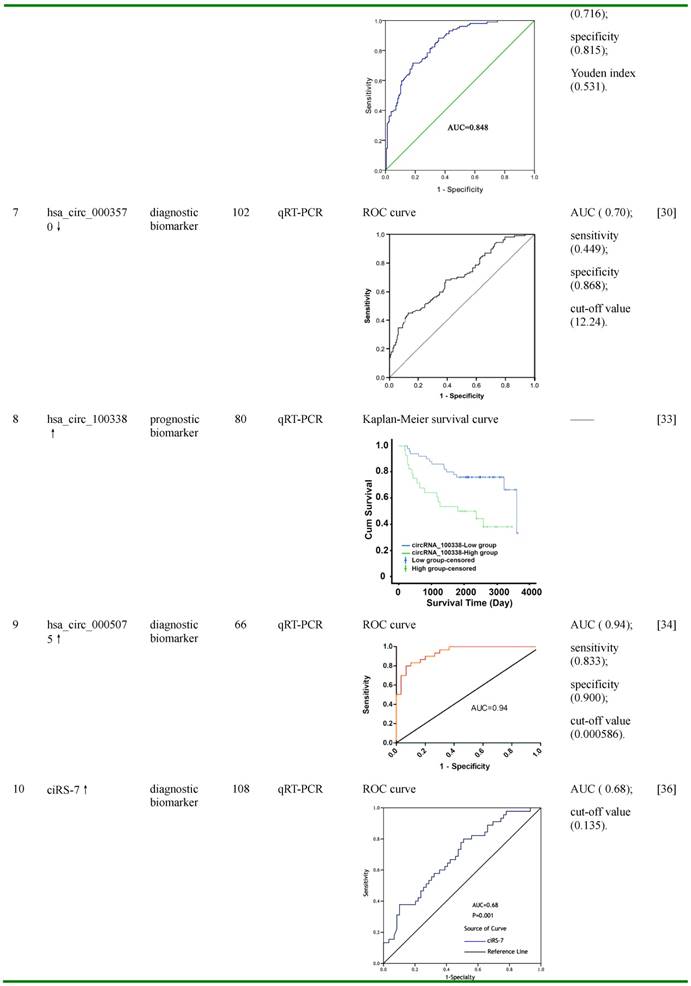
CircHIPK3 was generated from the exon 2 of HIPK3, a serine-threonine kinase which regulates transcription and apoptosis, and associates with multidrug resistance of cancer cells [43,64,65]. It was more abundant than its linear form in various tissues, such as brain, lung, heart, stomach, and colon, and particularly enriched in the brain [31]. QRT-PCR and FISH results suggested that this circRNA preferentially located in cytoplasm [31]. CircHIPK3 was found to be significantly up-regulated in HCC and bladder cancer tissues compared with matched normal tissues, and deregulation of this circRNA significantly impact on the proliferation, migration and invasion of cancer cells [31,32]. Bioinformatic prediction, RIP and luciferase reporter assays revealed that circHIPK3 may serve as a sponge for multiple miRNAs, such as miR-124, miR-558, miR-193a, and miR-379 [31,66]. Among them, miR-124, which was reported to inhibit cell proliferation and tumor growth in HCC by targeting phosphoinositide 3-kinase catalytic subunit alpha (PIK3CA) and signal transducer and activator of transcription 3 (STAT3), was found to be interacted with and co-localized with circHIPK3 in HeLa cells [31,67,68]. By acting as a miRNA sponge for miR-124, circHIPK3 significantly increased the expression of aquaporin 3 (AQP3), interleukin 6 receptor (IL6R), and distal-less homeobox 2 (DLX2), three proliferation- and migration-promoted targets of miR-124, resulting in enhanced proliferation and migration; whereas knocking down of circHIPK3 increased the expression of miR-124, leading to reduced expression of the three miR-124-target genes [31,32]. These suggested that circHIPK3 may function as a sponge of miR-124-3p, and may serve as a potential therapeutic target to reduce HCC tumor growth.
Hsa_circ_100338
Hsa_circ_100338, a circRNA screened by circRNA microarray and later validated by qRT-PCR, was found to be significantly up-regulated in HCC tissues compared with pericancerous liver tissues, and the expression of this circRNA was closely correlated with metastatic progression and the low cumulative survival rate in HCC patients [33]. Bioinformatic prediction, in silico analysis and dual-luciferase reporter assay demonstrated that miR-141, a tumor suppressor in various types of cancers, was down-regulated in HCC tissues, and was likely to interact with hsa_circ_100338 [33,69,70]. Further studies revealed that over-expression of hsa_circ_100338 increased migratiory and invasive ability of HCC cells at least partially by antagonizing miR-141-3p [33]. These results indicated that hsa_circ_100338 was a potential prognostic biomarker for survival rate and malignancy in HCC, and may also be involved in the regulation of metastatic progression of HCC.
Hsa_circ_0005075
Another up-regulated circRNA in HCC tissues, hsa_circ_0005075, was also originally selected by circRNA microarray and later validated by qRT-PCR [34]. Further analysis demonstrated that the expression of hsa_cir_0005075 was correlated with HCC tumor size, and showed good diagnostic potential for distingusing HCC tissues from adjacent non-tumor tissues [34]. The AUC of hsa_circ_0005075 was 0.94, with the cut-off value, sensitivity, and specificity of 0.000586, 0.833 and 0.900, respectively [34]. Bioinformatics analyses of GO and KEGG pathway indicate that hsa_circ_0005075 is associated strongly with cell adhesion, a biological process involved in cancer cell proliferation and metastasis [34]. Furthermore, using TargetScan and miRanda softwares, several miRNAs were predicted to be potentially interacted with hsa_circ_0005075, including hsa-miR-23b-5p, hsa-miR-93-3p, hsa-miR-581 and hsa-miR-23a-5p [34]. This suggested that hsa_circ_0005075 may function as a miRNA sponge regulating the more comprehensive circRNA-miRNA-mRNA network, and thus facilitate HCC progression [34]. These results indicate that hsa_cir_0005075 may serve as a potential biomarker with good sensitivity and specificity.
CircRNA_000839
MiR-200b is a metastasis suppressor miRNA, and has been reported to inhibit HCC progression by targeting B-cell specific moloney leukemia virus insertion site 1 (BMI1), Zinc finger E-box-binding homeobox 1 (ZEB1), DNA methyltransferase 3a (DNMT3a), Ras homologue A (RhoA), etc. [71-73]. Hsa_circ_000839 was predicted to function as miR-200b sponge impacting on the expression of RhoA [35]. In HCC tissues, the expression of hsa_circ_000839 and RhoA was significantly higher than in normal liver tissues, while miR-200b was significantly lower [35]. Furthermore, the expression of both hsa_circ_000839 and RhoA was inversely correlated with that of miR-200b, whereas the expression of has_circ_000839 was positively correlated with that of RhoA [35]. These data suggest that hsa_circ_000839 might be involved in miR-200b-mediated inhibition of HCC progression.
Miscellaneous CircRNAs in HCC
CiRS-7
CiRS-7 (circular RNA sponge for miR-7), also known as Cdr1as (cerebellar degeneration-related protein 1 antisense RNA) or hsa_circ_0001946, is a very special and unique circRNA, which is not derived from pre-mRNA and contains only repeat elements [74]. It harbors more than 70 convential binding sites and functions as a super sponge of miR-7, an important tumor suppressor which inhibits HCC progression by directly targeting multiple proteins, including phosphoinositide 3-kinase catalytic subunit delta (PIK3CD), ribosomal protein S6 kinase beta-1 (p70S6K, S6K1), voltage-dependent anion channel 1 (VDAC1), Cullin 5 (CUL5), cyclin E1 (CCNE1), epidermal growth factor receptor (EGFR), etc. [36,75-78]. CiRS-7 has been reported to participate in the progression of several cancers, including gastric cancer, colorectal cancer and HCC by sponging miR-7 [36,79,80]. Recently, the expression of ciRS-7 was detected in HCC tissues by qRT-PCR and microarray analysis. Yu et al. showed that ciRS-7 was up-regulated in 74% (26/35) HCC tissues compared with their matched non-tumor tissues [37], while results from Xu et al and Yang et al. demonstrated that this circRNA was down-regulated in 60.2% (65/108) and 73.2% (30/41) HCC tissues, respectively [36,38]. Although the deregulation of ciRS-7 in HCC tissues remains controversial, it was consistent that the higher expression of this circRNA was associated with the deterioration of HCC, such as serum AFP level (≥400 ng/μL) and hepatic microvascular invasion (AUC was 0.68 with the cut-off value of 0.135) [36-38]. In HCC tissues, higher expression of ciRS-7 was inversely correlated with that of miR-7, while it was positively correlated with miR-7-targeted genes PIK3CD and p70S6K, two components in PIK3CD/ p70S6K /mTOR pathway [36]. Further studies demonstrated that over-expression of ciRS-7 greatly promoted cell proliferation and G1/S phase transition; while knocking-down of ciRS-7 significantly reduced the proliferation and invasion of HCC cells [37,38]. CiRS-7 may function in HCC cell proliferation and invasion by inhibiting miR-7 levels to enhance the expression of miR-7-targeted genes CCNE1, PIK3CD and EGFR [37,38]. These data suggest that ciRS-7 may be used as a diagnostic biomarker of hepatic microvascular invasion, and inhibition of ciRS-7 may reduce HCC tumor growth and invasion.
CircARSP91
Androgen receptor (AR) is believed to play a role in HCC initiation and progression [81,82]. Most recently, AR and adenosine deaminase acting on RNA 1 (ADAR1), a negative regulator of circRNA expression, were found to be abnormally up-regulated and positively correlated in HCC tissues, and higher expression of ADAR1 was positively correlated with worse prognosis [39,83]. AR could globally inhibit circRNA expression by transcriptional activating the expression of ADAR1 p110 [39]. Among the down-regulated circRNAs by AR, circARSP91 (circRNA of AR suppressed PABPC1 91bp, hsa_circ_0085154), which originated from exon 9 of polyadenylate-binding protein 1 (PABPC1), was found to impact on HCC progression [39]. In in vitro studies using MTT and colony formation assays, over-expression of circARSP91 significantly reduced, whereas knocking-down of this circRNA greatly increased, the proliferation of HCC cells [39]. In in vivo studies using orthotopic implanted mouse model of HCC, over-expression of circARSP91 markedly reduced tumor volumes and weights [39]. These suggested that over-expression of circARSP91 may inhibit HCC progression by reducing the proliferation of HCC cells.
Conclusion and Future Perspectives
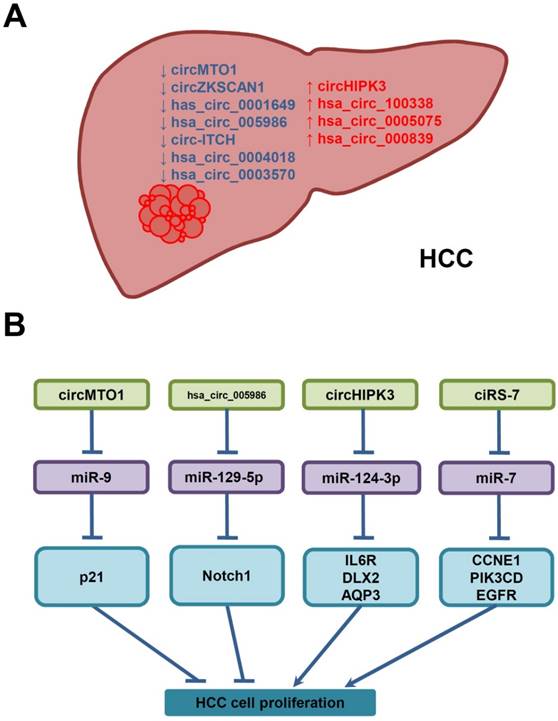
Growing evidences indicate that some circRNAs are differentially expressed in HCC tissues, and their deregulation is correlated with clinicopathological features in HCC patients. By functioning as miRNA sponges, RBP sponges, and transcriptional regulators, these circRNAs modulate the expression of target miRNAs and proteins which are associated with cell proliferation, invasion and metastasis, and contribute to HCC progression. Regarding to their unique advantages, such as better stability, higher abundance, and existing in different body fluids, circRNAs are promising diagnostic/prognosic biomarkers and good therapeutic targets for HCC. Specially, circRNAs are enriched and stable in serum-derived exosomes, which contain a specific cargo of protein and RNAs that could be transferred from tumor cells to recipient cells and thus modulate behaviors of recipient cells, suggesting that circRNAs may serve as non-invasive circulating biomarkers for cancer diagnosis, and may contribute to cancer progression [84-86]. Furthermore, a fusion-circRNA has been reported to play a role in therapy resistance of cancer, which is one of the main obstacles encountered in cancer chemotherapy and may contribute to the failure of existing therapies to eradicate malignant tumors [87]. However, it is still unknown whether any circRNA, including fusion-circRNA, is involved in therapy resistance of HCC. Compared with coding RNAs, mRNAs and lncRNAs, the studies of circRNAs in HCC are just on the way. Until now, only a small number of functional circRNAs have been identified and characterized in HCC (Figure 3A), Among these circRNAs, only part of them have been investigated mechanistically, while most of these studies focused on their functions as miRNA sponges in the proliferation of HCC cells (Figure 3B). For the vast majority of circRNAs waiting to be investigated, their biogenesis, degradation, cellular locations, functions and mechanisms of action still needed to be elucidated. Further understanding of the relationship between circRNAs and the aetiology of HCC, their functions and molecular mechanisms would deepen our understanding of human diseases, and also provide novel applications for better diagnosis/ prognosis and treatments of HCC.
Deregulated circRNAs in HCC, and their underlying molecular mechanisms in the proliferation of HCC cells. (A) CircRNAs are down- (↓, blue) or up-regulated (↑, red) in HCC. (B) Deregulated circRNAs may interfere with HCC cell proliferation by sponging miRNAs and increasing the expression of miRNA-target genes.
Abbreviations
HCC: hepatocellular carcinoma; ncRNA: non-coding RNA; lncRNA: long non-coding RNA; MRE: miRNA response element; circRNA: circular RNA; EcircRNA: exonic circRNA; ciRNA: circular intronic RNA; EIciRNA: exon-intron circular RNA; RBP: RNA-binding protein; CDK: cyclin-dependent kinase; AFP: alpha-fetoprotein; MMP: matrix metalloproteinase; ROC: receiver operating characteristic; AUC: area under ROC curve; SNP: single nucleotide polymorphism; BCLC stage: barcelona clinic liver cancer stage; TNM stage: tumor-node-metastasis stage.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81402840, 31572467, 31402016); the Natural Science Foundation of Jiangsu Province, China (BK20130495); Natural Science Foundation of Zhejiang Province, China (LY14H310010); the Training Project of Young Backbone Teachers of Jiangsu University, and Jiangsu University Senior Professional Science Foundation (11JDG120).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-87
2. Takayama T. Surgical treatment for hepatocellular carcinoma. Jpn J Clin Oncol. 2011;41:447-54
3. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
4. Zhao J, Sun BK, Erwin JA. et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750-56
5. Pandey RR, Mondal T, Mohammad F. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232-46
6. Wang X, Arai S, Song X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126-30
7. Khandelwal A, Bacolla A, Vasquez KM. et al. Long non-coding RNA: a new paradigm for lung cancer. Mol Carcinog. 2015;54:1235-51
8. Huang S, He X. The "Macro" world of microRNAs in hepatocellular carcinoma. Front Oncol. 2015;5:68
9. Xie K, Zhang Y, Liu J. et al. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176-92
10. Qiu L, Tang Q, Li G. et al. Long non-coding RNAs as biomarkers and therapeutic targets: recent insights into hepatocellular carcinoma. Life Sci. 2017;191:273-82
11. Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15:249-56
12. Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
13. Salzman J, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777
14. Zhang Y, Zhang XO, Chen T. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
15. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331-42
16. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
17. Yu B, Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus. 2016;7:155-66
18. Qu S, Yang X, Li X. et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-8
19. Zhang J, Liu H, Hou L. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151
20. Du WW, Yang W, Liu E. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acid Res. 2016;44:2846-58
21. Yang W, Du WW, Li X. et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-31
22. Gakiopoulou H, Korkolopoulou P, Levidou G. et al. Minichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: relationship with cell cycle regulators and prognostic implications. Br J Cancer. 2007;97:1124-34
23. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666-70
24. Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
25. Yao Z, Luo J, Hu K. et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422-37
26. Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161-9
27. Fu L, Chen Q, Yao T. et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878-88
28. Guo W, Zhang J, Zhang D. et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169-77
29. Fu L, Yao T, Chen Q. et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405-16
30. Fu L, Wu S, Yao T. et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2017 e22239
31. Zheng Q, Bao C, Guo W. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
32. Chen G, Shi Y, Liu M. et al. CircHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175
33. Huang XY, Huang ZL, Xu YH. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428
34. Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore). 2016;95:e3811
35. Wang BG, Li JS, Liu YF. et al. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol. 2017;39:1010428317719577
36. Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27
37. Yu L, Gong X, Sun L. et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One. 2016;11:e0158347
38. Yang X, Xiong Q, Wu Y. et al. Quantitative Proteomics Reveals the Regulatory Networks of Circular RNA CDR1as in Hepatocellular Carcinoma Cells. J Proteome Res. 2017;16:3891-902
39. Shi L, Yan P, Liang Y. et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171
40. Sun Z, Han Q, Zhou N. et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7:884-94
41. Cai L, Cai X. Up-regulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in patients with hepatocellular carcinoma. Diagn Pathol. 2014;9:1000
42. Drakaki A, Hatziapostolou M, Polytarchou C. et al. Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer. 2015;15:542
43. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-47
44. Unk I, Hajdu I, Fatyol K. et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103:18107-12
45. Lin JR, Zeman MK, Chen JY. et al. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42:237-49
46. Xu Y, Yao Y, Zhong X. et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455-61
47. Li WH, Song YC, Zhang H. et al. Decreased expression of hsa_circ_0001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698
48. Li T, Zhu Y, Han L. et al. VEGFR-1 activation-induced MMP-9-dependent invasion in hepatocellular carcinoma. Future Oncol. 2015;11:3143-57
49. Jin D, Tao J, Li D. et al. Golgi protein 73 activation of MMP-13 promotes hepatocellular carcinoma cell invasion. Oncotarget. 2015;6:33523-33
50. Sze KM, Chu GK, Lee JM. et al. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131-9
51. Giovannini C, Minguzzi M, Genovese F. et al. Molecular and proteomic insight into Notch1 characterization in hepatocellular carcinoma. Oncotarget. 2016;7:39609-26
52. Jue C, Lin C, Zhisheng Z. et al. Notch1 promotes vasculogenic mimicry in hepatocellular carcinoma by inducing EMT signaling. Oncotarget. 2017;8:2501-13
53. Gao J, Xiong Y, Wang Y. et al. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int J Oncol. 2016;48:329-37
54. Sui C, Zhuang C, Sun D. et al. Notch1 regulates the JNK signaling pathway and increases apoptosis in hepatocellular carcinoma. Oncotarget. 2017;8:45837-47
55. Perry WL, Hustad CM, Swing DA. et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143-146
56. Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010;22:717-27
57. Wei W, Li M, Wang J. et al. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol Cell Biol. 2012;32:3903-12
58. Zheng Y, Jiang L, Hu Y. et al. Metallothionein 1H (MT1H) functions as a tumor suppressor in hepatocellular carcinoma through regulating Wnt/beta-catenin signaling pathway. BMC Cancer. 2017;17:161
59. Huang M, Chen C, Geng J. et al. Targeting KDM1A attenuates Wnt/beta-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 2017;398:12-21
60. Li F, Zhang L, Li W. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001-13
61. Wan L, Zhang L, Fan K. et al. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/beta-Catenin Pathway. Biomed Res Int. 2016;2016:1579490
62. Huang G, Zhu H, Shi Y. et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225
63. Hu L, Zhu YT, Qi C. et al. Identification of Smyd4 as a potential tumor suppressor gene involved in breast cancer development. Cancer Res. 2009;69(9):4067-72
64. Begley DA, Berkenpas MB, Sampson KE. et al. Identification and sequence of human PKY, a putative kinase with increased expression in multidrug-resistant cells, with homology to yeast protein kinase Yak1. Gene. 1997;200:35-43
65. Curtin JF, Cotter TG. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J Biol Chem. 2004;279:17090-100
66. Li Y, Zheng F, Xiao X. et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646-59
67. Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426:247-52
68. Lu Y, Yue X, Cui Y. et al. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem Biophys Res Commun. 2013;441:873-9
69. Liep J, Kilic E, Meyer HA. et al. Cooperative Effect of miR-141-3p and miR-145-5p in the Regulation of Targets in Clear Cell Renal Cell Carcinoma. PLoS One. 2016;11:e0157801
70. Wang M, Hu M, Li Z. et al. miR-141-3p functions as a tumor suppressor modulating activating transcription factor 5 in glioma. Biochem Biophys Res Commun. 2017;490:1260-7
71. Tsai SC, Lin CC, Shih TC. et al. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol Carcinog. 2017;56:2035-47
72. Li XY, Feng XZ, Tang JZ. et al. MicroRNA-200b inhibits the proliferation of hepatocellular carcinoma by targeting DNA methyltransferase 3a. Mol Med Rep. 2016;13:3929-35
73. Wong CM, Wei L, Au SL. et al. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget. 2015;6:13658-70
74. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
75. Ma C, Qi Y, Shao L. et al. Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life. 2013;65:1026-34
76. Zhang X, Hu S, Wang L. et al. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2014;443:1078-84
77. Chen YJ, Chien PH, Chen WS. et al. Hepatitis B Virus-Encoded X Protein Downregulates EGFR Expression via Inducing MicroRNA-7 in Hepatocellular Carcinoma Cells. Evid Based Complement Alternat Med. 2013;2013:682380
78. Wang F, Qiang Y, Zhu L. et al. MicroRNA-7 downregulates the oncogene VDAC1 to influence hepatocellular carcinoma proliferation and metastasis. Tumour Biol. 2016;37:10235-46
79. Pan H, Li T, Jiang Y. et al. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem. 2018;119:440-6
80. Weng W, Wei Q, Toden S. et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:3918-28
81. De Maria N, Manno M, Villa E. Sex homones and liver cancer. Mol Cell Endocrinol. 2002;193:59-63
82. Kanda T, Jiang X, Yokosuka O. Androgen receptor signaling in hepatocellular carcinoma and pancreatic cancers. World J Gastroenterol. 2014;20:9229-36
83. Ivanov A, Memczak S, Wyler E. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170-7
84. Li Y, Zheng Q, Bao C. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
85. Dou Y, Cha DJ, Franklin JL. et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982
86. Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One. 2016;11:e0148407
87. Guarnerio J, Bezzi M, Jeong JC. et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289-302
Author contact
![]() Corresponding author: Li-Peng Qiu, E-mail: tulip_lipengcom and Ke-Ping Chen, E-mail: kpchenedu.cn.
Corresponding author: Li-Peng Qiu, E-mail: tulip_lipengcom and Ke-Ping Chen, E-mail: kpchenedu.cn.

 Global reach, higher impact
Global reach, higher impact