Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(10):1754-1764. doi:10.7150/jca.24569 This issue Cite
Research Paper
Alternative splicing events implicated in carcinogenesis and prognosis of colorectal cancer
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China.
Received 2017-12-26; Accepted 2018-3-3; Published 2018-4-19
Abstract
Background: Emerging evidence suggested that aberrant alternative splicing (AS) is pervasive event in development and progression of cancer. However, the information of aberrant splicing events involved in colorectal carcinogenesis and progression is still elusive.
Materials and Methods: In this study, splicing data of 499 colon adenocarcinoma cases (COAD) and 176 rectum adenocarcinoma (READ) with clinicopathological information were obtained from The Cancer Genome Atlas (TCGA) to explore the changes of alternative splicing events in relation to the carcinogenesis and prognosis of colorectal cancer (CRC). Gene interaction network construction, functional and pathway enrichment analysis were performed by multiple bioinformatics tools.
Results: Overall, most AS patterns were more active in CRC tissues than adjacent normal ones. We detected altogether 35391 AS events of 9084 genes in COAD and 34900 AS events of 9032 genes in READ, some of which were differentially spliced between cancer tissues and normal tissues including genes of SULT1A2, CALD1, DTNA, COL12A1 and TTLL12. Differentially spliced genes were enriched in biological process including muscle organ development, cytoskeleton organization, actin cytoskeleton organization, biological adhesion, and cell adhesion. The integrated predictor model of COAD showed an AUC of 0.805 (sensitivity: 0.734; specificity: 0.756) while READ predictor had an AUC of 0.738 (sensitivity: 0.614; specificity: 0.900). In addition, a number of prognosis-associated AS events were discovered, including genes of PSMD2, NOL8, ALDH4A1, SLC10A7 and PPAT.
Conclusion: We draw comprehensive profiles of alternative splicing events in the carcinogenesis and prognosis of CRC. The interaction network and functional connections were constructed to elucidate the underlying mechanisms of alternative splicing in CRC.
Keywords: alternative splicing, colorectal cancer, carcinogenesis, prognosis.
Introduction
A majority of genes within the human genome are alternatively spliced to generate multiple transcripts, often encoding proteins with different or opposite function[1, 2]. Under the circumstance of normal conditions, alternative splicing (AS) is precisely regulated to produce diverse protein isoforms for the demands of complex biological process[3, 4]. If disordered, however, tumour cells generate aberrant proteins with inserted, missing, or altered functional domains which lead to tumorigenesis[5]. There is accumulating evidence that aberrant AS is pervasive event in development and progression of cancer[6].
Colorectal cancer (CRC), one of the most frequently detected cancers in digestive tract, is the third leading cause of cancer-related deaths worldwide[7]. Genovariation has been proved to be involved in the CRC development[8]. In addition to commonly mutated genes such as APC [9] and TP53[10], aberrant pre-mRNA splicing becomes another event that reflect abnormalities of CRC cells. Therefore, medicine targeting pathologic splicing events which influence CRC occurrence and survival, or strategies to alter post-translational modifications of splicing regulatory proteins might shed new light on CRC treatment.
Given the importance of AS, a number of recent studies focused on the role of aberrant splicing in CRC. For instance, SRSF6 functions the essential roles in mediating CRC progression via modulating AS[11]. As a histone methyltransferase, SETD2 regulates alternative splicing through epigenetic regulation of RNA processing to inhibit intestinal tumorigenesis[12]. In addition, HNRNPLL has been reported to be a novel metastasis suppressor of CRC, and affects CD44 alternative splicing process of epithelial-mesenchymal transition[13]. PRPF6, a member of the trisnRNP (small ribonucleoprotein) spliceosome complex, induces CRC proliferation through preferential splicing of multiple genes related with growth regulation[14].
Despite recent advances of disordered AS in colorectal cancer, the entire picture of aberrant splicing events involved in colorectal carcinogenesis and progression is still elusive. Considering the enormous amount of uncharacterized AS events, the reported ones are probably only the tip of the iceberg of biologically relevant splicing events. Here, we draw comprehensive alternative splicing profiles of Colon Adenocarcinoma (COAD) and Rectum Adenocarcinoma (READ) by analysing RNA-seq data. Splicing network was constructed by integrated bioinformatics analysis in order to provide functional insight into the full repertoire of AS in the initiation and development of CRC.
Materials and Methods
Access of raw data
The detailed information of colorectal cancer patients was downloaded from The Cancer Genome Atlas (TCGA), a public available database (cancergenome.nih.gov) which is collaboration between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) that has generated comprehensive, multi-dimensional maps of the critical genomic changes in 33 types of cancer[15]. Raw data of more than 11,000 patients with tumor tissue and matched normal tissues were stored in TCGA dataset. In this study, data of 499 colon adenocarcinoma cases (TCGA-COAD) and 176 rectum adenocarcinoma (TCGA-READ) with clinicopathological information was obtained to explore the changes of alternative splicing events in relation to the carcinogenesis and prognosis of CRC. For TCGA samples, neoadjuvant treatment was not allowable: TCGA's goal was to accelerate the understanding of the underlying genomics of primary untreated tumors, and cancer treatment can involve mutagens or carcinogens which could cloud the origin of the cancer.
Identification of alternative splicing events
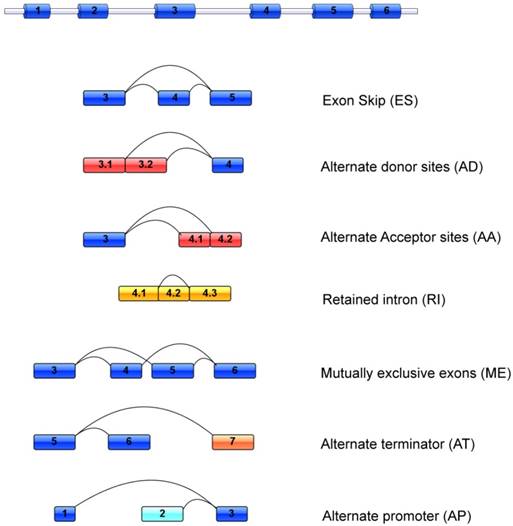
Seven common patterns of alternative splicing events include Alternate Acceptor site (AA), Alternate Donor site (AD), Alternate Promoter (AP), Alternate Terminator (AT), Exon Skip (ES), Mutually Exclusive Exons (ME), and Retained Intron (RI), which was visualized in Figure 1. In this study, the different splicing types of COAD and READ were classified by TCGA SpliceSeq, a resource for investigation of cross-tumor and tumor-normal alterations in mRNA splicing patterns of RNASeq data[16]. With an interactive, visual results viewer, SpliceSeq could be used to investigate the transcriptome of samples and perform comparative analysis to identify significant changes in alternative splicing. For each sample and every possible splice event, percent-splice-in (PSI) value was calculated, which is the ratio of normalized read counts indicating inclusion of a transcript element over the total normalized reads for that event (both inclusion and exclusion reads). Changes in average PSI values when comparing groups of samples mean a shift in splicing patterns between the groups or a splice event.
Gene interaction network construction
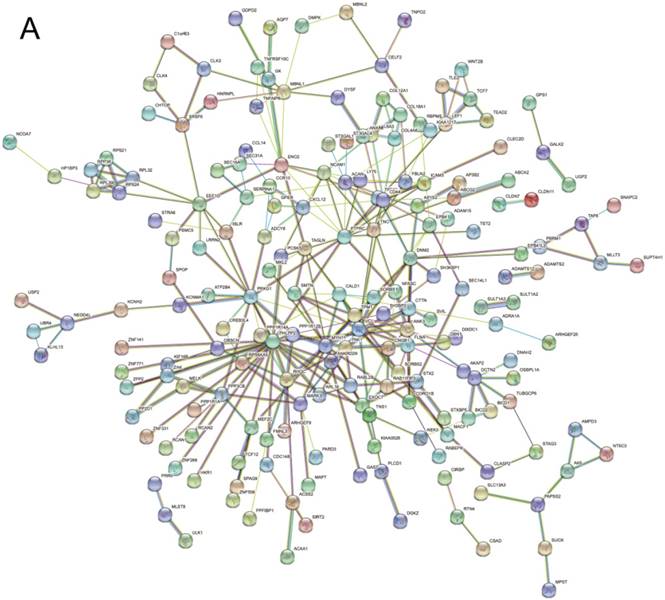
In order to further explore the functional interactions of the alternatively spliced genes in colorectal cancer, we constructed gene interaction network of the corresponding gene identifier names of alternative splicing events through Search Tool for the Retrieval of Interacting Genes (STRING) in the present study. STRING database (www.string-db.org/) could predict associations for a particular group of proteins and summarize the complex interactions in a network view.
Functional and pathway enrichment analysis
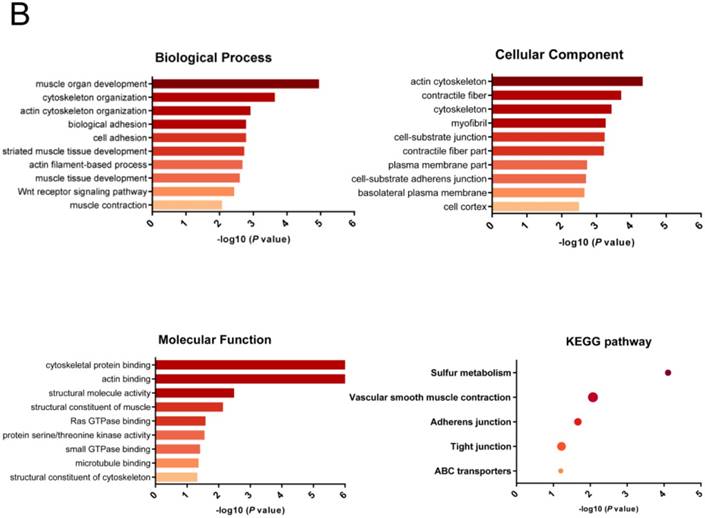
The biological function and pathway of the differentially spliced genes between COAD/READ and normal tissues would demonstrate instructive information. Therefore, we performed functional and pathway enrichment analysis via Database for Annotation, Visualization and Integrated Discovery (DAVID). DAVID (https://david.ncifcrf.gov/), a bioinformatics data resource with an integrative biology knowledge database and comprehensive analysis tools, benefits researchers to discover biological meaning behind large amount of genes[17]. Gene ontology (GO) analysis of the cellular component, molecular function, and biological process [18] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis [19] were conducted for the identified differentially spliced genes via DAVID. P value<0.05 indicates statistically significant.
Representative model of seven different alternative splicing types.
Statistical analysis
The statistical analysis was conducted by R language (Version 3.4.1). Instead of previously Venn diagram, we adopted UpSet plot in this study to visualize various combination of intersections of seven AS types, which can clearly show quantitative results of multiple interactive sets. In order to investigate the difference of AS preference between CRC tissues and normal tissues, the percentage of seven AS types was calculated (PSI<first quartile or PSI>third quartile). Student's t-test was performed to detect the difference of AS prevalence between cancer and normal tissues. Receiver operating characteristic (ROC) curve was used to assess the value of AS in predicting CRC occurrence. In addition, the individual splicing event was analysed in relation with the overall survival (OS) of CRC patients by cox model to identify promising prognostic biomarkers.
Results
Alternative splicing profiles in COAD and READ
After analysing all the raw data of COAD and READ, we detected altogether 35391 AS events of 9084 genes in COAD and 34900 AS events of 9032 genes in READ. In COAD, we observed 13087 ESs in 5635 genes, 7740 ATs in 3382 genes, 6653 APs in 2693 genes, 2917 AAs in 2125 genes, 2332 RIs in 1601 genes, 2524 ADs in 1834 genes and 138 MEs in 138 genes; in READ, we found 12913 ESs in 5576 genes, 7618 ATs in 3337 genes, 6554 APs in 2649 genes, 2883 AAs in 2092 genes, 2326 RIs in 1596 genes, 2476 ADs in 1804 genes and 130 MEs in 130 genes.
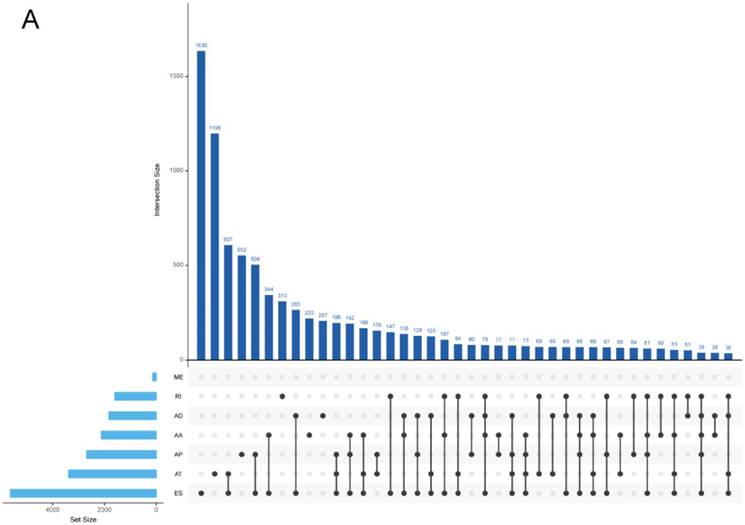
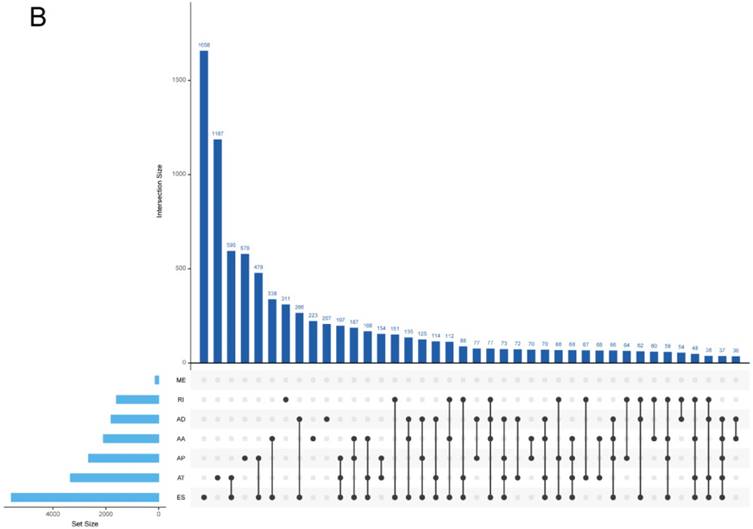
In both COAD and READ, ES was the most frequent AS events and ME was the rarest AS events. The prevalence of different AS events was similar between COAD and READ, indicating relevant pathogenesis of these two types of cancers. It is worthy that one gene might possess several alternative splicing patterns. And the detailed information about the specific AS types of genes was visualized in Upset plot (Figure 2), which can demonstrate quantitative results of multiple interactive sets more effectively than traditional Venn diagram.
Upset plot of different types of alternative splicing types. (A), COAD; (B), READ.
Differentially spliced events between CRC and normal tissues.
| COAD | READ | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Splice type | Exon arrangement | Cancer | Normal | Cancer | Normal | Change | |
| SULT1A2 | RI | 1.2:1.3 | 79.9 | 20.2 | 79.2 | 9.2 | Up | |
| CALD1 | ES | 8.3:9 | 24.4 | 77.8 | 27.1 | 90.3 | Down | |
| DTNA | ES | 31:32.1 | 79.5 | 16.8 | 73.9 | 11.4 | Up | |
| COL12A1 | ES | 3:4:5:6:7:8:9:10:11:12:13:14:15:16:17 | 98.9 | 48.6 | 99.3 | 37.6 | Up | |
| TTLL12 | AP | 1 | 80.8 | 19.2 | 81 | 21.3 | Up | |
| TTLL12 | AP | 12 | 19.2 | 80.8 | 19 | 78.7 | Down | |
| SERPINA1 | AA | 2.1:2.2:2.3 | 65 | 14.5 | 65.2 | 5.9 | Up | |
| FBLN2 | ES | 11 | 15.2 | 75.7 | 14.4 | 73.7 | Down | |
| SVIL | ES | 21 | 11.5 | 52 | 13.5 | 71.8 | Down | |
| ATP2B4 | ES | 21 | 21.4 | 63.3 | 24.4 | 81.7 | Down | |
| TMEM151B | AT | 4 | 86.9 | 31.3 | 88.8 | 32 | Up | |
| TMEM151B | AT | 3 | 13.1 | 68.7 | 11.2 | 68 | Down | |
| GAS7 | AP | 4 | 17.6 | 61.5 | 14.2 | 70.2 | Down | |
| CCR10 | AP | 2 | 48.8 | 88.7 | 35.6 | 89.7 | Down | |
| CCR10 | AP | 1 | 51.2 | 11.3 | 64.4 | 10.3 | Up | |
| TNS1 | ES | 23 | 19 | 62.2 | 24.1 | 77.8 | Down | |
| SULT2B1 | AP | 1 | 84.8 | 49.2 | 85.5 | 31.8 | Up | |
| SULT2B1 | AP | 2.1 | 15.2 | 50.8 | 14.5 | 68.2 | Down | |
| ISLR | AP | 2 | 64.4 | 14.9 | 63 | 9.7 | Up | |
| ISLR | AP | 1 | 35.6 | 85.1 | 37 | 90.3 | Down | |
Difference of alternative splicing in CRC tissues and normal ones. (A), COAD; (B), READ.
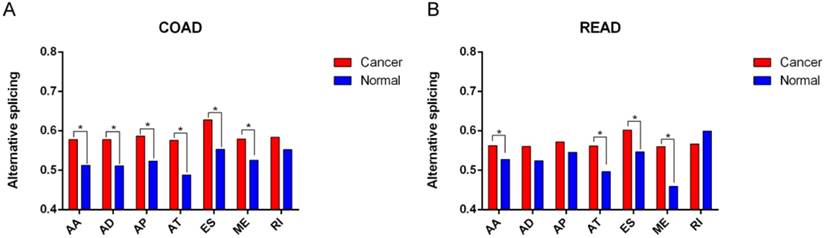
Difference of percent-splice-in (PSI) value between CRC and normal tissues according to seven alternative splicing types.
| COAD | READ | ||||||
|---|---|---|---|---|---|---|---|
| Splice type | Cancer | Normal | P | Cancer | Normal | P | |
| AA | 0.578±0.134 | 0.513±0.047 | 0.002 | 0.562±0.122 | 0.527±0.034 | 0.022 | |
| AD | 0.578±0.134 | 0.511±0.047 | 0.002 | 0.560±0.124 | 0.524±0.051 | 0.069 | |
| AP | 0.587±0.141 | 0.523±0.056 | 0.005 | 0.572±0.128 | 0.545±0.047 | 0.158 | |
| AT | 0.576±0.124 | 0.488±0.043 | <0.001 | 0.561±0.119 | 0.496±0.037 | <0.001 | |
| ES | 0.628±0.126 | 0.553±0.051 | <0.001 | 0.602±0.121 | 0.547±0.045 | 0.004 | |
| ME | 0.579±0.156 | 0.525±0.087 | 0.029 | 0.560±0.145 | 0.459±0.043 | <0.001 | |
| RI | 0.584±0.124 | 0.552±0.07 | 0.107 | 0.566±0.111 | 0.599±0.058 | 0.130 | |
Differentially spliced genes (DSGs) between CRC and normal tissues
Differentially spliced genes (DSGs) between CRC and normal tissues were analysed and the top 20 significantly altered AS events were summarized in Table 1. ESs and APs were dominant AS type, and several genes (TTLL12, TMEM151B, CCR10, SULT2B1, ISLR) demonstrated two AS events with opposite preference in CRC and normal tissues.
In order to reveal the role of different AS patterns in carcinogenesis of COAD and READ, we compared the difference of AS preference between CRC tissues and normal tissues. As was shown in Figure 3, six types of AS (AA, AD, AP, AT, ES, ME) were significantly increased in COAD than normal tissues, while four types of AS (AA, AT, ES, ME) were significantly enhanced in READ than normal tissues (Table 2). It is obvious that most AS patterns were more active in CRC tissues than adjacent normal ones. However, the results based on tumor stages demonstrated no significant difference of AS patterns among different stages of COAD and READ (all P>0.05), which indicate that certain alternative splicing patterns might show significant differences between CRC and normal, but might not distinguish different stages of CRC.
GO functional enrichment and KEGG pathway analysis
We then performed GO functional enrichment and KEGG pathway analysis of Differentially spliced genes (DSGs) between CRC and normal tissues (top 500 AS events). DSGs mainly enriched in biological process (BP) including muscle organ development, cytoskeleton organization, actin cytoskeleton organization, biological adhesion, and cell adhesion. Cellular component (CC) analysis indicated enrichment in cell adhesion, contractile fiber, cytoskeleton, myofibril, and cell-substrate junction. These DSGs showed significant enrichment in molecular function (MF) of cytoskeletal protein binding, actin binding, structural molecule activity, structural constituent of muscle and Ras GTPase binding. KEGG analysis enriched DSGs in pathways of Sulfur metabolism, vascular smooth muscle contraction, adherens junction, tight junction, and ABC transporters (Table 3). The gene interaction network was built and the visual bar results of GO or KEGG analysis was displayed (Figure 4).
Potentials of AS to predict CRC occurrence
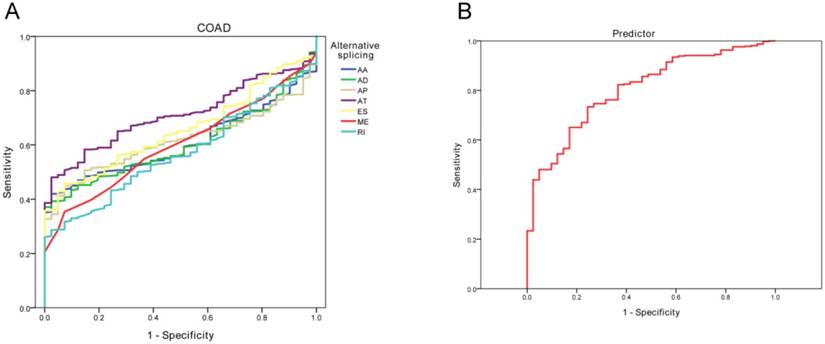
Considering the significantly increased AS events in CRC than normal tissues, we subsequently explored if AS patterns could serve as an early predictor of occurrence of COAD or READ by ROC curve. In COAD, AA, AD, AP, AT, ES, ME all demonstrated an AUC over 0.6, of which AT best predict the occurrence of COAD (sensitivity: 0.567; specificity: 0.976; AUC: 0.710) (Table 4, Figure 5). In READ, AT and ME suggested an AUC over 0.6, of which ME best predict the occurrence of READ (sensitivity: 0.651; specificity: 0.900; AUC: 0.742). The integrated predictor model of COAD showed an AUC of 0.805 (sensitivity: 0.734; specificity: 0.756) while READ predictor had an AUC of 0.738 (sensitivity: 0.614; specificity: 0.900). Overall, abnormally active alternative splicing was a specific event in CRC because most models demonstrated a relatively high specificity value.
Survival associated alternative splicing events in COAD and READ
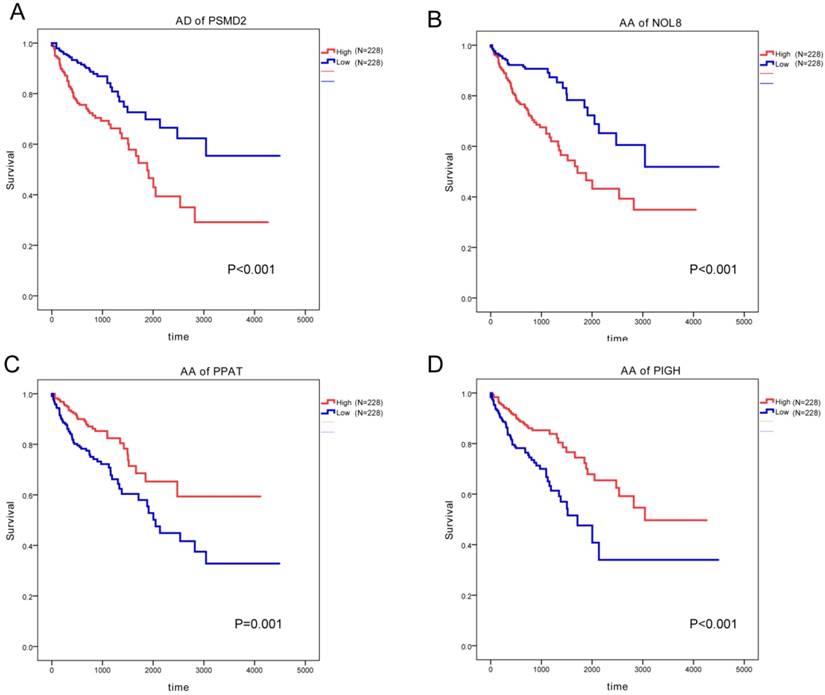
We next used multivariate Cox proportional hazards models to assess adjusted hazards ratios (HR) and 95% confidence intervals (CI) of AS event and estimate its effect on CRC survival with adjustment for age and sex. The top significant results were shown in Table 5. More prognostic results of COAD and READ were summarized in Supplementary Table 1. In COAD, AS events such as AP of LBH, AD of PSMD2, AA of NOL8, AD of HAUS4 and RI of ALS2CL were associated with worse survival of patients; AS events including AA of PPAT, AA of PIGH, AA of LSM7, AT of PSPC1 and AT of UPK3B were linked with favourable prognosis (Figure 6). In READ, AS events such as ES of ALDH4A1, AT of SLC10A7, ES of ANXA11, AT of ILF3 and AP of MCF2L predicted shorter survival time; AS events including ES of LRRC28, ES of IRF3, AT of PLA2R1, AP of BCAR1 and C16orf13 were related with better prognosis.
Functional GO analysis and KEGG analysis of differentially spliced genes between CRC and normal tissues.
| Category | Term | Count | % | P value |
|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0007517~muscle organ development | 16 | 5.1 | 1.1E-05 |
| GOTERM_BP_FAT | GO:0007010~cytoskeleton organization | 21 | 6.7 | 0.00023 |
| GOTERM_BP_FAT | GO:0030036~actin cytoskeleton organization | 13 | 4.1 | 0.00122 |
| GOTERM_BP_FAT | GO:0022610~biological adhesion | 26 | 8.3 | 0.00165 |
| GOTERM_BP_FAT | GO:0007155~cell adhesion | 26 | 8.3 | 0.00166 |
| GOTERM_CC_FAT | GO:0015629~actin cytoskeleton | 16 | 5.1 | 4.7E-05 |
| GOTERM_CC_FAT | GO:0043292~contractile fiber | 10 | 3.2 | 0.0002 |
| GOTERM_CC_FAT | GO:0005856~cytoskeleton | 41 | 13.1 | 0.00037 |
| GOTERM_CC_FAT | GO:0030016~myofibril | 9 | 2.9 | 0.00055 |
| GOTERM_CC_FAT | GO:0030055~cell-substrate junction | 9 | 2.9 | 0.00058 |
| GOTERM_MF_FAT | GO:0008092~cytoskeletal protein binding | 30 | 9.6 | 5E-08 |
| GOTERM_MF_FAT | GO:0003779~actin binding | 23 | 7.3 | 1.5E-07 |
| GOTERM_MF_FAT | GO:0005198~structural molecule activity | 23 | 7.3 | 0.00324 |
| GOTERM_MF_FAT | GO:0008307~structural constituent of muscle | 5 | 1.6 | 0.00725 |
| GOTERM_MF_FAT | GO:0017016~Ras GTPase binding | 6 | 1.9 | 0.02626 |
| KEGG_PATHWAY | hsa00920:Sulfur metabolism | 5 | 1.6 | 7.8E-05 |
| KEGG_PATHWAY | hsa04270:Vascular smooth muscle contraction | 8 | 2.5 | 0.00846 |
| KEGG_PATHWAY | hsa04520:Adherens junction | 6 | 1.9 | 0.0216 |
| KEGG_PATHWAY | hsa04530:Tight junction | 7 | 2.2 | 0.05998 |
| KEGG_PATHWAY | hsa02010:ABC transporters | 4 | 1.3 | 0.06299 |
Functional enrichment analysis results of differentially spliced genes in CRC. (A), gene-gene interaction network; (B), GO analysis and KEGG analysis.
ROC curve results of different alternative splicing types in predicting risks of COAD and READ.
| Category | Alternative splicing | Cut-off | Sensitivity | Specificity | AUC | Lower limit | Upper limit |
|---|---|---|---|---|---|---|---|
| COAD | AA | 0.598 | 0.419 | 0.976 | 0.606 | 0.551 | 0.661 |
| AD | 0.605 | 0.393 | 0.976 | 0.604 | 0.546 | 0.661 | |
| AP | 0.606 | 0.448 | 0.927 | 0.615 | 0.561 | 0.669 | |
| AT | 0.567 | 0.480 | 0.976 | 0.710 | 0.657 | 0.762 | |
| ES | 0.618 | 0.459 | 0.927 | 0.656 | 0.596 | 0.716 | |
| ME | 0.646 | 0.354 | 0.927 | 0.602 | 0.537 | 0.667 | |
| Predictor | 0.087 | 0.734 | 0.756 | 0.805 | 0.744 | 0.866 | |
| READ | AT | 0.545 | 0.518 | 0.900 | 0.649 | 0.562 | 0.736 |
| ME | 0.500 | 0.651 | 0.800 | 0.742 | 0.661 | 0.823 | |
| Predictor | 0.057 | 0.614 | 0.900 | 0.738 | 0.657 | 0.819 |
Survival-associated alternative splicing events in COAD and READ.
| 95%CI | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cancer | Gene | Splice type | Exon arrangement | Uniprot ID | Adjusted HR | Lower limit | Upper limit | P |
| COAD | LBH | AP | 2.1 | Q53QV2 | 3.45 | 1.96 | 6.06 | <0.001 |
| PSMD2 | AD | 1.2 | Q13200 | 3.09 | 1.95 | 4.90 | <0.001 | |
| NOL8 | AA | 16.1 | Q76FK4 | 2.96 | 1.86 | 4.69 | <0.001 | |
| HAUS4 | AD | 1.3 | Q9H6D7 | 2.68 | 1.67 | 4.31 | <0.001 | |
| ALS2CL | RI | 18.2:18.3 | Q60I27 | 2.67 | 1.68 | 4.24 | <0.001 | |
| PPAT | AA | 3.1 | Q06203 | 0.34 | 0.21 | 0.54 | <0.001 | |
| PIGH | AA | 1 | Q14442 | 0.36 | 0.23 | 0.57 | <0.001 | |
| LSM7 | AA | 3.1 | Q9UK45 | 0.36 | 0.23 | 0.58 | <0.001 | |
| PSPC1 | AT | 9 | Q8WXF1 | 0.37 | 0.23 | 0.60 | <0.001 | |
| UPK3B | AT | 6 | Q9BT76 | 0.38 | 0.24 | 0.61 | <0.001 | |
| READ | ALDH4A1 | ES | 13 | P30038 | 12.99 | 3.45 | 50.00 | <0.001 |
| SLC10A7 | AT | 3 | Q0GE19 | 8.93 | 2.75 | 28.57 | <0.001 | |
| ANXA11 | ES | 3.2:4 | P50995 | 9.01 | 1.75 | 45.45 | 0.008 | |
| ILF3 | AT | 20 | Q12906 | 3.69 | 1.29 | 10.53 | 0.015 | |
| MCF2L | AP | 14 | O15068 | 3.38 | 1.20 | 9.43 | 0.021 | |
| LRRC28 | ES | 7.1:7.2 | Q86X40 | 0.08 | 0.02 | 0.29 | <0.001 | |
| IRF3 | ES | 5.1:5.2 | Q14653 | 0.13 | 0.04 | 0.40 | <0.001 | |
| PLA2R1 | AT | 30 | Q13018 | 0.33 | 0.12 | 0.87 | 0.024 | |
| BCAR1 | AP | 4.1 | P56945 | 0.29 | 0.10 | 0.86 | 0.026 | |
| C16orf13 | ES | 3 | Q96S19 | 0.34 | 0.13 | 0.88 | 0.027 | |
ROC curve evaluating the potential of alternative splicing in prediction of COAD risk. (A), different alternative splicing types; (B), integrated predictor.
Representative survival-associated alternative splicing events in COAD. (A), AD of PSMD2; (B) AA of NOL8; (C), AA of PPAT; (D), AA of PIGH.
Discussion
Aberrant pre-mRNA alternative splicing has been widely accepted as a novel contributor to cancer development[20, 21]. Although a number of cancer-specific mRNA isoforms were identified, our understanding of the alternative splicing events profile and their functional pathways lags far behind. With the rapid development of high-throughput sequencing and bioinformatics means, more comprehensive overview of AS in colorectal cancer could be revealed. In this study, we described the alternative splicing profiles and built their interaction network in COAD and READ using TCGA data. A series of CRC-specific and survival-related alternative splicing events were discovered, which would offer promising intervention targets for CRC treatment.
Diverse splicing patterns in one genes lead to a variety of isoforms, which makes AS and its regulation mechanism more complex in cancer[22, 23]. In this study, totally 35391 AS events of 9084 genes in COAD and 34900 AS events of 9032 genes in READ were detected, indicating that alternative splicing is a common process in CRC. In addition, both COAD and READ generate largest number of ESs and smallest number of MEs. After comparing the difference of AS preference between CRC tissues and normal tissues, six types of AS (AA, AD, AP, AT, ES, ME) were significantly increased in COAD than normal tissues while four types of AS (AA, AT, ES, ME) were significantly enhanced in READ than normal tissues. It is obvious that most AS patterns were more active in CRC tissues than adjacent normal ones. The abnormal enhancement of AS and its misregulation mechanism are of great importance to clarify colorectal carcinogenesis.
Differentially spliced genes (DSGs) between CRC and normal tissues were analysed, of which APs, ESs and ATs were dominant alternative splicing type. It is worth noting that COAD and READ showed certain similarity of alternative splicing, which might indicate some similar tumorigenic process of these two types of cancers in the continuous intestinal tract. Of the top twenty DSGs, several genes (TTLL12, TMEM151B, CCR10, SULT2B1, ISLR) demonstrated two AS events with opposite preference in CRC and normal tissues. The inverse shift of AS patterns the same gene might function as valuable biomarkers in CRC development. TTLL12 (tubulin tyrosine ligase like 12), a posttranslational modificator of tubulins, has been reported to participate in tumorigenesis of prostate cancer via influencing the cytoskeleton, tubulin modification and chromosomal ploidy[24]. CCR10 is the receptor for chemokine CCL28. CCL28 attracts leukocytes expressing CCR10 as a mediator of antimicrobial effect and showed a significantly decreased protein level in colon cancers than in normal tissue[25]. Increased expression of SULT2B1b has been reported as an independent prognostic biomarker and promotes cell growth and invasion in CRC[26]. Both TMEM151B and ISLR have not yet been studied in cancer, neither are their biological functions clear. Therefore, misregulation of alternative promoters, alternative terminators and exon skip of these genes are promising research direction to elucidate novel etiology of CRC in future.
GO functional enrichment and KEGG pathway analysis of Differentially spliced genes (DSGs) between CRC and normal tissues provided helpful clues for the elucidation of colorectal tumorigenesis. DSGs mainly enriched in biological process of muscle organ development, cytoskeleton organization, biological adhesion and in molecular function of cytoskeletal protein binding, actin binding, structural molecule activity. From this point of view, CRC-related AS events mainly regulate cytoskeleton construction and cell adhesion. Indeed, loss of tight cellular junction and rearrangement of cell cytoskeleton has been regarded as critical hallmarks of cancer[27]. Further investigations into how alternative splicing modulates these procedures are required in the future. Besides, KEGG analysis enriched DSGs in pathways of Sulfur metabolism, vascular smooth muscle contraction, adherens junction, tight junction, and ABC transporters. ATP-binding cassette transporters (ABC transporters) are responsible for the translocation of various substrates across membranes, either for uptake or for export of the substrate[28]. Microbial pathways like sulfate-reducing bacteria (SRB) were likely to shape colonic sulfur metabolism as well as the component and availability of sulfated compounds[29]. Imbalance of microbe in intestinal tract has been considered as an inducement of malignant transformation of colorectal epithelium[30]. Future research concerning the specific alternations of AS events involved in pathways such as ABC transporters and Sulfur metabolism might offer novel therapeutic targets for CRC treatment.
In order to evaluate the potential of AS patterns as an early predictor of CRC occurrence and specific AS events as indicator of CRC prognosis, we conducted analysis of ROC curve and cox regression model, respectively. For CRC initiation, AT best predicted the occurrence of COAD while ME was the best predictor for READ occurrence. The integrated predictor model of COAD showed an AUC of 0.805 (sensitivity: 0.734; specificity: 0.756) while READ predictor had an AUC of 0.738 (sensitivity: 0.614; specificity: 0.900). Overall, abnormally active alternative splicing was a specific event in CRC because most models demonstrated a relatively high specificity value. Besides, we also identified a series of survival-associated alternative splicing events such as AD of PSMD2, AA of NOL8 in COAD and ES of ALDH4A1, AT of SLC10A7 in READ. The survival-related AS events would provide novel insight into the complex progression of colorectal cancer. It is worth noting that some AS events of the same gene might come up with opposite prognostic effect, like two different AT types of CEP68 and UPK3B. These alternative splicing events might be key regulator of CRC development, which demonstrate promising potential as therapeutic targets. The aberrant proteins of the differentially expressed and prognosis-associated alternative splicing events in COAD and READ were of great importance. In the future, more investigations including immunohistochemistry should be performed to confirm the significance of the AS events we found. The specific molecular mechanisms of the observed significance for these AS regulation also need further studies to elucidate.
Conclusion
In summary, we draw comprehensive profiles of alternative splicing events in the carcinogenesis and prognosis of CRC. A series of cancer-specific and prognosis-associated AS events were identified to provide potential therapeutic targets for CRC. The interaction network and functional connections were also constructed, which would enrich our understanding of the role of RNA alternative splicing in the tumorigenesis of CRC.
Supplementary Material
Supplementary table 1.
Acknowledgements
This study is supported by grants from the National Science and Technology Support Program (2015BAI13B07) and Public Welfare Foundation of Liaoning Province (No. 2015005002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Anczukow O, Krainer AR. Splicing-factor alterations in cancers. RNA (New York, NY). 2016;22(9):1285-1301
2. Bates DO, Morris JC, Oltean S, Donaldson LF. Pharmacology of Modulators of Alternative Splicing. Pharmacological reviews. 2017;69(1):63-79
3. Donaldson LF, Beazley-Long N. Alternative RNA splicing: contribution to pain and potential therapeutic strategy. Drug discovery today. 2016;21(11):1787-1798
4. Kozlovski I, Siegfried Z, Amar-Schwartz A, Karni R. The role of RNA alternative splicing in regulating cancer metabolism. Human genetics. 2017;136(9):1113-1127
5. Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nature medicine. 2016;22(9):976-986
6. Munkley J, Livermore K, Rajan P, Elliott DJ. RNA splicing and splicing regulator changes in prostate cancer pathology. Human genetics. 2017;136(9):1143-1154
7. Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, Kuipers EJ. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637-1649
8. Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology. 2015;149(5):1177-1190 e1173
9. Zhang L, Shay JW. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. Journal of the National Cancer Institute. 2017:109 (8)
10. Li XL, Zhou J, Chen ZR, Chng WJ. P53 mutations in colorectal cancer - molecular pathogenesis and pharmacological reactivation. World journal of gastroenterology. 2015;21(1):84-93
11. Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, Wu Y, Han F, Zhang L, Yu T. et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2017
12. Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q. et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. The Journal of clinical investigation. 2017;127(9):3375-3391
13. Baranwal S. Commentary: HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Frontiers in cell and developmental biology. 2017;5:91
14. Adler AS, McCleland ML, Yee S, Yaylaoglu M, Hussain S, Cosino E, Quinones G, Modrusan Z, Seshagiri S, Torres E. et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes & development. 2014;28(10):1068-1084
15. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490(7418):61-70.
16. Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic acids research. 2016;44(D1):1018-1022
17. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4(5):3
18. The Gene Ontology (GO) project in 2006. Nucleic acids research. 2006; 34(Database issue):322-326.
19. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27-30
20. Paronetto MP, Passacantilli I, Sette C. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell death and differentiation. 2016;23(12):1919-1929
21. Narayanan SP, Singh S, Shukla S. A saga of cancer epigenetics: linking epigenetics to alternative splicing. The Biochemical journal. 2017;474(6):885-896
22. Scotti MM, Swanson MS. RNA mis-splicing in disease. Nature reviews Genetics. 2016;17(1):19-32
23. Song X, Zeng Z, Wei H, Wang Z. Alternative splicing in cancers: From aberrant regulation to new therapeutics. Seminars in cell & developmental biology. 2017
24. Wasylyk C, Zambrano A, Zhao C, Brants J, Abecassis J, Schalken JA, Rogatsch H, Schaefer G, Pycha A, Klocker H. et al. Tubulin tyrosine ligase like 12 links to prostate cancer through tubulin posttranslational modification and chromosome ploidy. International journal of cancer. 2010;127(11):2542-2553
25. Muthuswamy RV, Sundstrom P, Borjesson L, Gustavsson B, Quiding-Jarbrink M. Impaired migration of IgA-secreting cells to colon adenocarcinomas. Cancer immunology, immunotherapy: CII. 2013;62(6):989-997
26. Hu L, Yang GZ, Zhang Y, Feng D, Zhai YX, Gong H, Qi CY, Fu H, Ye MM, Cai QP. et al. Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Laboratory investigation; a journal of technical methods and pathology. 2015;95(9):1005-1018
27. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674
28. Tanaka KJ, Song S, Mason K, Pinkett HW. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochimica et biophysica acta. 2017
29. Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Frontiers in physiology. 2012;3:448
30. Akin H, Tozun N. Diet, microbiota, and colorectal cancer. Journal of clinical gastroenterology. 2014;48(Suppl 1):67-69
Author contact
![]() Corresponding author: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening, Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: yuanyuanedu.cn
Corresponding author: Prof. Chengzhong Xing, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: xcz1966com and Prof. Yuan Yuan, Tumor Etiology and Screening, Department of Cancer Institute and General Surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact