3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(13):2275-2283. doi:10.7150/jca.24797 This issue Cite
Research Paper
Aberrant methylation of FAT4 and SOX11 in peripheral blood leukocytes and their association with gastric cancer risk
1. Department of Epidemiology, College of Public Health, Harbin Medical University, Harbin, Heilongjiang Province, P. R. China.
2. Department of Colorectal Cancer Surgery, The third affiliated hospital, Harbin Medical University, Harbin, Heilongjiang Province, P. R. China.
3. Xiangfang Center for Disease Control and Prevention, Harbin 150081, Heilongjiang Province, P. R. China.
* These authors contributed equally to this work
Received 2018-1-7; Accepted 2018-3-31; Published 2018-6-5
Abstract
Background: Aberrant DNA methylation, especially tumor suppressor gene hypermethylation, is a well-recognized biomarker of initial tumorogenesis stages. FAT4 and SOX11 are putative tumor suppressor genes and can be down-regulated by hypermethylation in various cancers tissues. However, in peripheral blood leukocytes, the association between these two genes methylation status, as well as the effects of gene-environment interactions, and gastric cancer (GC) risk remain unclear.
Methods: A hospital-based case-control study including 375 cases and 394 controls was conducted. Peripheral blood leukocytes DNA methylation status were detected by methylation-sensitive high-resolution melting (MS-HRM) assay. Logistic regression was adopted to analyze the relationship of FAT4 and SOX11 methylation with GC susceptibility.
Results: Positive methylation (Pm) and total positive methylation (Tpm) of FAT4 were significantly increased the risk of GC (OR = 2.204, 95% CI: 1.168-4.159, P = 0.015; OR = 1.583, 95% CI: 1.031-2.430, P = 0.036, respectively). Compared with controls, cases exhibited higher SOX11 Pm frequencies with OR of 2.530 (95% CI: 1.289-4.969, P = 0.007). Nonetheless, no statistically significant association between SOX11 Tpm and GC risk was observed. Additionally, interactions between FAT4 Tpm and increased consumption of freshwater fish (≥1 times/week) displayed an antagonistic effect on GC (OR = 0.328, 95% CI: 0.142-0.762, P = 0.009), and high salt intake interacted with SOX11 Tpm also showed statistically significant (OR = 0.490, 95% CI: 0.242-0.995, P = 0.048).
Conclusions: FAT4 aberrant methylation in peripheral blood leukocytes and gene-environment interactions were associated with the risk of GC, while SOX11 was controversial and needed to be more investigated.
Keywords: DNA methylation, peripheral blood leukocytes, gastric cancer, gene-environment interaction.
Introduction
Gastric cancer (GC), a neoplasm with aggressive and highly proliferative condition, contributes considerable healthy burden from a global perspective. According to the Globocan 2012, GC is the fifth most common malignancy and ranks as the third leading cause of cancer-related death in the world [1]. In China, GC has become the second leading cause of cancer death. According to the statistics, 679,100 new GC cases and 498,000 cancer deaths are estimated to occur in 2015 [2]. Although GC showed a decreasing incidence and mortality trend in recent years, population growth and aging will still lead to a large number of new cases in China [2].
It is well recognized that the development of GC is a multifactorial induction process, genetic and epigenetic alterations are involved in it as critical contributors. Nowadays, much attention has been paid to epigenetic alterations, which can induce heritable changes in gene expression without any alterations in the primary DNA sequence [3]. DNA methylation, which plays a key role in gene transcription and expression programming, is one of the most extensively studied epigenetic alterations. Aberrant DNA methylation frequently occurs at CpG islands within promoter regions, leading to transcription inhibition and gene inactivation, and is recognized as a critical component of the mechanism underlying tumorigenesis [4]. Several studies have demonstrated that aberrant DNA methylation can be a driver event in the pathogenesis of cancers GC [5-7].
FAT4, which located at the apical point of the Hippo signaling pathway, controls the size of organs by modulating cell growth, proliferation, and apoptosis [8, 9]. SOX11 encodes a member of the group C SOX (SRY-related HMG-box) transcription factor family that participates in cell proliferation, embryonic neurogenesis and tissue remodeling [10]. Both SOX11 and FAT4 are supposed to be potential tumor suppressor genes and can be down-regulated by promoter aberrant methylation modification in several cancers, including GC [11-14]. It has been also reported that FAT4 functions as a GC suppressor gene by modulating Wnt/β-catenin signaling [15], and aberrant SOX11 promoter methylation is related to the clinical outcome of GC [16, 17].
Multiple environmental factors, including H. pylori infection, can profoundly influence tumorigenesis [18]. Recently, growing evidences have revealed that environmental factors may induce epigenetic alterations to further increase cancer susceptibility [19-23]. In addition, previous studies frequently focused on tumor tissues to examine DNA methylation status. However, in comparison with tissues which usually display incomplete heterogeneity information, blood samples may capture the entire heterogeneity of cancer [24]. DNA methylation in the peripheral blood leukocytes is also linked to the susceptibility to several cancers according to recent studies [25-27]. Therefore, we carried out this case-control study to detect the methylation status of FAT4 and SOX11 in peripheral blood leukocytes and investigate the association between genes methylation, gene-environment interactions, and GC risk.
Materials and methods
Study subjects
A hospital-based case-control study with 375 GC cases and 394 cancer-free controls was carried out. All newly pathologically confirmed GC cases without other digestive system tumors were recruited from the Third Affiliated Clinical Hospital of Harbin Medical University from 2010 and 2012. The controls comprised 334 ophthalmic and orthopedic patients of the Second Affiliated Hospital of Harbin Medical University, 33 neurological patients of the Fourth Affiliated Hospital of Harbin Medical University and 27 healthy individuals who underwent physical examinations at the Center for Disease Control of Xiangfang District, Harbin between 2010 and 2013. All control subjects with a history of malignant tumors or gastrointestinal diseases were excluded. Approximately 5 ml blood samples were obtained and every participant completed a face-to-face questionnaire after obtaining informed written consent according to the protocols approved by the Human Research and Ethics Committee of Harbin Medical University. The overall response rate for cases and controls were approximately 90%.
DNA extraction and bisulfite modification
Genomic DNA was extracted and then bisulfite-modified using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and EpiTect Plus DNA Bisulfite Kit (Qiagen) according to the manufacturer's instructions, respectively. DNA quantity was measured by the Nanodrop 2000 Spectrophotometer (Thermo scientific) and bisulfite-modified DNA was stored at -20℃ until use.
Serologic tests of the H.pylory by ELISA
The serologic test for H.pylori IgG antibodies were performed in duplicate using an enzyme immunoassay kit (IBL, German). The sensitivity and specificity of the kit were all greater than 95% according to the manufacturer's instructions.
Methylation-sensitive high-resolution melting (MS-HRM) assay
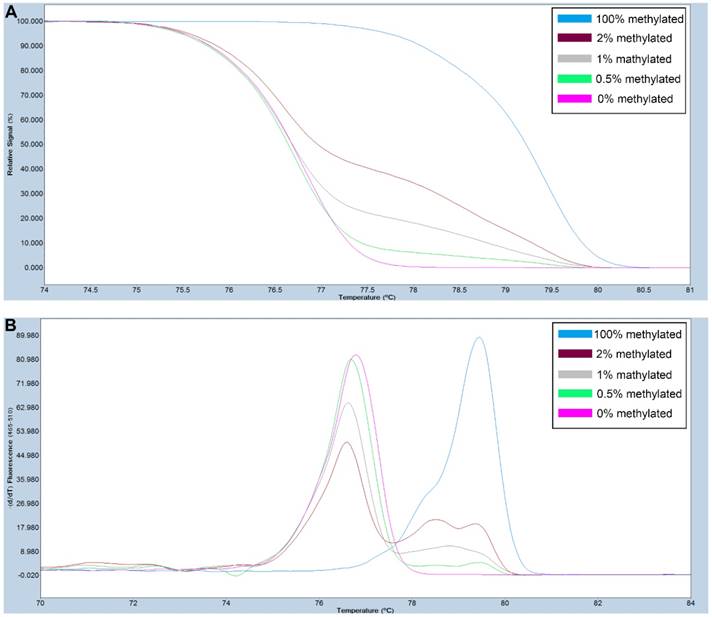
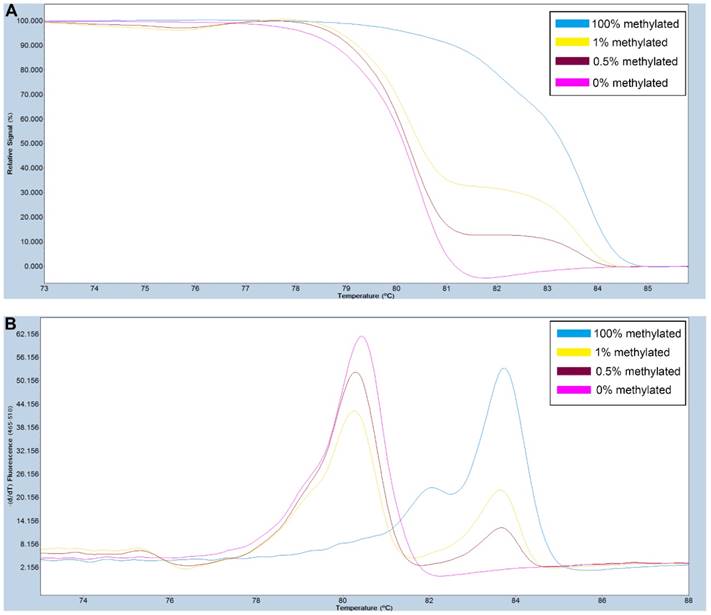
Primers used for amplifying FAT4 gene CpG rich regions were designed using Primer Premier 5.0 software. For SOX11, primers were selected from previously published study [28]. The primer sequences used are listed in Table S1. PCR amplification and MS-HRM assay was performed on a LightCycler480 (Roche Applied Science, Mannheim, Germany) equipped with Gene Scanning software (version 2.0) to detect and analyze the methylation status of genes. The final volume of each reaction system was 5µl, including 2.5µl of LightCycler480 High Resolution Melting Master Mix (Roche), 0.5µl of sodium bisulfite-modified template DNA, 0.1 µl of each forward and reverse primer, and 0.6 µl MgCl2. A series of methylated DNA standards (100%, 2%, 1%, 0.5% and 0% methylated DNA) were constructed by mixing universal unmethylated (0% methylated) and methylated (100% methylated) human whole genomic DNA samples (Zymo Research). The profiles of normalized melting curves and melting peaks of the MS-HRM assay for each gene were shown in Figure 1-2. The methylation status of genes was determined by comparing the curve of each sample with the standards curves. Then, 0% and 0.5% methylated DNA served as the cut-off values to distinguish negative methylation (Nm) and positive methylation (Pm), respectively. Heterogeneous methylation (Hm) was defined when samples melting curves presented a wider peak than the unmethylation peak or a methylation peak but before the real methylation melting profile. Total positive methylation (Tpm) was defined as Hm and Pm. In addition, DNA-free distilled water was used as blank control (no-template control) in each batch, and second trials were conducted for the unsure results.
A series of methylated DNA standards (100%, 2%, 1%, 0.5% and 0% methylated DNA) was used for FAT4. A: Normalized melting curves of the MS-HRM assay for FAT4. B: Melting peaks were generated by taking the negative derivative (d) of the melting curve data divided by the derivative with respect to time -(d/dT).
Statistical analysis
The chi-square test was applied to estimate categorical variables. Univariate and multivariate logistic regression analyses were used to assess odd ratios (OR) and 95% confidence interval (CI) for the associations between gene-specific DNA methylation, environmental factors and GC risk. Multivariable logistic regression model with backward conditional selection method was used to select GC-associated environmental risk factors (P values of 0.05 and 0.10 were specified as the thresholds for entry and removal of variables, respectively). The combined effects of environmental factors and gene-specific DNA methylation were calculated by crossover analysis. Gene-environment interactions on the risk of GC were evaluated on a multiplicative scale with a product-term coefficient using multivariable logistic regression. The relationships of gene methylation and environmental factors were further explored by multivariable logistic regression. All statistical analyses were performed using SPSS software version 19.0, with P-values of < 0.05 considered statistically significant.
A series of methylated DNA standards (100%, 1%, 0.5% and 0% methylated DNA) was used for SOX11. A: Normalized melting curves of the MS-HRM assay for SOX11. B: Melting peaks were generated by taking the negative derivative (d) of the melting curve data divided by the derivative with respect to time -(d/dT).
Results
Characteristics of subjects
A total of 375 cases and 394 controls were enrolled in this study. The basic characteristics of these subjects are presented in Table 1. The distribution of BMI and occupation between cases and controls was statistically different (P < 0.001). The proportion of members with family history of GC in cases (12.8%) was higher than that in controls (1.8%; P < 0.001).
Characteristics of subjects.
| Characteristics | Case (%) | Control (%) | P | |
|---|---|---|---|---|
| Gender | Male | 285 (76.0) | 298 (75.6) | 0.906 |
| Female | 90 (24.0) | 96 (24.4) | ||
| Age (years) | <60 | 200 (53.5) | 202 (51.3) | 0.540 |
| ≥60 | 174 (46.5) | 192 (48.7) | ||
| Educational level | Primary school or below | 110 (29.3) | 114 (28.9) | 0.996 |
| Middle school | 139 (37.1) | 147 (37.3) | ||
| Senior school | 78 (20.8) | 84 (21.3) | ||
| College or above | 48 (12.8) | 49 (12.4) | ||
| BMI (kg/m2) | ≤18.5 | 42 (11.3) | 24 (6.2) | <0.001* |
| 18.5-23.00 | 181 (48.5) | 133 (34.5) | ||
| ≥23.00 | 150 (40.2) | 229 (59.3) | ||
| Nation | Han | 361 (96.3) | 372 (94.9) | 0.357 |
| Other | 14 (3.7) | 20 (5.1) | ||
| Regions | Urban | 226 (60.6) | 238 (60.6) | 0.993 |
| Rural | 147 (39.4) | 155 (39.4) | ||
| Occupation | Non-manual worker | 135 (36.2) | 31 (8.1) | <0.001* |
| Light manual worker | 76 (20.4) | 157 (41.1) | ||
| Medium and heavy manual worker | 162 (43.4) | 194 (50.8) | ||
| Family history of GC | No | 326 (87.2) | 379 (98.2) | <0.001* |
| Yes | 48 (12.8) | 7 (1.8) | ||
BMI, body mass index; GC, gastric cancer. *Statistically significant.
Associations between environmental factors and GC risk
Multivariate logistic regression analyses displayed that several environmental factors, including irregular diet, high salt intake, overnight food, freshwater fish, fried food, smoking, drinking and H.pylori infection were significantly associated with increased GC risk (P < 0.01). In contrast, increased consumption of green vegetables, bean products and garlic could decrease GC risk with ORs of 0.270, 0.610, and 0.306 (Table S2). Backward conditional selection results suggested that irregular diet, salt, green vegetables, freshwater fish, garlic, H.pylori, drinking, overnight food and fried food were associated with GC risk (P < 0.05; Table S3).
Association between the methylation status of FAT4, SOX11 and GC risk
Compared with Nm, subjects with FAT4 Pm and Tpm showed 2.204-fold and 1.583-fold increased risk of GC (95% CI: 1.168-4.159, P = 0.015 and 95% CI: 1.031-2.430, P = 0.036, respectively). Individuals with FAT4 Hm could have a 1.695-fold increased risk of GC before adjustment (95% CI: 1.239-2.319, P = 0.001). Statistically significant association between SOX11 Pm and GC risk was observed (OR = 2.530, 95% CI: 1.289-4.969, P = 0.007), while no significant differences of SOX11 Hm and Tpm between cases and controls were found (Table 2).
Stratified analyses were further conducted in this research. The results of age-stratified analyses revealed that SOX11 Pm and Tpm was associated with increased risk of GC only in the older group (≥ 60 years, OR = 3.342, 95% CI: 1.480-7.545, P = 0.004 and OR = 1.721, 95% CI: 1.008-2.936, P = 0.047, respectively; Table 3). Moreover, in both the positive and negative H. pylori infection groups, individuals with SOX11 Pm conferred an increased risk of GC after adjusting for gender, BMI, occupation and family history of GC (OR = 2.284, 95% CI: 1.053-4.952, P=0.036 and OR = 3.323, 95% CI: 1.385-7.973, P = 0.007, respectively). Non-significant associations of FAT4 and SOX11 Hm with GC risk were observed in these two stratums (Table 4).
Relationships between FAT4, SOX11 methylation and environmental factors
The relationships between FAT4, SOX11 methylation and environmental factors were explored in all the 375 GC cases and 394 controls. As shown in Table S4, consumption of green vegetables (≥ 250g/day) and freshwater fish (≥ 1 times/week) decreased the risk of FAT4 methylation (OR = 0.450, 95% CI: 0.262-0.772, P = 0.004 and OR = 0.661, 95% CI: 0.458-0.953, P = 0.027, respectively), while drinking increased the risk of FAT4 and SOX11 methylation (OR = 1.480, 95% CI: 1.025-2.136, P = 0.036 and OR = 1.835, 95% CI: 1.272-2.646, P = 0.001, respectively).
Association between methylation status of FAT4, SOX11 and GC risk.
| Methylation status | Control (%) | Case (%) | OR (95% CI) | P | ORa (95% CI) | P a | ORb (95% CI) | P b | |
|---|---|---|---|---|---|---|---|---|---|
| FAT4 | Nm | 167 (43.5) | 114 (30.8) | 1.000 | 1.000 | 1.000 | |||
| Pm | 45 (11.7) | 57 (15.4) | 1.856 (1.174-2.932) | 0.008* | 1.751 (1.038-2.952) | 0.036* | 2.204 (1.168-4.159) | 0.015* | |
| Hm | 172 (44.8) | 199 (53.8) | 1.695 (1.239-2.319) | 0.001* | 1.418 (0.986-2.038) | 0.060 | 1.434 (0.913-2.251) | 0.117 | |
| Tpm | 217 (56.5) | 256 (69.2) | 1.728 (1.282-2.331) | <0.001* | 1.486 (1.052-2.099) | 0.024* | 1.583 (1.031-2.430) | 0.036* | |
| SOX11 | Nm | 162 (42.1) | 131 (36.4) | 1.000 | 1.000 | 1.000 | |||
| Pm | 32 (8.3) | 62 (17.2) | 2.396 (1.475-3.891) | <0.001* | 2.583 (1.467-4.546) | 0.001* | 2.530 (1.289-4.969) | 0.007* | |
| Hm | 191 (49.6) | 167 (46.4) | 1.081 (0.793-1.474) | 0.621 | 0.947 (0.659-1.362) | 0.770 | 0.945 (0.608-1.468) | 0.800 | |
| Tpm | 223 (57.9) | 229 (63.6) | 1.270 (0.945-1.706) | 0.112 | 1.159 (0.822-1.636) | 0.400 | 1.155 (0.760-1.756) | 0.499 | |
Nm, negative methylation; Pm, positive methylation; Hm, heterogeneous methylation; Tpm, total positive methylation
CI, confidence interval; OR, odds ratio. *Statistically significant.
a Adjusted for BMI, occupation, family history of GC, age, gender.
b Adjusted for BMI, occupation, family history of GC, age, gender, regular diet, salt, green vegetables, freshwater fish, garlic, H.pylori, drinking, overnight food, fried food.
Association between methylation status of genes and risk of GC by age.
| Methylation status | < 60 years | ≥ 60 years | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | ORa(95% CI) | P a | OR (95% CI) | P | ORa (95% CI) | P a | |||
| FAT4 | Nm | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| Pm | 1.849 (0.973-3.517) | 0.061 | 1.548 (0.759-3.160) | 0.230 | 1.866 (0.970-3.590) | 0.062 | 1.950 (0.893-4.260) | 0.094 | ||
| Hm | 1.878 (1.223-2.886) | 0.004* | 1.280 (0.788-2.078) | 0.318 | 1.513 (0.953-2.401) | 0.079 | 1.586 (0.905-2.778) | 0.107 | ||
| Tpm | 1.872 (1.244-2.817) | 0.003* | 1.336 (0.845-2.112) | 0.215 | 1.585 (1.020-2.463) | 0.041* | 1.661 (0.971-2.841) | 0.064 | ||
| SOX11 | Nm | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| Pm | 1.986 (0.987-3.997) | 0.054 | 2.105 (0.946-4.683) | 0.068 | 3.038 (1.540-5.993) | 0.001* | 3.342 (1.480-7.545) | 0.004* | ||
| Hm | 0.956 (0.626-1.458) | 0.833 | 0.725 (0.446-1.180) | 0.196 | 1.291 (0.814-2.046) | 0.278 | 1.405 (0.799-2.468) | 0.237 | ||
| Tpm | 1.092 (0.730-1.635) | 0.668 | 0.881 (0.555-1.398) | 0.591 | 1.561 (1.008-2.417) | 0.046* | 1.721 (1.008-2.936) | 0.047* | ||
Nm, negative methylation; Pm, positive methylation; Hm, heterogeneous methylation; Tpm, total positive methylation
CI, confidence interval; OR, odds ratio. *Statistically significant.
aAdjusted for BMI, occupation, family history of GC, gender.
Association between methylation status of genes and risk of GC by H.pylori infection.
| Methylation status | Negative H. pylori infection | Positive H. pylori infection | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | ORa(95% CI) | P a | OR (95% CI) | P | ORa (95% CI) | P a | |||
| FAT4 | Nm | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| Pm | 1.824 (0.951-3.497) | 0.071 | 1.781 (0.821-3.861) | 0.144 | 1.879 (0.979-3.607) | 0.058 | 1.714 (0.819-3.584) | 0.152 | ||
| Hm | 1.540 (0.974-2.435) | 0.065 | 1.168 (0.674-2.026) | 0.580 | 1.762 (1.138-2.728) | 0.011* | 1.522 (0.922-2.513) | 0.101 | ||
| Tpm | 1.602 (1.038-2.471) | 0.033* | 1.295 (0.773-2.169) | 0.326 | 1.785 (1.174-2.713) | 0.007* | 1.560 (0.966-2.521) | 0.069 | ||
| SOX11 | Nm | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| Pm | 2.537 (1.237-5.202) | 0.011* | 3.323 (1.385-7.973) | 0.007* | 2.255 (1.150-4.425) | 0.018* | 2.284 (1.053-4.952) | 0.036* | ||
| Hm | 1.342 (0.846-2.129) | 0.212 | 1.142 (0.648-2.011) | 0.647 | 0.916 (0.597-1.406) | 0.689 | 0.834 (0.510-1.365) | 0.470 | ||
| Tpm | 1.513 (0.973-2.352) | 0.066 | 1.409 (0.823-2.411) | 0.211 | 1.109 (0.739-1.666) | 0.617 | 1.023 (0.641-1.634) | 0.924 | ||
Nm, negative methylation; Pm, positive methylation; Hm, heterogeneous methylation; Tpm, total positive methylation
CI, confidence interval; OR, odds ratio. *Statistically significant.
a Adjusted for BMI, occupation, family history of GC, age, gender
Interactions between FAT4, SOX11 methylation and environmental factors on the risk of GC
According to the results of gene-environment interactions analyses, the significant interaction between FAT4 methylation and increased consumption of freshwater fish (≥ 1 times/week) displayed an antagonistic effect on the risk of GC (OR = 0.328, 95% CI: 0.142-0.762, P = 0.009). FAT4 methylation significantly increased risk of GC at high consumption of freshwater fish (≥ 1 times/week) and relatively slight increased risk at low consumption (< 1 times/week; OR = 7.446, 95% CI: 4.143-13.380 and OR = 2.215, 95% CI: 1.423-3.448, respectively; Table S5). As for SOX11 methylation, a marginally significant antagonistic interaction with high salt intake was observed (OR = 0.490, 95% CI: 0.242-0.995, P = 0.048; Table S6).
Discussion
Aberrant DNA methylation, which leads to inappropriate silencing of tumor suppressor and other cancer-related genes in cancer cells, is the most well-defined epigenetic change in the initiation and progression of GC [5]. Two considered potential tumor suppressor genes FAT4 and SOX11 involved in this study have been shown to participate in a series of cancer related molecular events. FAT4 exerts a tumor suppressor role in GC cell lines by modulating Wnt/β-catenin signaling [15]. Aberrant SOX11 promoter methylation has been demonstrated to be associated with poor prognosis in GC [17]. Results in this research indicated that compared with Nm, subjects with FAT4 Pm and Tpm exhibited 2.204-fold and 1.583-fold increased risk of GC, respectively. SOX11 Pm associated with the risk of GC (OR = 2.530), while Tpm and Hm had no association with GC risk. Inconsistent with our findings, Tomomitsu Tahara et al. failed to find any association between SOX11 methylation in blood-derived DNA and GC risk [25], perhaps due to relative small sample size (72 GC patients and 69 controls) and different detection methods.
It has been uncovered that aging can alter DNA methylation, and H. pylori infection can trigger inflammatory response and then also induce accumulated aberrant DNA methylation [22, 29]. Based on above mentioned points, we performed stratified analyses by age and H. pylori infection. The result of age-stratified analysis indicated that individuals with SOX11 methylation had a higher risk of GC only in elder, which might help explain the late onset of GC. Moreover, significant associations between SOX11 Pm and GC risk retained in both the H. pylori positive and negative subjects.
Considerable evidence indicated that methylation changes in cancer patients occur systematically and can be measured in surrogate nontumor tissues [26, 30]. Different cancers elicit a clearly recognizable immune response in peripheral blood [31]. The differences in blood-derived DNA methylation in cancer cases can be largely explained by systematic differences in the methylation characteristics of the leukocyte subsets [32]. However, the exact mechanism underlying the alterations in the methylation of peripheral blood-derived DNA among GC-sensitive individuals remains unclear. It is also worth noting that since the negligible density, circulating tumor cell DNA is unlikely to interfere with the results of blood-derived DNA methylation [33-35]. Several previous studies have demonstrated that DNA methylation changes in peripheral blood leukocytes is a detectable biological indicator for GC risk assessment [35-37]. Our research once again verified that blood-derived DNA methylation changes can be detected. Furthermore, compared with cancer biopsy, the detection of peripheral blood leukocytes DNA methylation has the advantages of noninvasively and relatively inexpensive.
Heterogeneous methylation refers to multiple epialleles, each with a different pattern of methylated and unmethylated CpG sites, can co-exist for a given region [38]. Many previous studies have reported that heterogeneous methylation could occur in different cancers [39-41]. However, fewer researches explored it in a relative large sample for GC. Our data showed that both FAT4 and SOX11 had heterogeneous methylation in GC patients and cancer-free controls. FAT4 Hm affected 53.8% of GC patients, significant higher than 44.8% of cancer-free controls, while no significant association between SOX11 Hm and GC risk was found. Speculative comments advanced by Azhikina et al. that heterogeneous methylation may disturb transcription process but not be sufficient to abolish transcription of the gene could partly explain these results [39]. Nevertheless, the physiological and clinical significance of heterogeneous methylation is still debated and need to be further researched.
MS-HRM was used in this research to assess FAT4 and SOX11 methylation status. It is a fast, cost- and labor-efficient technology that allowed us to quantify methylation or recognize heterogeneous methylation in a large sample panel [39, 42]. It is noteworthy that heterogeneous methylation can only be qualitative through MS-HRM, quantifying the levels of heterogeneous methylation usually need sequencing methodologies [38]. However, due to the relatively high cost of pyrosequencing, it is unsuitable for this large sample size research.
In this study, univariate and multivariate logistic regression analysis showed that freshwater fish was a risk factor for GC, which was somewhat unexpected. Several researches have reported that freshwater fish raised in contaminated water may have a relatively high level of toxic chemicals that could increase susceptibility to cancer [43-45], which to some extent explained our unexpected result. Besides, an antagonistic interaction between FAT4 methylation and increased consumption of freshwater fish was also observed. Freshwater fish is rich in methionine, which serves as the substrate for S-adenosyl methionine (SAM) and plays a critical role in maintaining the flux of methyl groups for remethylation [46]. Long-term low dietary methionine intake can disturb methionine cycle and then induce normally non-methylated DNA situation to be aberrantly hypermethylated [47], which could provide a biologically plausible basis to FAT4-fish interactions. Furthermore, analyses of the relationship between FAT4 methylation and environmental factors in all subjects showed that freshwater fish could decrease the risk of FAT4 methylation, which might further help account for this interaction.
High salt intake was found to be another GC risk factor in this study. In addition, SOX11 methylation interacted with high salt intake displayed an antagonistic effect on GC risk was also observed. Although no significant association between SOX11 methylation and high salt intake existed in all subjects, negative association was observed in cases (P = 0.043; data not shown), which might provide another way to support the observed antagonistic interaction. Environmental factors may influence tumorigenesis by affecting the process of DNA methylation of certain key genes [48, 49]. Making changes in environmental exposures might provide an opportunity to inhibit or reverse this process and then counteract cancer [50]. However, the potential biological mechanisms underlying environmental factors interfering with DNA methylation in the initiation and progression of GC are complex and variable, further explorations are needed.
Additionally, gene polymorphisms also play an important role in the occurrence of GC [51, 52]. Previous exome sequencing identified a greater than expected number of non-synonymous mutations of FAT4 in GC [53], but no studies have reported the associations between the polymorphisms of SOX11 and GC risk. Moreover, gene polymorphisms could also interacted with gene methylation status to affect the occurrence and development of GC [54, 55]. Further studies are needed to investigate the relationship between FAT4, SOX11 gene polymorphisms and GC risk, as well as the interactions between methylation and polymorphisms of these two genes on the risk of GC.
There are two main limitations in this case-control study. First, recall bias might still be inevitable in the process of collecting information on environmental factors, although measures were taken to control this bias. Second, the observed methylation changes in GC patients occurred before or after tumorigenesis remain unclear.
In conclusion, this study suggested that FAT4 methylation in peripheral blood leukocytes was associated with the risk of GC, while SOX11 was controversial and needs to be more investigated. Gene methylation status interacted with environmental factors might influence GC susceptibility.
Abbreviations
GC: gastric cancer; FAT4: FAT atypical cadherin 4; SOX11: SPY-Box11; MS-HRM: Methylation-sensitive high resolution melting; H. pylori: Helicobacter pylori; ELISA: enzyme-linked immunosorbent assay; BMI: body mass index; OR: odds ratio; CI: confidence interval; SAM: S-adenosyl-methionine; Nm: negative methylation; Pm: positive methylation; Hm: heterogeneous methylation; Tpm: total positive methylation.
Supplementary Material
Supplementary tables.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (2016-2019, Grant No. 81573219) and Innovation Research Program of Harbin Medical University (2016JCZX22).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Human Research and Ethics Committee of Harbin Medical University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
3. Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-59
4. Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol. 2006;41:401-7
5. Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53-65
6. Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2015;141:211-20
7. Yu Y, Yan W, Liu X, Jia Y, Cao B, Yu Y. et al. DACT2 is frequently methylated in human gastric cancer and methylation of DACT2 activated Wnt signaling. Am J Cancer Res. 2014;4:710-24
8. Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182-91
9. Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129
10. Roisman A, Slavutsky I. [Expression of SOX11 transcription factor. Its implication in mantle cell lymphoma]. Medicina (B Aires). 2014;74:140-6
11. Gustavsson E, Sernbo S, Andersson E, Brennan DJ, Dictor M, Jerkeman M. et al. SOX11 expression correlates to promoter methylation and regulates tumor growth in hematopoietic malignancies. Mol Cancer. 2010;9:187
12. Yoshida S, Yamashita S, Niwa T, Mori A, Ito S, Ichinose M. et al. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2017;20:136-45
13. Hide T, Takezaki T, Nakatani Y, Nakamura H, Kuratsu J, Kondo T. Sox11 prevents tumorigenesis of glioma-initiating cells by inducing neuronal differentiation. Cancer Res. 2009;69:7953-9
14. Sernbo S, Gustavsson E, Brennan DJ, Gallagher WM, Rexhepaj E, Rydnert F. et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405
15. Cai J, Feng D, Hu L, Chen H, Yang G, Cai Q. et al. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/beta-catenin signalling. Br J Cancer. 2015;113:1720-9
16. Qu Y, Zhou C, Zhang J, Cai Q, Li J, Du T. et al. The metastasis suppressor SOX11 is an independent prognostic factor for improved survival in gastric cancer. Int J Oncol. 2014;44:1512-20
17. Xu X, Chang X, Li Z, Wang J, Deng P, Zhu X. et al. Aberrant SOX11 promoter methylation is associated with poor prognosis in gastric cancer. Cell Oncol (Dordr). 2015;38:183-94
18. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-13
19. Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci. 2005;96:206-11
20. Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101-18
21. Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359-76
22. Maeda M, Moro H, Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 2017;20:8-15
23. Ahmed FE. Colorectal cancer epigenetics: the role of environmental factors and the search for molecular biomarkers. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2007;25:101-54
24. Ellinger J, Muller SC, Dietrich D. Epigenetic biomarkers in the blood of patients with urological malignancies. Expert Rev Mol Diagn. 2015;15:505-16
25. Tahara T, Maegawa S, Chung W, Garriga J, Jelinek J, Estecio MR. et al. Examination of whole blood DNA methylation as a potential risk marker for gastric cancer. Cancer Prev Res (Phila). 2013;6:1093-100
26. Dauksa A, Gulbinas A, Endzinas Z, Oldenburg J, El-Maarri O. DNA methylation at selected CpG sites in peripheral blood leukocytes is predictive of gastric cancer. Anticancer Res. 2014;34:5381-8
27. Cho YH, McCullough LE, Gammon MD, Wu HC, Zhang YJ, Wang Q. et al. Promoter Hypermethylation in White Blood Cell DNA and Breast Cancer Risk. J Cancer. 2015;6:819-24
28. Putnik M, Brodin D, Wojdacz TK, Fagerstrom-Billai F, Dahlman-Wright K, Wallberg AE. The transcriptional coregulator MAML1 affects DNA methylation and gene expression patterns in human embryonic kidney cells. Mol Biol Rep. 2016;43:141-50
29. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413-8
30. Wang BH, Li YY, Han JZ, Zhou LY, Lv YQ, Zhang HL. et al. Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget. 2017;8:31692-704
31. Whiteside TL. 22. Immune responses to malignancies. J Allergy Clin Immunol. 2003;111:S677-86
32. Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA. et al. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev. 2012;21:1293-302
33. Beeharry MK, Liu WT, Yan M, Zhu ZG. New blood markers detection technology: A leap in the diagnosis of gastric cancer. World J Gastroenterol. 2016;22:1202-12
34. Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-904
35. Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat. 2011;129:69-77
36. Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. J Clin Oncol. 2011;29:1133-9
37. Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V. et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866-74
38. Candiloro IL, Mikeska T, Dobrovic A. Assessing combined methylation-sensitive high resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics. 2011;6:500-7
39. Wojdacz TK, Windelov JA, Thestrup BB, Damsgaard TE, Overgaard J, Hansen L. Identification and characterization of locus-specific methylation patterns within novel loci undergoing hypermethylation during breast cancer pathogenesis. Breast Cancer Res. 2014;16:R17
40. Candiloro IL, Mikeska T, Hokland P, Dobrovic A. Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenetics Chromatin. 2008;1:7
41. Azhikina T, Kozlova A, Skvortsov T, Sverdlov E. Heterogeneity and degree of TIMP4, GATA4, SOX18, and EGFL7 gene promoter methylation in non-small cell lung cancer and surrounding tissues. Cancer Genet. 2011;204:492-500
42. Wojdacz TK, Moller TH, Thestrup BB, Kristensen LS, Hansen LL. Limitations and advantages of MS-HRM and bisulfite sequencing for single locus methylation studies. Expert Rev Mol Diagn. 2010;10:575-80
43. Hou H, She Y, Ma Y, Hu C, Zheng M, Zhang S. Investigations on methyl mercury contamination of fishes in the Second Songhua River. Biomed Environ Sci. 1988;1:79-82
44. Wu WZ, Schramm KW, Kettrup A. Bioaccumulation of polychlorinated dibenzo-p-dioxins and dibenzofurans in the foodweb of Ya-Er Lake area, China. Water Res. 2001;35:1141-8
45. Cheng Z, Chen KC, Li KB, Nie XP, Wu SC, Wong CK. et al. Arsenic contamination in the freshwater fish ponds of Pearl River Delta: bioaccumulation and health risk assessment. Environ Sci Pollut Res Int. 2013;20:4484-95
46. Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr. 2012;3:21-38
47. Zhu K, Davidson NE, Hunter S, Yang X, Payne-Wilks K, Roland CL. et al. Methyl-group dietary intake and risk of breast cancer among African-American women: a case-control study by methylation status of the estrogen receptor alpha genes. Cancer Causes Control. 2003;14:827-36
48. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245-54
49. Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141-51
50. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253-62
51. He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP. et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235-44
52. He J, Wang MY, Qiu LX, Zhu ML, Shi TY, Zhou XY. et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog. 2013;52(Suppl 1):E70-9
53. Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J. et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570-4
54. Sobota RS, Kodaman N, Mera R, Piazuelo MB, Bravo LE, Pazos A. et al. Epigenetic and genetic variation in GATA5 is associated with gastric disease risk. Hum Genet. 2016;135:895-906
55. Tian T, Xiao L, Du J, Zhu X, Gu Y, Qin N. et al. Polymorphisms in CARS are associated with gastric cancer risk: a two-stage case-control study in the Chinese population. Gastric Cancer. 2017;20:940-7
Author contact
![]() Corresponding authors: Wenjing Tian, M.D., Ph.D., Department of Epidemiology, College of Public Health, Harbin Medical University, 197 Xuefu Road, Harbin 150081, Heilongjiang Province, P. R. China. Tel: +86-451-87502685; Fax: +86-451-87502885; E-mail: twj8267com; Postal codes: 150081. Yashuang Zhao, M.D., Ph.D., Department of Epidemiology, College of Public Health, Harbin Medical University, 197 Xuefu Road, Harbin 150081, Heilongjiang Province, P. R. China. Tel: +86-451-87502823; Fax: +86-451-87502885; E-mail: zhao_yashuangnet; Postal codes: 150081.
Corresponding authors: Wenjing Tian, M.D., Ph.D., Department of Epidemiology, College of Public Health, Harbin Medical University, 197 Xuefu Road, Harbin 150081, Heilongjiang Province, P. R. China. Tel: +86-451-87502685; Fax: +86-451-87502885; E-mail: twj8267com; Postal codes: 150081. Yashuang Zhao, M.D., Ph.D., Department of Epidemiology, College of Public Health, Harbin Medical University, 197 Xuefu Road, Harbin 150081, Heilongjiang Province, P. R. China. Tel: +86-451-87502823; Fax: +86-451-87502885; E-mail: zhao_yashuangnet; Postal codes: 150081.

 Global reach, higher impact
Global reach, higher impact