3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(13):2334-2340. doi:10.7150/jca.25829 This issue Cite
Review
Enhancer RNAs (eRNAs): New Insights into Gene Transcription and Disease Treatment
1. Department of Urology, Shenzhen Second People′s Hospital, the First Affiliated Hospital of Shenzhen University, Clinical Medicine College of Anhui Medical University, Shenzhen 518000, Guangdong, China.
2. Anhui Medical University, Hefei 230032, Anhui Province, China
3. Department of Urology, Shenzhen Second People′s Hospital, the First Affiliated Hospital of Shenzhen University, Shenzhen 518000, China
* Equal contributors
Received 2018-3-1; Accepted 2018-3-30; Published 2018-6-6
Abstract
Enhancers are cis-acting elements that have the ability to increase the expression of target genes. Recent studies have shown that enhancers can act as transcriptional units for the production of enhancer RNAs (eRNAs), which are hallmarks of activity enhancers and are involved in the regulation of gene transcription. The in-depth study of eRNAs is of great significance for us to better understand enhancer function and transcriptional regulation in various diseases. Therefore, eRNAs may be a potential therapeutic target for diseases. Here, we review the current knowledge of the characteristics of eRNAs, the molecular mechanisms of eRNAs action, as well as diseases related to dysregulation of eRNAs.
Keywords: Enhancer RNAs, Enhancers, Gene transcription, Associated diseases
Introduction
According to whether it carries the information encoding the proteins, RNA molecules in vivo could be divided into two categories - messenger RNAs (mRNAs) and non-coding RNAs (ncRNAs)[1]. The GENCODE consortium has discovered that non-protein coding regions (more than 98%) vastly outnumber the protein coding regions in the human genome[2, 3]. It is possible to reveal a novel RNA-mediated genetic information regulatory network to annotate and elucidate the structure and function of the biological genome at an angle different from that of protein-coding genes.
The enhancer element was first observed when a 72bp-long DNA fragment located upstream of simian virus SV40 early genes significantly increased the expression of β-globin gene[4]. Subsequent decades of research have made enhancers well be annotated. Enhancers are cis-regulatory elements that bind to specific transcription factors to enhance corresponding gene transcription regardless of their orientation and location relative to the promoters[5]. It is generally believed that enhancers are highly conserved sequences, essential for developmental regulatory gene expression and tissue specificity[6, 7]. Enhancers possess some classic epigenetic features, including in histone modifications (such as, histone 3 Lysine 4 monomethylation, H3K4me1; histone 3 lysine 27 acetylation, H3K27ac), histone variants (such as, H2AZ, H3.3), co-activators (such as, Mediator complex) and an open chromatin architectures (such as, DNase I hypersensitivity) that facilitates interaction with transcriptional regulators[6, 8-11]. The development of sequencing technology has identified more than 400,000 enhancers based on the above characteristics[3, 12, 13].
In 2010, two remarkable studies demonstrated that a subset of enhancers were occupied by RNA polymerase II (RNAP II) and transcribed into a class of ncRNAs termed enhancer RNAs (eRNAs)[14, 15]. Subsequently, more and more eRNA-producing enhancer regions have been confirmed and eRNAs seem to play a noticeable role in gene transcriptional regulation. Although we have not yet fully understood the molecular mechanisms of eRNAs and their roles in gene transcription, mounting evidence suggests that the abnormal expressions of eRNAs are closely related to various human diseases. This article reviews the latest advances in eRNAs research to provide a reference for further study.
Active enhancers transcribe into eRNAs bi-directionally.
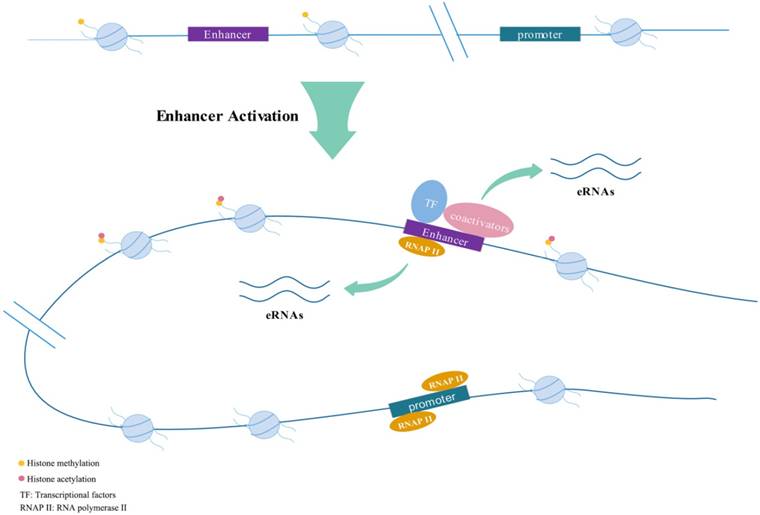
Features of eRNAs
Enhancers could respond to various stimuli (e.g., estrogen; androgen) to generate eRNAs[16-18]. They are often appeared quickly after induction, with a peak in about 1 hour, then declined rapidly[15]. In early stages of research, it was generally believed that eRNAs were 5'capped, unstable, short, usually nonpolyadenylated and unspliced and had short half-lives which were completely different from long non-coding RNAs (lncRNAs)[11, 16, 19-21]. The length of lncRNA is usually greater than 200 nucleotides (nt). Non-coding RNA-activating (ncRNA-a) is a set of lncRNA that possess enhancer-like function[22]. However, later research shows the boundaries between eRNAs and lncRNAs are not so absolute. Paralkar et al.[23] confirmed that a 434nt-long lncRNA named Locked was actually an eRNA by exploring the effect on relevant gene expression after deleting its genomic region or terminating its transcription. They discovered the transcription start site (TSS) of Locked physically interacted with the promoter of related genes like an enhancer element. Brazao et al.[24] classified a portion of lncRNAs in B-lymphocytes as eRNAs by combining chromatin immunoprecipitation (ChIP) assays with sequencing (ChIP-seq). Briefly, eRNAs have been universally discovered in a variety of cell lineages.
As enhancers tend to be tissue-specific, eRNAs derived from the same enhancer across tissues may be different[25] and the same stimulation could induce the production of various eRNAs through diversified signaling pathways[26]. Kim et al.[14] showed that not all, but only about half of the intergenic enhancers could be transcribed into eRNAs and another study indicated only 30% of typical enhancer regions overlapped with eRNAs[27]. Pulakanti et al.[28] demonstrated the increase of H3K27ac, abundant DNA hydroxylase Tet1 and DNA hypomethylation are the signatures of eRNA-generating enhancers. Dorighi et al.[29] indicated that histone methyltransferases Mll3 and Mll4 are essential for RNAP II accumulation at enhancers to activate transcription. Loss of Mll3/4 from enhancers decreased the binding rate of RNAP II and thus eRNA production. Other studies have suggested condensins could modulate the binding of co-repressors or co-activators (like p300 and RIP140) by the recruitment of an E3 ubiquitin ligase to regulate eRNA transcription[30]. Zhu et al.[31] conducted a logistic regression model and revealed that a group of four chromatin modifications was adequate to exactly estimate eRNA transcription.
Rahman et al.[32] showed that eRNAs were merely located in the nucleus and possessed similar dynamics as target mRNAs when was stimulated by estrogen in MCF-7 cells. However, Heward et al[26] demonstrated eRNAs were not limited to the nucleus, suggesting the function of eRNAs may not only be cis-mediated and some eRNAs may play a role in the cytoplasm. Multiple studies showed eRNAs transcription and enhancer-promoter looping (E:P looping) took precedence over the transcriptional activation of target genes[33, 34]. Similarly, enhancer transcription was also prior to the methylation of the histone H3 lysine 4 (H3K4me)[35]. The promoter and enhancer activity of the interferon-β gene (IFNB1) were completely preserved in the absence of eRNAs produced by viral inducible enhancer L2[36]. Blocking the production of eRNAs by flavopiridol did not affect the function of enhancers, indicating that the loading of active enhancers happened before eRNA synthesis[16].
Function of eRNAs
eRNAs are hallmarks of active enhancers
A study of estrogen-associated eRNAs showed transcriptional enhancers possessed epigenetic characteristics as we mentioned above, however, these characteristics were not observed in the absence of eRNAs[16]. The presence of eRNAs can distinguish whether the enhancer is active or silent[25, 37]. Wu et al.[38] demonstrated that the presence of tissue-specific eRNAs was a hallmark of active enhancers and they could be available to forecast the corresponding enhancers independent of known epigenetic markers through RNA sequencing. Zhu et al.[31] compared the ability of H3K27ac and eRNA transcription to predict enhancer activity and revealed the latter was more representative. Sandor et al.[39] determined whether the enhancers were functional by measuring the production of eRNAs in thymocytes. Previous study showed P53 regulated hundreds of eRNAs and Leveille et al. localized p53-regulated enhancers by eRNA production[40, 41].
eRNAs involved in transcriptional regulation of target genes
The function of eRNAs was controversial at an early stage, whether they were merely by-products of transcription events under high concentrations of RNAP II or indeed played a critical role in transcriptional activation. In recent years mounting studies suggests that eRNAs play a functional role in orchestrating gene expression which allows for the regulation of multiple gene expression through one gene binding site[42]. Large-scale analysis demonstrated a positive correlation between the expression level of eRNAs and target[43] or nearby genes[14, 20]. p53-bound enhancer regions (p53BERs) which is necessary for transcriptional enhancement produced eRNAs in a p53 dependent way[42]. Knockdown of eRNAs by siRNAs or LNAs or terminating transcription of corresponding enhancers by drugs would decrease the target gene expression[16, 17, 21]. A large body of functional genomic data over the past few years had brought substantial evidence that eRNAs worked through diverse pathways to regulate gene transcription.
eRNAs act on RNAP II with multiple means
eRNAs were synthesized prior to the target gene transcription and interact with the negative elongation factor (NELF) complex as a decoy to release the paused RNAP II to trigger transcription. Knockdown of eRNAs blocked the efficiently release of NELF, which was consistent with the decrease in mRNA expression, but had no effect on the enhancer-promoter looping[34, 44]. In addition, prostate-specific antigen (PSA) eRNA activated positive transcription elongation factor (P-TEFb) by binding to CYCLIN T1, and then increased phosphorylation of serine-2 of RNAP II to facilitate target gene transcription[45]. Other knockdown studies of eRNAs have also demonstrated that eRNAs may facilitate RNAP II recruitment specifically to the gene promoter of the myogenic differentiation antigen (MyoD)[46]. eRNA E2-3 derived from the distal enhancer of heme oxygenase-1 gene human (HO-1) selectively regulated HO-1 gene induction by affecting RNAP II binding to enhancer regions and the promoter[47]. In conclusion, eRNAs may not only be the products, but also the reason for the high concentration of RNAP II occupied by enhancers[46].
Enhancer-promoter looping (E:P looping)
Enhancers could physically interact with promoters to form E:P looping, thereby regulating gene expression independent of distance and orientation[48, 49]. Studies have shown that inducible eRNAs are extensively transcribed by short treatments with AR or ER, thus strengthening specific E:P looping to upregulate relevant coding genes selectively[17]. Huang et al.[50] discovered that eRNA-driven looping enhanced the expression of the lncRNA DHRS4-AS1. Interactions of enhancers with promoters of the gonadotropin gene are lost after eRNA knockout[51]. The E:P looping is mediated at least in part by mediator complex and cohesins[17, 18, 22, 52]. But single-cell profiling demonstrated that accumulation of eRNAs at chromosomal looping was not essential for mRNA continuous transcription[32].
The molecular mechanism of eRNAs action include: eRNAs interact with looping factors, such as cohesions and mediators, to strengthen enhancer-promotor looping to promote transcription; eRNAs act as decoys of the negative elongation factor (NELF) to eliminate transcriptional repression or activate positive transcription elongation factor (P-TEFb) to facilitate gene transcription; eRNAs modify histones to promote chromatin accessibility to influence gene expression.
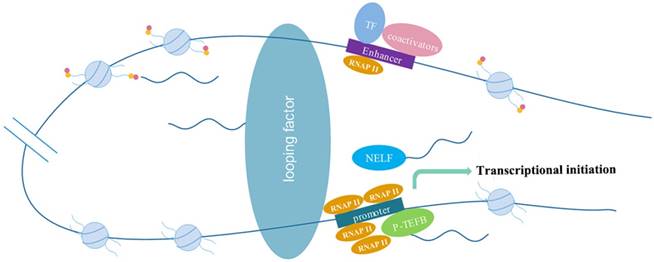
Histone modifications
Creb-binding protein (CBP) interacts with a large number of transcription factors to form transcriptional regulatory networks, allowing for the recruitment of almost all enhancers[53]. This makes CBP one of the hallmarks of enhancers[54, 55]. eRNAs could bind to CBP to regulate the acetyltransferase activity and then increased H3K27ac to promote transcription of the corresponding genes[56]. Bromodomain-containing protein 4 (BRD4), a member of the BET (Bromodomain and Extra Terminal) family proteins, assisted elongation of both coding and enhancer RNAs by interacting with acetylated histones[57]. The treatment of LPS resulted in increased H3K4me2 and H3K27ac at enhancer regions of the primary monocytes through ChIP-seq and knockdown of eRNAs by antisense oligonucleotides mainly affected histone modifications[34].
eRNAs can also alter histone modifications to regulate transcriptional repression. Nuclear receptor Rev-Erbs inhibited transcription in assistance of co-repressor histone deacetylase (NCoR)-HDAC3 complexes. After binding with enhancers, the levels of eRNAs and histone 3 lysine9 acetylation (H3K9Ac) were both reduced. Knockout of Rev-Erbs significantly derepressed eRNA production at relevant enhancers[19].
Abnormalities of eRNAs are closely associated with a variety of diseases
Many previous studies on gene-related diseases have focused on mutations in coding genes, because they contain a blueprint for protein synthesis, which directly affects cellular biological functions. However, a large number of studies have shown that ncRNAs play a non-redundant role in regulating gene transcription and protein generation, such as eRNAs[58]. eRNAs can participate in the development of various disease by regulating the expression of multiple genes.
eRNAs and inflammation
NE et al.[59] identified 76 LPS-induced eRNAs in monocytes. Among them, IL1β-eRNA mediated by the NF-KB signaling pathway was required for proinflammatory responses. Knockdown of IL1β-eRNA reduced the corresponding mRNA expression and the inflammatory media IL1β production, exhibiting the ability of eRNAs to regulate LPS-induced immune response. Furthermore, demethylase Kdm6a promoted IFN-β transcription through promoting full activation of IFN-β-specific enhancers to produce eRNAs in macrophages upon innate stimuli[60]. Bmal1 encoded by Arntl gene is a core circadian clock gene that regulates various genes involved in circadian rhythm. Bmal1 regulates inflammatory responses in macrophages by regulating the epigenetic states of enhancers, such as the level of H3K27 acetylation and modulating enhancer RNA transcription[61]. Inducible eRNAs may participate in intra- and inter-chromosomal gene regulation in the innate immune response[62]. In consequence, the close relationship between eRNAs and immune response in vivo provides a new perspective for the anti-inflammatory treatment.
As a crucial NF-kB regulatory factor, BRD4 regulates the expression of many NF-kB-associated inflammatory genes. BRD4 occupies distal enhancer regions where its presence is associated with enhancer activity and eRNAs transcription[63]. Helicobacter pylori (H. pylori) recruits BRD4 to the promoters and enhancers of inflammatory genes to propel transcription. BRD4 inhibitor JQ1 attenuates the immune response by inhibiting H. pylori-induced eRNAs synthesis, in concomitant with decreased target gene synthesis. These provides potential therapeutic options for Helicobacter pylori associated gastric diseases, including gastritis and gastric cancer[27].
eRNAs and neurodegenerative disorders
Pathogenic mutations carried on enhancers are easily overlooked in traditional genetic studies, but they play an important role in the development of hereditary neurodevelopmental and neuropsychiatric disorders. More than 100 enhancers have been found in brain tissue and they are more active than other tissues. Le et al.[64] discovered that most of the enhancers in mouse striatum were transcribed into eRNAs. In huntington's disease (HD) mouse, relevant eRNAs were downregulated due to the reduction of RNAP II binding sites at HD mouse enhancers, leading to down-regulation of target genes compared with healthy individuals. Another study showed that enhancer regions producing eRNAs have genetic variants and were related to autism spectrum disorders (ASD)[65], indicating enhancer-derived eRNAs as pivotal factors in neurodevelopmental impairments.
eRNAs and tumors
Zhao et al.[66] identified hundreds of differentially expressed AR-eRNAs in enzalutamide-resistant prostate cancer cells through AR ChIP-seq analyses. eRNAs involved in regulating the biological behavior of enzalutamide-resistant prostate cancer cells. What's more, PSA eRNA was up-regulated in castration resistant prostate cancer (CRPC) cells, patient-derived xenografts (PDX), and patient tissues, and played its role as we described previously[45], which may become a potential therapeutic target for CRPC. In another hormone-dependent tumor breast cancer cell MCF-7, Li et al.[21] had reported widely estrogen-induced eRNA transcription with upregulated corresponding coding genes. Further studies showed that eRNAs enhanced E:P looping to increase estrogen-associated gene expression, and this was dependent on cohesins.
Mutations located in enhancer regions in thyroid cancer are extremely rare and have a high genetic risk. He et al.[67] found that the capacity of mutated enhancer binding to transcription factor POU2F and YY1 was significantly weakened, accompanying by a large number of reduced eRNAs. Mutations in enhancer regions were also observed in a type of basal cell carcinomas (BCC), which was called Bazex-Dupré-Christol syndrome (BDCS). The activity of mutant enhancer around ACTRT1 was reduced, as was the expression of ACTRT1. Act in related protein Tl (ARP-Tl) was encoded by ACTRT1 and the loss of ARP-Tl could abnormally activate the Hedgehog pathway, leading to the occurrence of BCC[68].
eRNAs and diabetes
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor (NR). Rosiglitazone activates PPARγ potently with highly selective to improve insulin sensitivity in the body to control blood glucose effectively. Rosi-up-regulated eRNAs occurred at sites of strong PPARγ binding to which co-activators such as MED1, CBP and p300 were recruited. Remarkably, however, downregulation of eRNA transcription occurred at sites that were devoid of PPARγ but enriched for C/EBP and AP-1 family members[69]. Rosi-induced eRNAs occured only at PPARγ binding sites and could activate or inhibit adipocyte gene transcription in conjunction with co-activators or some transcription factors.
Inducible eRNAs related to diseases
| Inducible eRNAs | Corresponding enhancers | Inducible factors | Target genes | References |
|---|---|---|---|---|
| Inflammatory associated eRNAs | ||||
| L2 enhancer, IFN-β-specific enhancers | Sendai virus, LPS | IFNB1 | 35, 58 | |
| MnSOD intronic enhancer | Pro-inflammatory stimuli, the TEAD1/p65 complex | MnSOD | 60 | |
| enhancers of inflammatory genes | Helicobacter pylori, LPS | NF-kB-dependent inflammatory genes | 25, 26, 57 | |
| Tumor-associated eRNAs | ||||
| AR-regulated enhancers | Androgen | ARHGEF26, FTO, NKX3.1, et al. | 17, 43, 64 | |
| ER-regulated enhancers | Estradiol | GREB1, P2RY2, SMAD7, et al. | 16, 21 | |
| p53-bound enhancers | Retroviruses, nutlin-3 | p53-dependent genes | 38, 39, 40 | |
| Brain region-specific eRNAs | ||||
| Arc enhancers | Neural activity | NELF complex | 42 | |
| Diabetes-associated eRNAs | ||||
| PPAR γ-binding enhancer sites | Rosiglitazone | Fabp4, Pdk4 et.al | 67 |
LPS: lipopolysaccharide; IFNB1: the interferon-β gene; MnSOD: manganese superoxide dismutase; AR: androgen receptor; ER: estrogen receptor; Arc: activity-regulated cytoskeletal protein; PPAR γ: peroxisome proliferator-activated receptor γ.
Conclusions and perspectives
In summary, the discovery of eRNAs provides a new perspective for better understanding of enhancers. eRNAs play an important role in the transcriptional process of target genes and the development of various diseases, and may be used to guide the treatment of disease safely and efficiently in the future. Nevertheless, the remaining puzzles of eRNAs require more extensive and thorough investigations. First of all, our understanding of eRNAs is largely based on high-throughput sequencing technologies. Given unstable and short half-life characteristics of eRNAs, tools to better detect changes intemporal and spatial with greater sensitivity are yet to be developed. In addition, opening up the methods for delaying the degradation of eRNAs is beneficial to our research. Next, generally identifying and documenting various patterns of eRNAs motion in different cell types is essential, as well as the function of silent enhancers that can't transcribe into eRNAs. Finally, we should expand the scope of eRNAs study. For instance, verifying abnormal expression of eRNAs in human tissues in case of illness is contribute to guide us in developing agents for eRNAs. Therefore, further research needs to be done to better understand the biology functions and mechanisms of eRNAs in gene regulation, human diseases and homeostasis.
Acknowledgements
This work was supported by the National Key Basic Research Program of China (973 Program) (2014CB745201), National Natural Science Foundation of China (81772737), the Shenzhen Municipal Government of China (JCYJ20170413161749433, JSGG20160301161836370), The Sanming Project of Shenzhen Health and Family Planning Commission, SZSM201412018, SZSM201512037. The high level university's medical discipline construction 2016031638.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Eddy SR. Non-coding RNA genes and the modern RNA world. Nature reviews Genetics. 2001;2:919-929
2. Djebali S, Davis CA, Merkel A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-108
3. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
4. Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299-308
5. Lee K, Hsiung CC, Huang P. et al. Dynamic enhancer-gene body contacts during transcription elongation. Genes & development. 2015;29:1992-1997
6. Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327-339
7. Plank JL, Dean A. Enhancer function: mechanistic and genome-wide insights come together. Molecular cell. 2014;55:5-14
8. Levine M. Transcriptional enhancers in animal development and evolution. Current biology: CB. 2010;20:R754-763
9. Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60-63
10. Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome biology. 2011;12:R113
11. Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annual review of genetics. 2012;46:1-19
12. Maurano MT, Humbert R, Rynes E. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190-1195
13. Thurman RE, Rynes E, Humbert R. et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75-82
14. Kim TK, Hemberg M, Gray JM. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182-187
15. De Santa F, Barozzi I, Mietton F. et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS biology. 2010;8:e1000384
16. Hah N, Murakami S, Nagari A. et al. Enhancer transcripts mark active estrogen receptor binding sites. Genome research. 2013;23:1210-1223
17. Hsieh CL, Fei T, Chen Y. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7319-7324
18. Wang D, Garcia-Bassets I, Benner C. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390-394
19. Lam MT, Cho H, Lesch HP. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511-515
20. Andersson R, Gebhard C, Miguel-Escalada I. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455-461
21. Li W, Notani D, Ma Q. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516-520
22. Lai F, Orom UA, Cesaroni M. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497-501
23. Paralkar VR, Taborda CC, Huang P. et al. Unlinking an lncRNA from Its Associated cis Element. Molecular cell. 2016;62:104-110
24. Brazao TF, Johnson JS, Muller J. et al. Long noncoding RNAs in B-cell development and activation. Blood. 2016;128:e10-19
25. Cheng JH, Pan DZ, Tsai ZT. et al. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Scientific reports. 2015;5:12648
26. Heward JA, Roux BT, Lindsay MA. Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells. FEBS letters. 2015;589:396-406
27. Chen J, Wang Z, Hu X. et al. BET Inhibition AttenuatesHelicobacter pylori-Induced Inflammatory Response by Suppressing Inflammatory Gene Transcription and Enhancer Activation. The Journal of Immunology. 2016;196:4132-4142
28. Pulakanti K, Pinello L, Stelloh C. et al. Enhancer transcribed RNAs arise from hypomethylated, Tet-occupied genomic regions. Epigenetics. 2013;8:1303-1320
29. Dorighi KM, Swigut T, Henriques T. et al. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Molecular cell. 2017;66:568-576 e564
30. Li W, Hu Y, Oh S. et al. Condensin I and II Complexes License Full Estrogen Receptor alpha-Dependent Enhancer Activation. Molecular cell. 2015;59:188-202
31. Zhu Y, Sun L, Chen Z. et al. Predicting enhancer transcription and activity from chromatin modifications. Nucleic acids research. 2013;41:10032-10043
32. Rahman S, Zorca CE, Traboulsi T. et al. Single-cell profiling reveals that eRNA accumulation at enhancer-promoter loops is not required to sustain transcription. Nucleic acids research. 2017;45:3017-3030
33. Kim YW, Lee S, Yun J. et al. Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the beta-globin locus. Bioscience reports. 2015;35:1-9
34. Shii L, Song L, Maurer K. et al. SERPINB2 is regulated by dynamic interactions with pause-release proteins and enhancer RNAs. Molecular immunology. 2017;88:20-31
35. Kaikkonen MU, Spann NJ, Heinz S. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310-325
36. Banerjee AR, Kim YJ, Kim TH. A novel virus-inducible enhancer of the interferon-beta gene with tightly linked promoter and enhancer activities. Nucleic acids research. 2014;42:12537-12554
37. Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nature reviews Genetics. 2016;17:207-223
38. Wu H, Nord AS, Akiyama JA. et al. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS genetics. 2014;10:e1004610
39. Sandor K, Daniel B, Kiss B. et al. Transcriptional control of transglutaminase 2 expression in mouse apoptotic thymocytes. Biochimica et biophysica acta. 2016;1859:964-974
40. Leveille N, Melo CA, Rooijers K. et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nature communications. 2015;6:6520
41. Allen MA, Andrysik Z, Dengler VL. et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200
42. Melo CA, Drost J, Wijchers PJ. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524-535
43. Sanyal A, Lajoie BR, Jain G. et al. The long-range interaction landscape of gene promoters. Nature. 2012;489:109-113
44. Schaukowitch K, Joo JY, Liu X. et al. Enhancer RNA facilitates NELF release from immediate early genes. Molecular cell. 2014;56:29-42
45. Zhao Y, Wang L, Ren S. et al. Activation of P-TEFb by Androgen Receptor-Regulated Enhancer RNAs in Castration-Resistant Prostate Cancer. Cell reports. 2016;15:599-610
46. Mousavi K, Zare H, Dell'orso S. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606-617
47. Maruyama A, Mimura J, Itoh K. Non-coding RNA derived from the region adjacent to the human HO-1 E2 enhancer selectively regulates HO-1 gene induction by modulating Pol II binding. Nucleic acids research. 2014;42:13599-13614
48. Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Molecular cell. 2002;10:1467-1477
49. Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Molecular cell. 2005;19:631-642
50. Yang Y, Su Z, Song X. et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Scientific reports. 2016;6:20961
51. Pnueli L, Rudnizky S, Yosefzon Y. et al. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4369-4374
52. Kagey MH, Newman JJ, Bilodeau S. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430-435
53. Bedford DC, Kasper LH, Fukuyama T. et al. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9-15
54. Creyghton MP, Cheng AW, Welstead GG. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931-21936
55. May D, Blow MJ, Kaplan T. et al. Large-scale discovery of enhancers from human heart tissue. Nature genetics. 2011;44:89-93
56. Bose DA, Berger SL. eRNA binding produces tailored CBP activity profiles to regulate gene expression. RNA biology. 20170
57. Kanno T, Kanno Y, LeRoy G. et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nature structural & molecular biology. 2014;21:1047-1057
58. Murakawa Y, Yoshihara M, Kawaji H. et al. Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights into Diseases. Trends in genetics: TIG. 2016;32:76-88
59. NE II, Heward JA, Roux B. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nature communications. 2014;5:3979
60. Li X, Zhang Q, Shi Q. et al. Demethylase Kdm6a epigenetically promotes IL-6 and IFN-beta production in macrophages. Journal of autoimmunity. 2017;80:85-94
61. Oishi Y, Hayashi S, Isagawa T. et al. Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Scientific reports. 2017;7:7086
62. Chokas AL, Bickford JS, Barilovits SJ. et al. A TEAD1/p65 complex regulates the eutherian-conserved MnSOD intronic enhancer, eRNA transcription and the innate immune response. Biochimica et biophysica acta. 2014;1839:1205-1216
63. Najafova Z, Tirado-Magallanes R, Subramaniam M. et al. BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic acids research. 2017;45:127-141
64. Le Gras S, Keime C, Anthony A. et al. Altered enhancer transcription underlies Huntington's disease striatal transcriptional signature. Scientific reports. 2017;7:42875
65. Yao P, Lin P, Gokoolparsadh A. et al. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nature neuroscience. 2015;18:1168-1174
66. Zhao J, Zhao Y, Wang L. et al. Alterations of androgen receptor-regulated enhancer RNAs (eRNAs) contribute to enzalutamide resistance in castration-resistant prostate cancer. Oncotarget. 2016;7:38551-38565
67. He H, Li W, Wu D. et al. Ultra-rare mutation in long-range enhancer predisposes to thyroid carcinoma with high penetrance. PloS one. 2013;8:e61920
68. Bal E, Park HS, Belaid-Choucair Z. et al. Mutations in ACTRT1 and its enhancer RNA elements lead to aberrant activation of Hedgehog signaling in inherited and sporadic basal cell carcinomas. Nature medicine. 2017;23:1226-1233
69. Step SE, Lim HW, Marinis JM. et al. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARgamma-driven enhancers. Genes & development. 2014;28:1018-1028
Author contact
![]() Corresponding authors: Yuchen Liu, Weiren Huang. E-mail: liuyuchenmdcgcom (Yuchen Liu); pony8980com (Weiren Huang)
Corresponding authors: Yuchen Liu, Weiren Huang. E-mail: liuyuchenmdcgcom (Yuchen Liu); pony8980com (Weiren Huang)

 Global reach, higher impact
Global reach, higher impact