Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(12):2082-2092. doi:10.7150/jca.23681 This issue Cite
Research Paper
Copy number variation and high expression of DNA topoisomerase II alpha predict worse prognosis of cancer: a meta-analysis
Department of anorectal surgery, the First Affiliated Hospital of China Medical University
#, these authors contribute equally to this work.
Received 2017-11-6; Accepted 2018-3-26; Published 2018-5-24
Abstract
Background: Increasing numbers of literatures have investigated the association between TOP2A and cancer prognosis. But the results of the relationship between the two were inconclusive. The aim of this meta-analysis was to elucidate whether TOP2A could predict prognosis of cancer.
Materials and Methods: A systematically searching for potentially valuable literature was conducted through electronic databases containing PubMed and Web of Science. Hazard Ratio (HR) and their 95% confidence interval (CI) were used to assess the strength of association between TOP2A and cancer prognosis.
Results: Finally twenty-five studies were included in this meta-analysis. High expression of TOP2A was associated with shorter disease free survival (DFS) of cancer prognosis compared with low expression of TOP2A (HR= 1.36, 95% CI= 1.18-1.57, P<0.001). Amplification of TOP2A gene showed no significant association with overall survival (OS), disease free survival (DFS) or relapse free survival (RFS) compared with non-amplification of TOP2A (OS: HR= 0.96, 95%CI= 0.75-1.22, P= 0.735; DFS: HR= 0.93, 95%CI= 0.70-1.23, P= 0.621; RFS: HR= 0.97, 95%CI= 0.71-1.34, P= 0.867). In the subgroup of regions, TOP2A amplification was associated with longer overall survival (HR= 0.66, 95%CI= 0.46-0.96, P= 0.029) in Australia. Alteration (amplification or deletion) of TOP2A gene demonstrated shorter survival according to OS and RFS compared with those with normal TOP2A status (OS: HR= 1.37, 95%CI= 1.22-1.55, P<0.001; RFS: HR= 1.26, 95%CI= 1.12-1.41, P<0.001).
Conclusion: High TOP2A expression suggested significant relationship with worse cancer prognosis. Alteration (amplification or deletion) of TOP2A gene was also significantly related to shorter survival of cancer patients. Therefore, TOP2A might be used as an indicator for poor prognosis of cancer in the future.
Keywords: TOP2A, cancer, copy number variation, prognosis.
Introduction
Malignant tumor, characterized by strengthened and unlimited cell division during the cellular genetic process [1], is the leading cause of death in the world. As the genetic material inside every cell nucleus, DNA is indispensable for the maintenance of genetic stability and integrity. Because of the structure of duplex DNA, it inevitably leads the consequences of the topology such as supercoils [2]. DNA topoisomerases, ubiquitously present in eukaryotes, archaebacteria and Eubacteria, are necessary for the regulation of DNA topology in various cellular procedures [3, 4]. A number of studies have indicated that DNA topoisomerases play an essential role in the DNA world through allowing DNA double helices or strands to cut across each other [4, 5]. According to their different acting mechanisms, DNA topoisomerases can be classified as type I and type II enzymes [3, 6]. TOP2A is one of the isoenzymes which can mediate the catalytic activity of type II topoisomerases [6].
TOP2A (DNA topoisomerase II alpha) gene, mapped to chromosome 17q12-q21, covers approximately 27.5 kb and includes 35 exons, encoding a 170 kDa protein [7]. TOP2A encodes an enzyme which is implicated in almost any process of DNA metabolism including transcription, replication, movement and untangling [3, 8, 9], which catalyze the passage of two DNA duplexes across each other to resolve the entanglements and coiling of cellular DNA [10]. It modulates the topological states of DNA by transient cleavage, strand passing and religation of double-stranded DNA resulting in decatenation of intertwined DNA molecules and relaxation of supercoiled DNA [8, 9]. Due to its critical function in chromosome condensation and segregation in proliferation and division of cell [8], TOP2A has been widely investigated in multiple diseases including cancer.
In a variety of the malignant tumors, TOP2A protein expression and TOP2A gene status are usually abnormal. However, the relationships between TOP2A and the prognosis of malignant tumor were not consistent. For example, Ito F et al. revealed that the patients with high expression of TOP2A who suffering from endometrial cancer had a poor prognosis in Japanese, which maybe because that TOP2A immunoexpression significantly correlated with advanced stages and tumor aggressiveness [11]. While in another study, Won HS et al. suggested that high expression of TOP2A in breast cancer had no significant predictive value for disease free survival (DFS), which showed limited application value of TOP2A as a prognostic marker [12]. However, another article pointed out that overexpression of TOP2A in HER2-positive and HER2-negative breast tumors had an obviously opposite prognostic impact in Australia, which indicated possible relationship between TOP2A and HER2 [13].
In addition, the role of TOP2A copy number variation in the development and prognosis of tumors was still unclear. In invasive carcinoma with adjacent ductal carcinoma in situ (DCIS-AIC), TOP2A showed a lower frequency of copy number increase in males compared to females, which indicate probable differences in breast carcinogenesis between the different sexes[14]. Kaya I et al. found no significant association between TOP2A deletion and abnormal cytologic findings in cervical cell lesions [15]. As for the clinical implications, the amplification of TOP2A in the patients who underwent HER2 amplified breast tumors predicted a better overall survival (OS) and disease free survival (DFS) in British, offering a possibility that amplification of TOP2A might be a useful marker to predict cancer prognosis [16]. But another investigation found that TOP2A amplification demonstrated no association with disease free survival (DFS) and overall survival (OS) in Australia, which showed limited association between amplification of TOP2A and the prognosis of breast cancer [17].
Until now, no clear conclusion on the association between TOP2A and the prognosis of malignant tumor has been drawn. In order to explore the relationship between the two, we made a retrospective meta-analysis in this study to elucidate the prognostic role of TOP2A in cancer.
Materials and methods
Identification and eligibility of relevant studies
We conducted a systematically literature search on the electronic databases including PubMed and Web of Science. Different combinations of the following key words were used including “TOP2A/topoisomerase II alpha”, “cancer/malignancy/malignant tumor”, and “survival/prognosis”. In case the data provided in the article were not sufficiently enough, the authors were contacted for specific raw data. When overlapping data were detected, only the latest and largest sample could be adopted for this meta-analysis. July 21th, 2017 was the last search date.
Inclusion and exclusion criteria
Studies included in this meta-analysis must pass the inclusion criteria as follows: studies concerning the relationship between TOP2A and the prognosis of malignant tumor; studies should be published in English; studies should contain sufficient raw data to assess Hazard Ratio (HR) and their 95% confidence interval (CI). The principle for exclusion criteria were reviews or letters; meta-analysis; no relevance; animal experiments for TOP2A; drug sensitivity studies; functional studies of TOP2A; duplicate publications; and studies not about TOP2A.
Data extraction
Two authors (Ling Ren and Jingwei Liu) extracted the data independently from the included studies. From each individual study, the following information was extracted: first author's name, year of publication, ethnicity and region of the population, the classification of cancer, numbers of patient, the detection methods of TOP2A, Hazard Ratio (HR) and their 95% confidence interval (CI). After discussion, the conflict was resolved, and all the extracted information has reached a consensus.
Statistical analysis
The statistical analysis of this study was carried out by Stata software (Version 11.0; Stata Corp, College Station, TX). Hazard Ratio (HR) and their 95% confidence interval (CI) were applied to assess the strength of the association between TOP2A and the prognosis of malignant tumor. P value <0.05 was considered as statistical significance. Heterogeneity was valued by using Q statistic (P < 0.05 means significant heterogeneity between studies) and I-squared (I2) value [18]. When the heterogeneity between the studies showed no significance, the pooled ORs were calculated using the fixed-effects model of the Mantel-Haenszel method [19]. On the contrary, a random-effects model using DerSimonian and Laird method [20] was used. Subgroup analyses were performed to investigate the effects of ethnicity. In addition, we evaluated publication bias quantitatively by Begg's test [21] and Egger's test [22], respectively. P value <0.05 for Begg's and Egger's tests represents significant publication bias.
Results
Study characteristics
Using different combinations of key words, a total of 237 literatures were initially selected from the PubMed and Web of Science after duplicates removed. Through reading the titles and abstracts of these potential useful literatures, 165 literatures were excluded mainly by the reason of irrelevant literatures, reviews or meta-analysis, animal experiments, functional research, drug sensitivity study, not raw data. Then, the left 72 full-text literatures were further valued for eligibility. Finally, we adopted 25 full-text literatures [8, 11-13, 16, 17, 23-41] with eligibility in our meta-analysis. The details of the flow chart of literatures selection was shown in Figure 1.
The flowchart of literature inclusion and exclusion
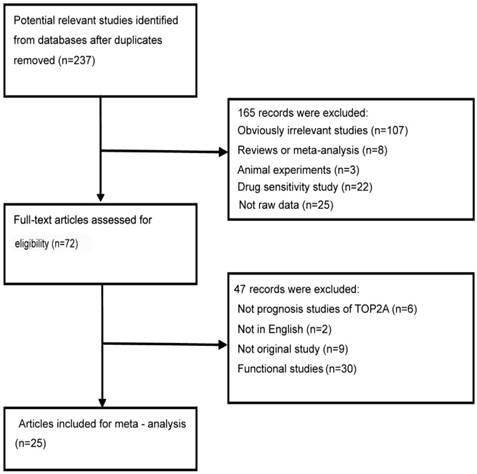
We summarized the major characteristics of these eligible literatures in this meta-analysis in Table 1. All the included literatures were published in English. Twelve articles [11-13, 23-29, 32, 38] investigated the association between TOP2A expression and the prognosis of malignant tumor for HR; thirteen articles [8, 13, 16, 17, 28, 30, 31, 33-37, 40] researched the relationship between the amplification of TOP2A gene and the cancer prognosis for HR; four articles [34, 39-41] studied the association between the alteration of TOP2A gene and the prognosis of cancer for HR. The types of cancers covered breast cancer, endometrial cancer, adrenocortical cancer, of which breast cancer accounts for the vast majority. The detection methods of TOP2A contained IHC, qPCR, and FISH/CISH/SISH. The regions of the population were divided into Asia, Europe, America and Australia. In the subgroup analysis, data concerning different regions were separated as individual studies.
Characteristics of eligible studies in this meta-analysis.
| Author | Year | Cancer type | Region | Ethnicity | Number | U/M | Expression | Method |
|---|---|---|---|---|---|---|---|---|
| High expression and OS | ||||||||
| Chen, J. R. | 2017 | Breast cancer | Asia | Taiwan | 309 | U | Protein | IHC |
| Wachter, D. L. | 2013 | Breast cancer | Europe | German | 100 | U | Protein | IHC |
| Ito, F. | 2016 | Endometrial cancer | Asia | Japanese | 56 | M | Protein | IHC |
| Ip, J. C. | 2015 | Adrenocortical carcinoma | Australia | Australian | 61 | M | Protein | IHC |
| Fountzilas, G. | 2012 | HER-2+breast cancer | Australia | Australian | 57 | U | Protein | IHC |
| Fountzilas, G. | 2012 | HER-2-breast cancer | Australia | Australian | 37 | U | Protein | IHC |
| Nikolenyi, A. | 2012 | Breast cancer | Europe | Hungarian | 106 | U | Protein | IHC |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 314 | M | RNA | qPCR |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 273 | U | Protein | IHC |
| O'Malley, F. P. | 2011 | Breast cancer | North America | Canadian | 477 | U | Protein | IHC |
| Roca, E. | 2017 | Adrenocortical cancer | Europe | European | 98 | M | Protein | IHC |
| Zaczek, A. J. | 2012 | Breast cancer | Europe | Polish | 322 | M | DNA | qPCR |
| High expression and DFS | ||||||||
| Milde-Langosch, K. | 2013 | Triple-negative breast cancer | Europe | German | 95 | U | RNA | Microarray |
| Milde-Langosch, K. | 2013 | HER2-positive breast cancer | Europe | German | 69 | U | RNA | Microarray |
| Milde-Langosch, K. | 2013 | Luminal breast cancer | Europe | German | 397 | M | RNA | Microarray |
| Roca, E. | 2017 | Adrenocortical cancer | Europe | European | 98 | M | Protein | IHC |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 273 | U | Protein | IHC |
| Ito, F. | 2016 | Endometrial cancer | Asia | Japanese | 56 | M | Protein | IHC |
| Ip, J. C. | 2015 | Adrenocortical carcinoma | Australia | Australian | 77 | U | Protein | IHC |
| Won, H. S. | 2014 | Breast cancer | Asia | Korean | 70 | M | Protein | IHC |
| Wachter, D. L. | 2013 | Breast cancer | Europe | German | 100 | U | Protein | IHC |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 314 | U | RNA | qPCR |
| Zaczek, A. J. | 2012 | Breast cancer | Europe | Polish | 322 | M | DNA | qPCR |
| Amplification and OS | ||||||||
| Gogas, H. | 2016 | Breast cancer | Europe | Greece | 119 | M | DNA | FISH |
| Fasching, P. A. | 2014 | Breast cancer | Europe | German | 628 | M | DNA | FISH |
| Fountzilas, G. | 2012 | HER-2+breast cancer | Australia | Australian | 50 | U | DNA | FISH |
| Kim, A. | 2012 | Breast cancer | Asia | Korean | 567 | U | DNA | SISH |
| Tubbs, R. | 2009 | Breast cancer | America | American | 1626 | M | DNA | FISH |
| Nielsen, K. V. | 2008 | Breast cancer | Europe | Danish | 773 | M | DNA | FISH |
| Arriola, E. | 2007 | Breast cancer | Europe | British | 232 | M | DNA | CISH |
| Engstrom, M. J. | 2014 | Breast cancer | Europe | Norwegian | 670 | U | DNA | FISH |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 266 | U | DNA | CISH |
| Fountzilas, G. | 2013 | Breast cancer | Australia | Australian | 979 | M | DNA | FISH |
| Bartlett, J. M. | 2015 | Breast cancer | Europe | British | 3098 | U | DNA | FISH |
| Bartlett, J. M. | 2010 | Breast cancer | Europe | British | 1762 | U | DNA | FISH |
| Lamy, P. J. | 2011 | HER2-amplified breast cancer | Europe | French | 86 | U | DNA | qPCR |
| Amplification and DFS | ||||||||
| Fountzilas, G | 2013 | Breast cancer | Australia | Australian | 979 | M | DNA | FISH |
| Kim, A. | 2012 | Breast cancer | Asia | Korean | 567 | U | DNA | SISH |
| Tubbs, R. | 2009 | Breast cancer | America | American | 1626 | M | DNA | FISH |
| Arriola, E. | 2007 | Breast cancer | Europe | British | 232 | M | DNA | CISH |
| Fountzilas, G. | 2012 | Breast cancer | Australia | Australian | 266 | M | DNA | CISH |
| Amplification and RFS | ||||||||
| Bartlett, J. M. | 2010 | Breast cancer | Europe | British | 1762 | U | DNA | FISH |
| Lamy, P. J. | 2011 | HER2-amplified breast cancer | Europe | French | 86 | U | DNA | qPCR |
| Nielsen, K. V. | 2008 | Breast cancer | Europe | Danish | 773 | M | DNA | FISH |
| Bartlett, J. M. | 2015 | Breast cancer | Europe | British | 3098 | U | DNA | FISH |
| Alteration and OS | ||||||||
| Pritchard, K. I. | 2012 | Breast cancer | North America | Canadian | 430 | M | DNA | FISH |
| Bartlett, J. M. | 2010 | Breast cancer | Europe | British | 1762 | M | DNA | FISH |
| O'Malley, F P | 2009 | Breast cancer | North America | Canadian | 438 | M | DNA | FISH |
| Bartlett, J. M. | 2015 | Breast cancer | Europe | British | 3098 | U | DNA | FISH |
| Alteration and RFS | ||||||||
| Pritchard, K. I. | 2012 | Breast cancer | North America | Canadian | 430 | M | DNA | FISH |
| Bartlett, J. M. | 2010 | Breast cancer | Europe | British | 1762 | M | DNA | FISH |
| O'Malley, F P | 2009 | Breast cancer | North America | Canadian | 438 | M | DNA | FISH |
| Bartlett, J. M. | 2015 | Breast cancer | Europe | British | 3098 | U | DNA | FISH |
IHC: Immunohistochemistry; FISH: Fluorescence in situ hybridization; CISH: Chromogenic in situ hybridization; SISH: Silver-enhanced in situ hybridization; qPCR: Quantitative real time polymerase chain reaction
Association between TOP2A expression and cancer prognosis
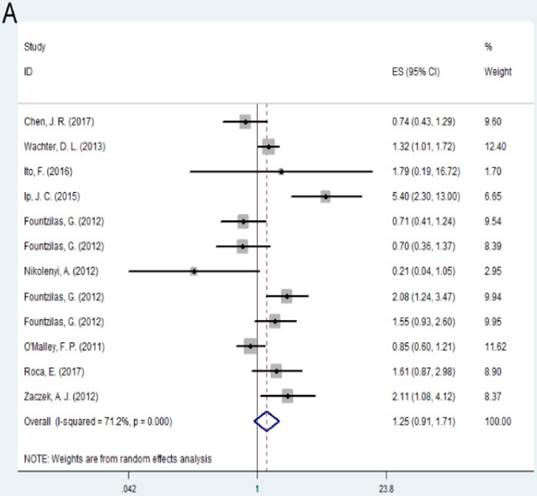
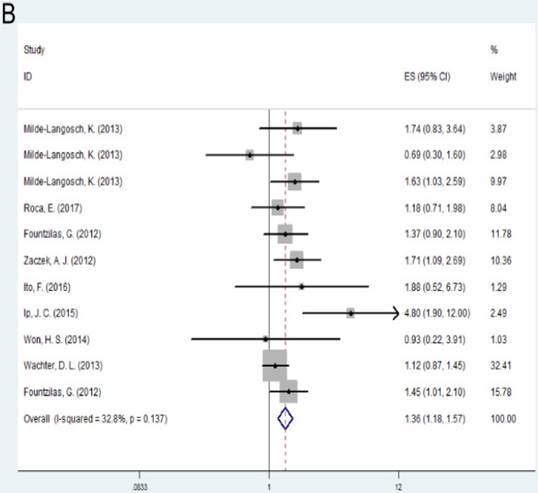
Individuals with high expression of TOP2A was observed to be associated with shorter disease free survival (DFS) of the cancer prognosis compared with low expression of TOP2A (HR= 1.36, 95% CI= 1.18-1.57, P<0.001). No significant association was found between TOP2A expression and overall survival (OS) (HR = 1.25, 95% CI= 0.91-1.71, P = 0.163). Subgroup analysis based on regions suggested that high expression of TOP2A was consistently related with worse OS and DFS in Europe (OS: HR = 1.38, 95% CI= 1.01-1.73, P=0.005; DFS: HR= 1.28, 95% CI= 1.07-1.52, P=0.007). As for Australia, high expression indicated unfavorable DFS (HR= 1.80, 95% CI = 1.07-3.04, P = 0.028), while there was no significant association between TOP2A expression and OS (HR= 1.48, 95% CI= 0.79-2.78, P= 0.218).
Association between amplification of TOP2A gene and cancer prognosis
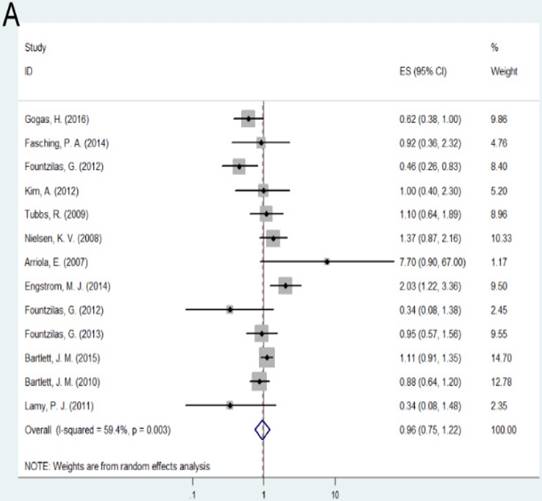
No significant association was found between amplification of TOP2A gene OS, DFS or relapse free survival (RFS) compared with non-amplification of TOP2A (OS: HR= 0.96, 95% CI= 0.75-1.22, P= 0.735; DFS: HR= 0.93, 95%CI= 0.70-1.23, P= 0.621; RFS: HR = 0.97, 95%CI = 0.71-1.34, P= 0.867). When considering the effect of regions, TOP2A amplification predicted longer overall survival (HR= 0.66, 95%CI= 0.46-0.96, P= 0.029) in Australia, while no significant association was observed between TOP2A amplification and OS in the subgroup of Europe (HR= 1.07, 95%CI= 0.80-1.45, P= 0.644). Due to the small sample numbers, no subgroup analysis was performed by different regions in the two groups: amplification of TOP2A with DFS and RFS.
Association between alteration of TOP2A gene and cancer prognosis
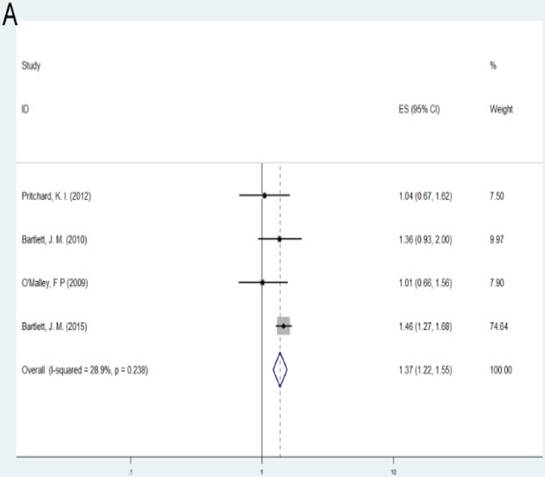
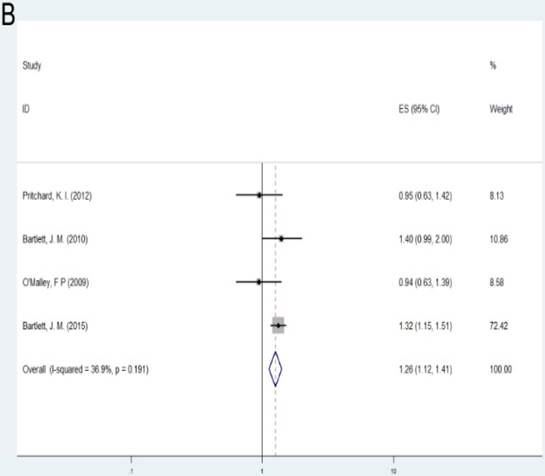
Patients with alteration (amplification or deletion) of TOP2A gene demonstrated shorter survival according to OS and RFS compared with those with normal TOP2A status (OS: HR= 1.37, 95%CI= 1.22-1.55, P<0.001; RFS: HR= 1.26, 95%CI= 1.12-1.41, P<0.001). As the number of samples is small, we did not conduct subgroup investigations in these two groups.
Heterogeneity
The heterogeneity results were summarized in Table 2, of which only some comparisons showed significant heterogeneity. This heterogeneity could not be completely eliminated by subgroup analysis. In addition, due to the limited study number, we did not make a meta-regression to explore the source of heterogeneity. For certain relationship such as alteration of TOP2A and cancer prognosis based on OS and RFS, no significant heterogeneity was detected (P>0.05). Excluding each study did not significantly alter the overall outcome, indicating that the results of this meta-analysis were robust.
Publication Bias
The Begg's test and Egger's test were adopted to quantitatively assess the publication bias in the literatures. No significant publication bias was observed for meta-analyses between high expression of TOP2A, amplification or alteration of TOP2A and the prognosis of malignant tumor. We summarized the results for publication bias test in Table 3.
Meta-analysis results of the association between expression of TOP2A, amplification or alteration of TOP2A and cancer prognosis for pooled HR.
| Comparison | Categories | Group/subgroup | Data set number | HR(95%CI) | P value | Model | P value | I² (%) |
|---|---|---|---|---|---|---|---|---|
| Expression (High vs. Low) | OS | Overall | 12 | 1.25(0.91-1.71) | 0.163 | R | <0.001 | 71.20% |
| Europe | 4 | 1.38(1.10-1.73) | 0.005 | F | 0.068 | 57.90% | ||
| Australia | 5 | 1.48(0.79-2.78) | 0.218 | R | <0.001 | 81.50% | ||
| DFS | Overall | 11 | 1.36(1.18-1.57) | <0.001 | F | 0.137 | 32.80% | |
| Europe | 6 | 1.28(1.07-1.52) | 0.007 | F | 0.257 | 23.60% | ||
| Australia | 3 | 1.80(1.07-3.04) | 0.028 | R | 0.044 | 67.90% | ||
| Amplification(Amp vs. non-Amp) | OS | Overall | 13 | 0.96(0.75-1.22) | 0.735 | R | 0.003 | 59.40% |
| Europe | 8 | 1.07(0.80-1.45) | 0.644 | R | 0.007 | 64.10% | ||
| Australia | 3 | 0.66(0.46-0.96) | 0.029 | F | 0.115 | 53.80% | ||
| DFS | Overall | 5 | 0.93(0.70-1.23) | 0.621 | F | 0.072 | 53.60% | |
| RFS | Overall | 4 | 0.97(0.71-1.34) | 0.867 | R | 0.036 | 64.80% | |
| Alteration (Altered vs. Normal) | OS | Overall | 4 | 1.37(1.22-1.55) | <0.001 | F | 0.238 | 28.90% |
| RFS | Overall | 4 | 1.26(1.12-1.41) | <0.001 | F | 0.191 | 36.90% |
R: random effect model; F: fixed effect model; OS: overall survival; DFS: disease free survival; RFS: relapse free survival
Publication bias.
| Begg's test | Egger's test | |||||
|---|---|---|---|---|---|---|
| Comparison | Group/subgroup | Categories | z value | P value | t value | P value |
| Expression(High vs. Low) | Overall | OS | 0.41 | 0.681 | 0.15 | 0.883 |
| DFS | 0.39 | 0.697 | 1.04 | 0.324 | ||
| Amplification(Amp vs. non-Amp) | Overall | OS | -0.37 | 0.714 | -0.61 | 0.556 |
| DFS | -0.49 | 0.624 | 0.10 | 0.926 | ||
| RFS | 0.00 | 1.000 | -0.26 | 0.817 | ||
| Alteration(Altered vs. Normal) | Overall | OS | -1.36 | 0.174 | -2.72 | 0.113 |
| RFS | -1.36 | 0.174 | -1.41 | 0.293 | ||
A: Forest plot for the association between TOP2A expression and cancer prognosis by OS; B: Forest plot for the association between TOP2A expression and cancer prognosis by DFS
A: Forest plot for the association between amplification of TOP2A and cancer prognosis by OS; B: Forest plot for the association between amplification of TOP2A and cancer prognosis by DFS
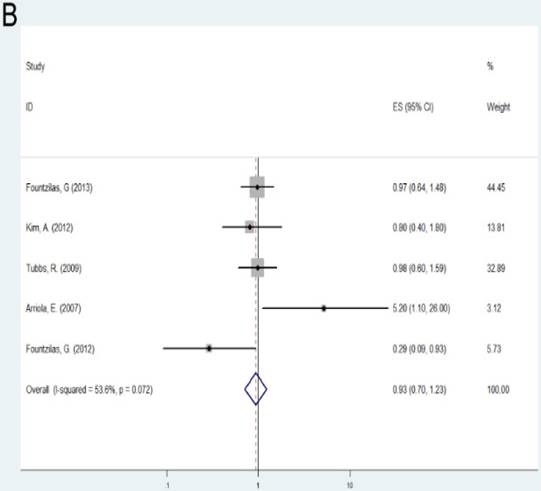
A: Forest plot for the association between alteration of TOP2A and cancer prognosis by OS; B: Forest plot for the association between alteration of TOP2A and cancer prognosis by RFS
Discussion
Although TOP2A has been researched extensively in various malignant tumors, the relationship between TOP2A and prognosis of cancer were inconclusive according previous individual literatures. Studies mainly focused on the influence of either TOP2A expression or copy number variation on clinical outcome of cancer. In this study, we performed a meta-analysis to research the association between the two above with cancer prognosis separately. As far as we know, this is the first comprehensive meta-analysis exploring the role of TOP2A in tumor prognosis. After analyzing the data extracted from 25 full-text case-control publications, we unraveled that both high TOP2A expression and alteration of TOP2A gene may indicated worse prognosis of cancer.
Uncontrollable high proliferation rates proved to be the main characters of malignant tumor [1]. In proliferating cells, DNA is the key regulator as genetic material. During replication, transcription, recombination and repair of DNA, DNA helix would inevitably occur unwinding or rewinding, bringing out DNA entanglement [42]. DNA topoisomerases are a family of nature's tools which can resolve these problems via the introduction of temporary single or double strand breaks in DNA [5]. As one of the isoenzymes of DNA topoisomerases, TOP2A is obviously up-regulated during these processes [6]. Therefore, TOP2A could perform a predominant role in proliferating cells and may probably be implicated in carcinogenesis.
In this meta-analysis, we observed that high TOP2A expression predicted worse prognosis of cancer. The relationship of high TOP2A expression with shorter DFS in both Europe and Australia remained significant, which indicated that ethnicity had little influence on the predictive role of TOP2A. A number of mechanism studies have explored the effect of TOP2A on cancer development [23, 43-46], which might, at least in part, explain the findings of our investigation. It was found in prostate cancer cells that knockdown of TOP2A decreased proliferation and tumorigenicity [43]. Meanwhile, TOP2A was upregulated in recurrence/metastasis prostate cancer [43]. Therefore, the close relationship between TOP2A up-regulation and increased proliferation of cancer cells account for its effect on poorer prognosis. These findings also provided significant implications for prostate cancer therapy that treatments to kill TOP2A positive cells may provide a better method to eradicate primary prostate cancer [43]. As an oncogene, HER2 resides on the long arm of chromosome 17, which is the same location with TOP2A [45, 46]. Accordingly, TOP2A amplification always came with HER2 amplification because amplification of one gene locus could simultaneously overexpress both of these genes [45]. As most well-known cancer suppressor genes, p53 and pRB are negative regulators of TOP2A [23, 44, 45]. The commonly inactivated and deleted of these two protective genes in cancer partly lead to the overexpression of TOP2A [45]. The phenomenon of oncogene up-regulation and cancer suppressor gene down-regulation upon high TOP2A expression in cancer would probably contribute to the unfavourable survival. TOP2A also determine the outcome of tumor chemotherapy. Colorectal cell-line SW620 overexpressing TOP2A demonstrated significant resistance to chemotherapeutic treatment with irinotecan and etoposide, which resulted from suppression of apoptosis in TOP2A over-expressed cells [47]. Resistant to chemotherapeutic regimen make it reasonable why patients with TOP2A overexpression suffered shorter survival. Our results in this meta-analysis concerning the correlation between high expression of TOP2A and poorer survival outcomes enhance its role in cancer development. As for clinical applications, TOP2A might serve as indicator for poor prognosis. In addition, targeting TOP2A high expression cells might be a novel treatment for malignant tumors.
Copy number variation (CNV), caused by the genome rearrangement, refers to the length of 1 kb or more large fragments of the genome copy number amplification or deletion [48]. Alternation of TOP2A copy number variation (amplification and deletion) has been found to be significantly increased in high histologic grade cancers whereas no TOP2A copy number variation was detected in well-differentiated tumors [49]. According to the results of our meta-analysis, alternation of TOP2A gene copy number variations (amplification and deletion) significantly correlated with shorter survival from aspects of both OS and RFS. It seems that as a key regulator of genetic process of cells, neither amplification nor deletion of TOP2A gene benefits the clinical outcome of cancer patients. Results of some investigations could explain these findings: both amplification and deletion of TOP2A gene were associated with polysomy of chromosome 17 [50]. Chromosome 17 polysomy was one of the frequent major abnormality events and correlated with aggressive biological behavior of breast cancer [51, 52]. In addition, Knoop et al. reported that significant relationship of TOP2A-amplified/TOP2A-deleted breast cancers with tumor size, nodal involvement and ER positivity [53]. Furthermore, TOP2A gene is located in close proximity to oncogene HER-2 locus on chromosome 17. Some investigators suggested that TOP2A amplification and deletion may not be two different ends of a continuum, but rather be regarded as an abnormal status to be distinguished from the normal status of HER-2 protein [54]. As a result, the examination of the abnormal status of TOP2A gene might serve as a helpful biomarker which could indicate severity of patients suffering cancer in the future.
In this study, we revealed no relationship between amplification of TOP2A gene and prognosis of cancer, except for the subgroup of Australia population. The findings that TOP2A amplification predicted longer overall survival in Australia might due to the different background of ethnicity. One study performed in Taiwanese invasive female breast cancers by tissue microarrays suggested no prognostic value of TOP2A amplification [32]. Besides, studies have indicated potential of TOP2A amplification as a useful clinical biomarker of sensitivity to adjuvant anthracycline-based chemotherapy in patients with breast cancers pertaining to the HER2 positive subgroup in British [16]. Patients with TOP2A amplifications showed a 51% reduction in the risk of death in breast cancer if allocated to CEF (cyclophosphamide, epirubicin, and fluorouracil) compared with TOP2A normal patients [8]. Study of TOP2A gene amplification in malignant cancer patients may affect the decision of taking adjuvant chemotherapy and thus protect patients from therapy-induced complications. Considering the limited sample size, the correlation between TOP2A gene amplification with survival time of cancer still need further investigations to confirm due to the limited sample size in specific subgroup.
We should acknowledge some limitations in this meta-analysis. First, only literatures published in English were included. Second, the sample capacity in the pooled analysis and some subgroup analyses was relatively small. Therefore, the results still need large-scale studies to confirm later. Third, other important personal data as age, sex, and family history were not applicable for each study, so we could not get results with adjustments by other co-variates. Fourth, the combination of different sequencing methods and different types of cancers may lead to heterogeneity of the population and reduce the strength of the study.
Conclusion
To be concluded, our meta-analysis suggested that high TOP2A expression predicted worse prognosis of cancer. Alteration (amplification or deletion) of TOP2A gene also showed significant relation with shorter survival for cancer patients. Therefore, TOP2A might serve as novel prognostic indicator for the prognosis of malignant tumor. Further well-designed and large-scale investigations concerning different regions are still necessary to prove the conclusion of our meta-analysis.
Acknowledgements
This study is supported by grants from Public Welfare Foundation of Liaoning Province (No. 2015005002).
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
2. Bush NG, Evans-Roberts K, Maxwell A. DNA Topoisomerases. EcoSal Plus. 2015:6
3. Watt PM, Hickson ID. Structure and function of type II DNA topoisomerases. The Biochemical journal. 1994;303( Pt 3):681-95
4. Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews Molecular cell biology. 2002;3:430-40
5. Bakshi RP, Galande S, Muniyappa K. Functional and regulatory characteristics of eukaryotic type II DNA topoisomerase. Critical reviews in biochemistry and molecular biology. 2001;36:1-37
6. Hajduk M. Topoisomerase II alpha-a fundamental prognostic factor in breast carcinoma. Polish journal of pathology: official journal of the Polish Society of Pathologists. 2009;60:67-75
7. Sng JH, Heaton VJ, Bell M, Maini P, Austin CA, Fisher LM. Molecular cloning and characterization of the human topoisomerase IIalpha and IIbeta genes: evidence for isoform evolution through gene duplication. Biochimica et biophysica acta. 1999;1444:395-406
8. Nielsen KV, Ejlertsen B, Moller S, Jorgensen JT, Knoop A, Knudsen H. et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta oncologica (Stockholm, Sweden). 2008;47:725-34
9. Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic acids research. 2009;37:738-48
10. McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutation research. 2007;623:83-97
11. Ito F, Furukawa N, Nakai T. Evaluation of TOP2A as a Predictive Marker for Endometrial Cancer With Taxane-Containing Adjuvant Chemotherapy. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2016;26:325-30
12. Won HS, Lee KE, Sung SH, Choi MY, Jo JY, Nam EM. et al. Topoisomerase II alpha and microtubule-associated protein-tau as a predictive marker in axillary lymph node positive breast cancer. Tumori. 2014;100:80-6
13. Fountzilas G, Valavanis C, Kotoula V, Eleftheraki AG, Kalogeras KT, Tzaida O. et al. HER2 and TOP2A in high-risk early breast cancer patients treated with adjuvant epirubicin-based dose-dense sequential chemotherapy. Journal of translational medicine. 2012;10:10
14. Vermeulen MA, Doebar S, van Deurzen C, Martens J, van Diest PJ, Moelans CB. Copy number profiling of oncogenes in ductal carcinoma in situ of the male breast. Endocrine-related cancer. 2017
15. Kaya I, Sivrikoz ON, Etlik O, Gok A. Associations between epidermal growth factor receptor and topoisomerase II-alpha gene copy number variations, human papillomavirus positivity, and cytologic analysis in cervical cell lesions. Indian journal of pathology & microbiology. 2017;60:328-35
16. Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K. et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast cancer research and treatment. 2007;106:181-9
17. Fountzilas G, Dafni U, Bobos M, Kotoula V, Batistatou A, Xanthakis I. et al. Evaluation of the prognostic role of centromere 17 gain and HER2/topoisomerase II alpha gene status and protein expression in patients with breast cancer treated with anthracycline-containing adjuvant chemotherapy: pooled analysis of two Hellenic Cooperative Oncology Group (HeCOG) phase III trials. BMC cancer. 2013;13:163
18. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539-58
19. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22:719-48
20. DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7:177-88
21. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
22. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315:629-34
23. Ip JC, Pang TC, Glover AR, Soon P, Zhao JT, Clarke S. et al. Immunohistochemical validation of overexpressed genes identified by global expression microarrays in adrenocortical carcinoma reveals potential predictive and prognostic biomarkers. The oncologist. 2015;20:247-56
24. Milde-Langosch K, Karn T, Muller V, Witzel I, Rody A, Schmidt M. et al. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast cancer research and treatment. 2013;137:57-67
25. Wachter DL, Fasching PA, Haeberle L, Schulz-Wendtland R, Dimmler A, Koscheck T. et al. Prognostic molecular markers and neoadjuvant therapy response in anthracycline-treated breast cancer patients. Archives of gynecology and obstetrics. 2013;287:337-44
26. Zaczek AJ, Markiewicz A, Seroczynska B, Skokowski J, Jaskiewicz J, Pienkowski T. et al. Prognostic significance of TOP2A gene dosage in HER-2-negative breast cancer. The oncologist. 2012;17:1246-55
27. Nikolenyi A, Uhercsak G, Csenki M, Hamar S, Csorgo E, Tanczos E. et al. Tumour topoisomerase II alpha protein expression and outcome after adjuvant dose-dense anthracycline-based chemotherapy. Pathology oncology research: POR. 2012;18:61-8
28. Fountzilas G, Christodoulou C, Bobos M, Kotoula V, Eleftheraki AG, Xanthakis I. et al. Topoisomerase II alpha gene amplification is a favorable prognostic factor in patients with HER2-positive metastatic breast cancer treated with trastuzumab. Journal of translational medicine. 2012;10:212
29. O'Malley FP, Chia S, Tu D, Shepherd LE, Levine MN, Huntsman D. et al. Topoisomerase II alpha protein and responsiveness of breast cancer to adjuvant chemotherapy with CEF compared to CMF in the NCIC CTG randomized MA.5 adjuvant trial. Breast cancer research and treatment. 2011;128:401-9
30. Engstrom MJ, Ytterhus B, Vatten LJ, Opdahl S, Bofin AM. TOP2A gene copy number change in breast cancer. Journal of clinical pathology. 2014;67:420-5
31. Tubbs R, Barlow WE, Budd GT, Swain E, Porter P, Gown A. et al. Outcome of patients with early-stage breast cancer treated with doxorubicin-based adjuvant chemotherapy as a function of HER2 and TOP2A status. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:3881-6
32. Chen JR, Chien HP, Chen KS, Hwang CC, Chen HY, Yeh KY. et al. Amplification of HER2 and TOP2A and deletion of TOP2A genes in a series of Taiwanese breast cancer. Medicine. 2017;96:e5582
33. Gogas H, Kotoula V, Alexopoulou Z, Christodoulou C, Kostopoulos I, Bobos M. et al. MYC copy gain, chromosomal instability and PI3K activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer. Journal of translational medicine. 2016;14:136
34. Bartlett JM, McConkey CC, Munro AF, Desmedt C, Dunn JA, Larsimont DP. et al. Predicting Anthracycline Benefit: TOP2A and CEP17-Not Only but Also. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:1680-7
35. Fasching PA, Weihbrecht S, Haeberle L, Gasparyan A, Villalobos IE, Ma Y. et al. HER2 and TOP2A amplification in a hospital-based cohort of breast cancer patients: associations with patient and tumor characteristics. Breast cancer research and treatment. 2014;145:193-203
36. Lamy PJ, Fina F, Bascoul-Mollevi C, Laberenne AC, Martin PM, Ouafik L. et al. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast cancer research: BCR. 2011;13:R15
37. Kim A, Shin HC, Bae YK, Kim MK, Kang SH, Lee SJ. et al. Multiplication of Chromosome 17 Centromere Is Associated with Prognosis in Patients with Invasive Breast Cancers Exhibiting Normal HER2 and TOP2A Status. Journal of breast cancer. 2012;15:24-33
38. Roca E, Berruti A, Sbiera S, Rapa I, Oneda E, Sperone P. et al. Topoisomerase 2alpha and thymidylate synthase expression in adrenocortical cancer. Endocrine-related cancer. 2017;24:299-307
39. O'Malley FP, Chia S, Tu D, Shepherd LE, Levine MN, Bramwell VH. et al. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. Journal of the National Cancer Institute. 2009;101:644-50
40. Bartlett JM, Munro AF, Dunn JA, McConkey C, Jordan S, Twelves CJ. et al. Predictive markers of anthracycline benefit: a prospectively planned analysis of the UK National Epirubicin Adjuvant Trial (NEAT/BR9601). The Lancet Oncology. 2010;11:266-74
41. Pritchard KI, Munro A, O'Malley FP, Tu D, Li X, Levine MN. et al. Chromosome 17 centromere (CEP17) duplication as a predictor of anthracycline response: evidence from the NCIC Clinical Trials Group (NCIC CTG) MA.5 Trial. Breast cancer research and treatment. 2012;131:541-51
42. Mondal N, Parvin JD. Transcription from the perspective of the DNA: twists and bumps in the road. Critical reviews in eukaryotic gene expression. 2003;13:1-8
43. Li X, Liu Y, Chen W, Fang Y, Xu H, Zhu HH. et al. TOP2Ahigh is the phenotype of recurrence and metastasis whereas TOP2Aneg cells represent cancer stem cells in prostate cancer. Oncotarget. 2014;5:9498-513
44. Iino K, Sasano H, Yabuki N, Oki Y, Kikuchi A, Yoshimi T. et al. DNA topoisomerase II alpha and Ki-67 in human adrenocortical neoplasms: a possible marker of differentiation between adenomas and carcinomas. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 1997;10:901-7
45. O'Connor JK, Hazard LJ, Avent JM, Lee RJ, Fischbach J, Gaffney DK. Topoisomerase II alpha expression correlates with diminished disease-free survival in invasive breast cancer. International journal of radiation oncology, biology, physics. 2006;65:1411-5
46. Hannemann J, Kristel P, van Tinteren H, Bontenbal M, van Hoesel QG, Smit WM. et al. Molecular subtypes of breast cancer and amplification of topoisomerase II alpha: predictive role in dose intensive adjuvant chemotherapy. British journal of cancer. 2006;95:1334-41
47. Coss A, Tosetto M, Fox EJ, Sapetto-Rebow B, Gorman S, Kennedy BN. et al. Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer letters. 2009;276:228-38
48. Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP. et al. Challenges and standards in integrating surveys of structural variation. Nature genetics. 2007;39:S7-15
49. Park K, Han S, Gwak GH, Kim HJ, Kim J, Kim KM. Topoisomerase II-alpha gene deletion is not frequent as its amplification in breast cancer. Breast cancer research and treatment. 2006;98:337-42
50. Badawy OM, Loay I. FISH Analysis of TOP2A and HER-2 Aberrations in Female Breast Carcinoma on Archived Material: Egyptian NCI Experience. Applied immunohistochemistry & molecular morphology: AIMM. 2017
51. Reinholz MM, Bruzek AK, Visscher DW, Lingle WL, Schroeder MJ, Perez EA. et al. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. The Lancet Oncology. 2009;10:267-77
52. Zaczek A, Markiewicz A, Supernat A, Bednarz-Knoll N, Brandt B, Seroczynska B. et al. Prognostic value of TOP2A gene amplification and chromosome 17 polysomy in early breast cancer. Pathology oncology research: POR. 2012;18:885-94
53. Knoop AS, Knudsen H, Balslev E, Rasmussen BB, Overgaard J, Nielsen KV. et al. retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:7483-90
54. Koscielny S, Terrier P, Spielmann M, Delarue JC. Prognostic importance of low c-erbB2 expression in breast tumors. Journal of the National Cancer Institute. 1998;90:712
Author contact
![]() Corresponding author: Prof. Chengzhong Xing, Department of anorectal surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: xcz1966com
Corresponding author: Prof. Chengzhong Xing, Department of anorectal surgery, the First Affiliated Hospital of China Medical University, 155# North Nanjing Street, Heping District, Shenyang City, 110001, Liaoning Province, China. E-mail address: xcz1966com

 Global reach, higher impact
Global reach, higher impact