Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(14):2502-2509. doi:10.7150/jca.25264 This issue Cite
Research Paper
The significant prognostic value of ZEB1-AS1 up-regulation in patients with cancer.
1. Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Jiangsu University, 301 Xuefu Road, Zhenjiang, Jiangsu 212013, China.
2. Department of Transfusion, The first affiliated hospital of Anhui medical university, Hefei, Anhui 230000, China.
3. Department of Cardiology, Affiliated Hospital of Jiangsu University, Zhenjiang, Jiangsu 212001, China
* These authors contributed equally to this work and should be considered co-first authors.
Received 2018-1-31; Accepted 2018-5-9; Published 2018-6-15
Abstract
Zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1) is a long non-coding RNA, which has found to unregulated in various kinds of cancer. This meta-analysis was conducted to demonstrate the association between ZEB1-AS1 expression levels and clinical outcome or prognosis of cancer patients.10 studies with 783 cancer patients were included in this meta-analysis by retrieving 5 databases (PubMed Central, EMBASE, Cochrane Library, Wiley Online Library and Medline).The result showed that overexpression of ZEB1-AS1 is significantly correlated with poor OS (Hazard ratio, HR=2.45, 95% confidence interval, CI: 1.89-3.16). ZEB1-AS1 expression levels were also associated with clinicopathological parameters including lymph node metastasis (Yes vs. No; OR=4.00, 95%CI: 2.23-7.17, P<0.00001), histologic differentiation (Moderate + poor vs. Well; OR=2.72, 95% CI: 1.69-4.37, p<0.0001), tumor metastasis and invasion (Yes vs. No; OR =2.52, 95%CI: 1.12-5.68, P=0.03) and TNM stage (III+IV vs. I+II; OR=2.76, 95 %CI 1.46-5.21, P=0.002). However, ZEB1-AS1 expression was not significantly associated with patients' gender (Male vs. Female; OR=1.20, 95% CI: 0.87-1.66; P=0.27).This meta-analysis indicated the potential value of ZEB1-AS1 as a biomarker for predicting a poor prognosis in patients with cancer.
Keywords: ZEB1-AS1, cancer, OS, meta-analysis, poor prognosis
Introduction
Cancer is now a major challenge in field of public health because it has become a leading cause of morbidity and mortality worldwide [1]. According to 2017 Cancer Statistics[2], a total of estimated 1.68 million new cancer cases and more than 600,000 cancer deaths are expected to occur in the United States in 2017. Tumor biomarkers have been considered as novel research targets to predict tumor prognosis, which is of great importance for disease surveillance and clinical treatment, since the 5-year survival rate remains poor for majority patients [1, 2].
Long noncoding RNAs (lncRNAs) are types of noncoding RNA which functionally defined as transcripts >200 nt in length with no protein-coding potential[3]. In recent years, increasing studies have shown that lncRNAs are involved in numerous important biological phenomena and uniquely expressed in differentiated tissues or specific cancer types, while it was considered as genomic “junk” and “noise” initially [4]. A variety of lncRNAs have appeared to deem as biomarkers for diagnosis and prognosis of patients with cancers. For example,high NEAT1 expression resulting in shorter OS may be a useful biomarker for predicting a poor prognosis [5] and high ZFAS1 expression may relate to poor clinical outcome in various cancers [6]. LncRNAs might have served as attractive targets in malignant progression and clinical application for cancer treatment [7] since their aberrant expression often results in tumor biological processes and is associated with poor prognosis of human cancer.
Zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1) is a non-coding antisense transcript emanating from the promoters of ZEB1, which is a critical transcription factor functioning in many tumors [8]. As a well-characterized oncogen-related lncRNA, ZEB1-AS1 is unregulated in various kinds of cancer, including esophageal squamous cell carcinoma (ESCC), hepatocellular carcinoma (HCC), osteosarcoma, glioma, colorectal cancer (CRC), bladder cancer (BC), prostate cancer (PC), B-lineage acute lymphoblastic leukemia (BALL) and gastric cancer(GC) [9-19].
It was observed that high ZEB1-AS1 expression was associated with metastasis and prognosis, thus ZEB1-AS1 may be a potential biomarker for prognosis. However, no meta-analysis has been conducted to date on the correlation of ZEB1-AS1 with the survival of cancer patients. This current meta-analysis is the first one to evaluate the overall clinical value of lncRNA ZEB1-AS1 in different kinds of cancers by analyzing the correlation between ZEB1-AS1 expression level and clinical parameters, and further determine whether ZEB1-AS1 could act as an effective diagnostic biomarker and prognostic factor.
Materials and Methods
Literature search strategies
PubMed Central, EMBASE, Cochrane Library, Wiley Online Library and Medline were searched by using “long non-coding RNA ZEB1-AS1” or “ZEB1 antisense RNA 1” or “LncZEB1-AS1” or “ZEB1-AS1” as keywords. Reference lists of relevant articles and review papers were also searched manually to identify potential studies. The latest searching time was March 5, 2018.
Inclusion and exclusion criteria
Inclusion criteria are as followed: (1) Studies were published in English, (2) patients were groupedaccording to the expression level of ZEB1-AS1 in cancer tissues, (3) Clinical data or clinicopathologic parameters including overall survival (OS), gender, LNM, TMN stage or et al. were described, and (4) containing sufficient data for calculating odds ratios (OR) and corresponding 95 % confidence intervals (CI) or a P. value. Exclusion criteria were: (1) animal studies, case reports, meta-analyses, and review articles and (2) papers lacking all raw data or inability to calculate its HR, 95% CI, and p values.
Data extraction and study quality assessment
According to Inclusion and exclusion criteria eligible studies included the first author's name, publication year, study regions, sample size, cancer type, sample type and method of ZEB1-AS1 testing. 8 out 10 studies also contain the HR statistics for outcome. For the survival data extraction, the data was directly applied if a study reported the detailed HRs and 95% CIs for survival. Otherwise, the HRs and 95% CIs was retrieved with Engauge Digitizer version 10.1 if a study only provided survival curves [20].
Study quality was assessed by using the Newcastle-Ottawa quality assessment scale (NOS). The NOS score items included selection, outcome, and comparability, and ranged between 0 and 9.
Statistical analysis
The heterogeneity among studies was identified via I2 statistics and chi-square Q test. The random effects model was adopted if there were significant heterogeneity between-studies (I2≥50%), otherwise the fixed effects model was used [21]. Sensitivity analysis was performed to assess whether the individual study affected the overall results. Statistical significance was defined when the p-value was less than 0.05.
All the statistical analysis above was performed with Stata statistical software version 12.0 (Stata Corporation, College Station, Texas, USA) and RevMan5.3 software (Cochrane Collaboration, http://www.cc-ims.net/RevMan/ relnotes.htm/).
Results
Literature retrieval and characteristics of eligible studies
A flow diagram of literature retrieval process was presented as Figure 1. After screening of the title and abstract carefully, 64 irrelevant articles were excluded and 12 potential eligible studies were selected. After further review of the full articles, 2 papers were eliminated due to lack of information regarding survival outcomes or clinicopathological parameters. Based on the standard of article selection, finally a total of 10 publications were included in this meta-analysis [9-18]. They have been all written in English and the publication time of all included articles has been ranged from 2015 to 2017.
A total of 783 patients were included, with sample size ranging from 30 to 124. Nine different types of cancer were included in present analysis, with 1 esophageal squamous cell carcinoma (ESCC), 1 hepatocellular carcinoma (HCC), 1 osteosarcoma, 1 glioma, 1 colorectal cancer (CRC), 1 bladder cancer (BC), 1 prostate cancer (PC), 1 B-lineage acute lymphoblastic leukemia (BALL) and 2 gastric cancer (GC). The main characteristics were summarized in Table 1. Among the 10 studies, 9 involved tissue collection, and 1 involved bone marrow and plasma analysis. The expression level of lncRNA ZEB1-AS1 was determined with quantitative polymerase chain reaction and normalized to GAPDH or β-actin.
Correlation of high ZEB1-AS1 expression with OS in human cancer
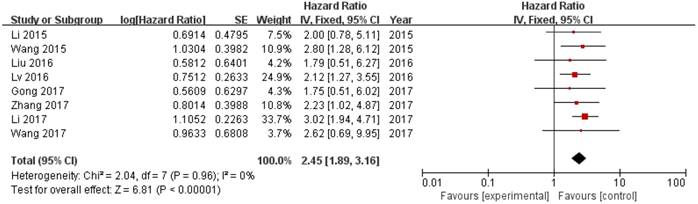
8 out of 10 studies showed OS according toZEB1-AS1 expression levels among the 614 patients. The pooled hazard ratio (HR) was 2.45 (95% confidence interval [CI]: 1.89-3.16) for the high ZEB1-AS1 expression group versus low expression group (P<0.00001; Fig. 2). There was no significant heterogeneity (I2=0%, PQ=0.96), and the fixed-effects model was chosen to estimate the pooled HR. This suggested that high expression of ZEB1-AS1 was a predicator of poor prognosis among human cancers.
Correlation of ZEB1-AS1 expression withlymph node metastasis
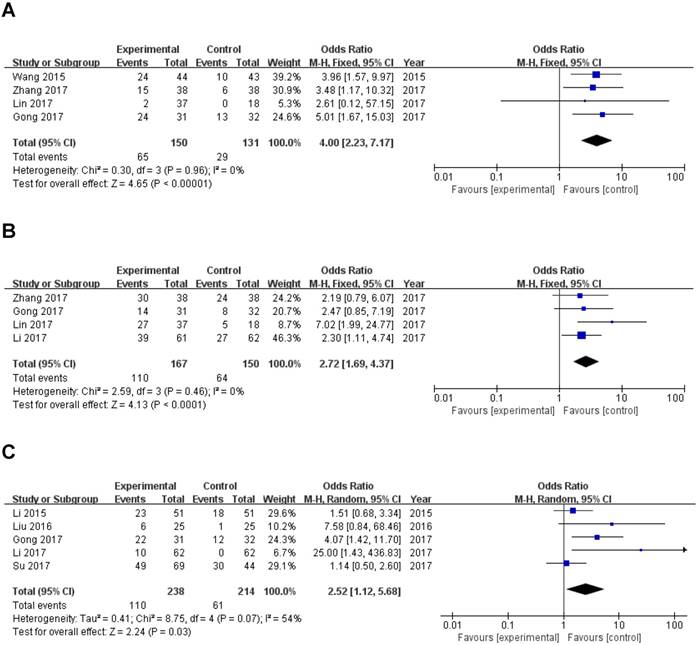
Four studies reported the lymph node metastasis (LNM) of 281 patients based on different ZEB1-AS1 expression levels. Analysis showed a pooled OR=4.00 (95%CI: 2.23-7.17, P<0.00001) in a fixed-effects model with no significant heterogeneity among the studies (I2=0%, PQ=0.96) (Fig. 3A). The result indicated that overexpression of ZEB1-AS1 was significantly related to LNM.
Flow chart of studies selection procedure
Forest plot of HR for ZEB1-AS1 high expression and overall survival
Characteristics of studies included in this meta-analysis
| Author | Year | Country | Sample size | Sample type | Cancer type | LNM | TNM stage | Follow-up | Method | Outcome | HR statistics | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes Vs No | I/II Vs III/IV | (Month) | ||||||||||
| Wang | 2015 | China | 87 | Tissue | ESCC | 34/53 | 36/51 | 60 | qRT-PCR | OS,DFS | Reported | 8 |
| Li | 2015 | China | 102 | Tissue | HCC | NA | 78/24 | 50 | qRT-PCR | OS,RFS | Reported | 8 |
| Liu | 2016 | China | 50 | Tissue | Osteosarcoma | 7/43 | 41/9 | 40 | qRT-PCR | OS,RFS | Survival curve | 8 |
| Lv | 2016 | China | 82 | Tissue | Glioma | NA | 37/45 | 50 | qRT-PCR | OS | Reported | 7 |
| Gong | 2017 | China | 63 | Tissue | CRC | 37/26 | 19/44 | 60 | qRT-PCR | OS,RFS | Survival curve | 8 |
| Lin | 2017 | China | 55 | Tissue | BC | 2/53 | 38/17 | NA | qRT-PCR | NA | NA | 7 |
| Su | 2017 | China | 114 | Tissue | PC | NA | 27/87 | NA | qRT-PCR | NA | NA | 7 |
| Wang | 2017 | China | 30 | bone marrow and plasma | B-ALL | NA | NA | >25 | qRT-PCR | OS | Survival curve | 7 |
| Li | 2017 | China | 124 | Tissue | GC | 58/66 | 75/49 | 72 | qRT-PCR | OS | Reported | 8 |
| Zhang | 2017 | China | 76 | Tissue | GC | 21/55 | 37/39 | 60 | qRT-PCR | OS | Survival curve | 7 |
ESCC, esophageal squamous cell carcinoma; HCC, hepatocellular carcinoma; CRC, colorectal cancer; BC, bladder cance; PC, prostate cancer; BALL, B-lineage acute lymphoblastic leukemia GC, gastric cancer; NA, not available
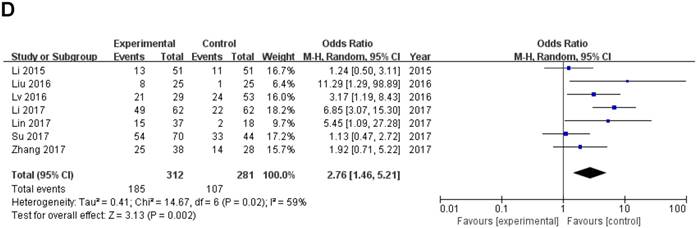
Forest plot of ZEB1-AS1 expression and OR for clinicopathological parameters. The investigated clinicopathological parameters are (A) LNM, (B) histologic differentiation, (C) tumor metastasis and invasion, (D) TNM stage
Correlation of ZEB1-AS1 expression with histologic differentiation
A total of 317 patients in four studies were included to detect the relationship between ZEB1-AS1 expression levels and histologic differentiation in this meta-analysis. We observed a significant association between histologic differentiation and ZEB1-AS1 expression (OR=2.72, 95% CI: 1.69-4.37, P<0.0001) (Fig. 3B). In addition, we also did not observe any significant heterogeneity among the studies (I2 =0%, PQ =0.46). This result demonstrated that high ZEB1-AS1 expression might be significantly associated with moderate or poor histologic differentiation.
Correlation of ZEB1-AS1 expression with tumor metastasis and invasion
Five studies with 171 cancer patients were included in the analysis about the association between ZEB1-AS1 expression and. It was showed that tumor metastasis and invasion (Yes vs. No; OR =2.52, 95%CI: 1.12-5.68; P=0.03; Fig. 3C) is related to ZEB1-AS1 up-regulation in patients with cancer.
Correlation of ZEB1-AS1 expression with TNM stage
Data of total OR and 95%CI of TNM stage were collected from the 7 eligible studies. Analysis showed a pooled OR of 2.76 with (95%CI 1.46-5.21, P=0.002) by a random-effects model as the significant heterogeneity (I2=59%, PQ =0.02) (Fig. 3D).
A sensitivity analysis was used to figure out the source of heterogeneity. After excluding the Li study [14], the heterogeneity declined (I2 =34%, PQ =0.18) and the results did not change (OR=2.11, 95 % CI: 1.20-3.71, P=0.009) (Sup-1, 2).The results also revealed a significant difference in the TNM stage between the two groups. Therefore, the results demonstrated that high ZEB1-AS1 expression might significantly related to higher tumor grade in patients with cancer.
Correlation of ZEB1-AS1 expression with other clinicopathological parameters and cancer type
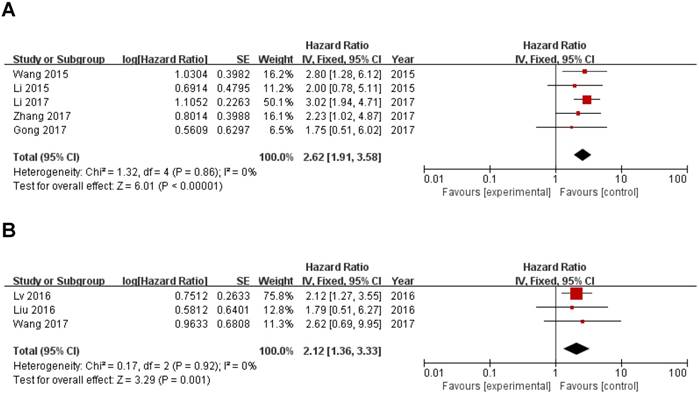
In this meta-analysis, we also investigated the correlation of ZEB1-AS1 expression with digestive system tumor and other tumors and other clinicopathological parameters, which are illustrated in Figure 3 and Table 2. High ZEB1-AS1 expression correlated with poor prognosis of digestive system tumor (HR=2.62, 95%CI: 1.91-3.58; Fig. 4A) and other tumors (HR=2.12, 95%CI: 1.36-3.33; Fig. 4B) patients. In addition, ZEB1-AS1 expression was not significantly associated with patients' gender (Male vs. Female; OR=1.20, 95% CI: 0.87-1.66; p=0.27; Sup-3).
ZEB1-AS1 expression in other cancer types
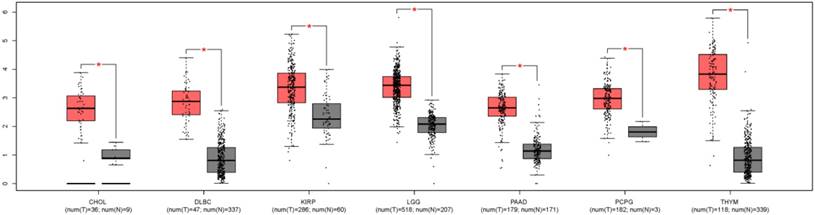
In order to understand the expression level of ZEB1-AS1 in other cancer types, we used GEPIA, a newly developed interactive web server for analyzing the RNA sequencing expression data from the TCGA and the GTEx projects, to analyze the expression of ZEB1-AS1 in different cancer types[22]. The results indicated that the expression of ZEB1-AS1 was higher in the tumor tissues than the corresponding normal issues (Fig. 5).
Results of the association between ZEB1-AS1 and clinicopathological parameters and cancer type.
| Outcome | Studies(n) | OR(HR) | 95%CI | P Value | Modle | Heterogeneity |
|---|---|---|---|---|---|---|
| Chi2, I2, P Value | ||||||
| Lymph node metastasis(Yes vs. No) | 4 | 4 | 2.23-7.17 | <0.00001 | Fixed | 0.30, 0%, 0.96 |
| Histologic differentiation (Moderate + poor vs. Well) | 4 | 2.72 | 1.69-4.37 | <0.0001 | Fixed | 2.59, 0%, 0.46 |
| Tumor metastasis and invasion (Yes vs. No) | 5 | 2.52 | 1.12-5.68 | 0.03 | Random | 8.75, 54%, 0.07 |
| TNM stage (III+IV vs. I+II) | 7 | 2.76 | 1.46-5.21 | 0.002 | Random | 14.67, 59%, 0.02 |
| Gender (Male vs. Female) | 8 | 1.2 | 0.87-1.66 | 0.27 | Fixed | 7.02, 0%, 0.43 |
| Digestive system malignancy | 5 | 2.62 | 1.91-3.58 | <0.00001 | Fixed | 1.32, 0%, 0.86 |
| Other tumors | 3 | 2.12 | 1.36-3.33 | 0.001 | Fixed | 0.17, 0%, 0.92 |
Forest plot of ZEB1-AS1 expression with OS in digestive system tumor and other tumors patients. (A) digestive system tumor, (B) other tumors
The ZEB1-AS1 gene expression profile across tumor samples and paired normal tissues. CHOL, Cholangio carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; KIRP, Kidney renal papillary cell carcinoma; LGG, Brain Lower Grade Glioma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; THYM, Thymoma.
Sensitivity analysis of the effect of the individual study on the pooled HRs for the correlation between ZEB1-AS1 expression and overall survival (OS)
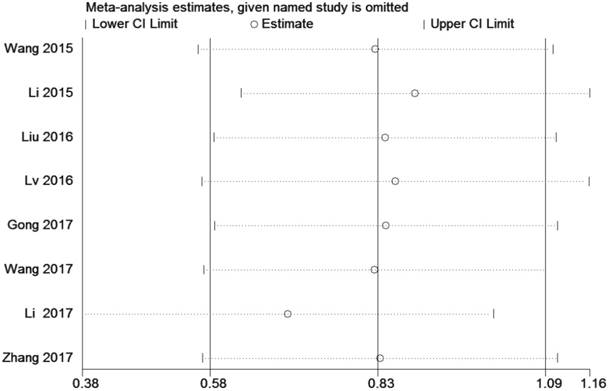
Sensitivity analysis
A sensitivity analysis was conducted by using Stata 12.0 to analyze the association between ZEB1-AS1 and OS by deleting one study at a time from the pooled analysis to examine the influence of the removed data set to the overall HR. The result was not influenced by the exclusion of each study, suggesting that the result of the synthetic analysis was robust (Fig. 6).
Discussion
LncRNA ZEB1-AS1, first identified as the most efficient lncRNA to boost cellular proliferation in human HCC by Li in 2015, is considered as a cancer-related lncRNA, which has been detected to be up-regulated in many malignancies and may act as a novel oncogene. Li et al [10] confirmed that ZEB1-AS1 could promote EMT and HCC metastasis through increasing the activity of ZEB1 promoter. Su et al [16] identified that ZEB1-AS1could also increased ZEB1 by indirectly inhibited miR200c to facilitate proliferation and migration of prostate cancer cells, which firstly suggested an impaired regulatory network in prostate cancer. Gong et al [13] reported that ZEB1-AS1 expression level of CRC cells can be increased through repressing p15 expression and first confirmed ZEB1-AS1 as an indicator of facilitating tumorigenesisfor colorectal cancer. Lv et al [12] demonstrated that ZEB1-AS1 overexpression could modulate the expression of Bax, Bcl-2, Cyclin D1 and CDK2 to influence apoptosis and migration of glioma cell, suggesting a potential prognostic marker for human gliomas. Li et al [14] found ZEB1-AS1 expression is associated with ZEB1 protein and demonstrated ZEB1-AS1 served as oncogenic roles in the regulation of EMT process through modulating ZEB1 in gastric cancer, which is similar to previous studies in human HCC. In addition, ZEB1-AS1 expression was found negatively correlated with miR-335-5p expression in gastric cancer tissues, which indicated ZEB1-AS1 may promoted cell proliferation and invasion through suppressing the tumor suppressor miR-335-5p [18].Based on these studies and owing to its function, therefore, the significant prognostic value of ZEB1-AS1 up-regulation is confirmed in patients with cancer.
Our meta-analysis was performed to explore the relationship between the expression level of ZEB1-AS1 and clinical outcomes in cancer patients. To our knowledge, this is the first meta-analysis providing comprehensive insights into the clinical value of lncRNAZEB1-AS1 in cancers. The results indicated that high expression of ZEB1-AS1 was correlated with poor OS in different types of cancers, suggesting that ZEB1-AS1 may act as an effective diagnostic biomarker and prognostic factor in various cancers. In addition, subgroup analyses showed that ZEB1-AS1 expression levels were significantly associated with clinicopathological parameters including tumor metastasis and invasion, lymph node metastasis, histologic differentiation and TNM stage.
Heterogeneity was existed between ZEB1-AS1 expression level and TNM stage, which is a potential problem in interpreting the results of any meta-analysis. To resolve this problem, we performed a sensitivity analysis, and the existing heterogeneity obviously declined from 59% to 34% after excluding the Li study [14], but the results did not change. This means Li study might have a significant influence on overall heterogeneity. Through our further analysis, we thought the potential sources of heterogeneity for this result may include the sample size, RT-qPCR reaction conditions and systems, different tumor clinical pathological parameters and the inconsistent cut-off value.
The meta-analysis above indicated that high expression of ZEB1-AS1 was associated with poor survival outcome, lymph node metastasis, moderate or poor histologic differentiation, positive tumor metastasis and invasion and high grades of clinical stage. However, several limits should not be ignored in the present analysis. First, both the number of included articles and the sample size in each article were limited. Only 10 studies with 9 types of cancer were included in this meta-analysis. Second, different types of cancers increased the heterogeneity which might influence accuracy of the results. Third, differences may occur between ethnic groups because all the studies were conducted in China. Additionally, there were 4 studies in which HR were calculated through survival curves, thus they may not have provided the most accurate HR estimate. Our results should be confirmed in further studies.
In conclusion, despite the limitations described above, the present study provides the evidence that high ZEB1-AS1 expression may result in unfavorable prognosis in human cancers since it is a risk factor for shorter OS time, LNM, poor histologic differentiation, tumor metastasis and invasion and clinical stage. Moreover, ZEB1-AS1 might be a useful biomarker to predict poorer prognosis in human cancers. However, more clinical studies and data are needed to further confirm the findings of this analysis.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 81772157), Natural Science Foundation of Jiangsu Province, China (grant number BK20161345) and Key Science Research Project in Colleges and Universities of Jiangsu Province, China (Grant number 17KJA320001).
Supplementary Material
Supplementary figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. et al. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians. 2016;66:271-89
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
3. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46-58
4. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature reviews Genetics. 2016;17:47-62
5. Fang J, Qiao F, Tu J, Xu J, Ding F, Liu Y. et al. High expression of long non-coding RNA NEAT1 indicates poor prognosis of human cancer. Oncotarget. 2017;8:45918-27
6. Liu F, Gao H, Li S, Ni X, Zhu Z. Long non-coding RNA ZFAS1 correlates with clinical progression and prognosis in cancer patients. Oncotarget. 2017;8:61561-9
7. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452-63
8. Li J, Li Z, Leng K, Xu Y, Ji D, Huang L. et al. ZEB1-AS1: A crucial cancer-related long non-coding RNA. Cell proliferation. 2017
9. Wang YL, Bai Y, Yao WJ, Guo L, Wang ZM. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. International journal of clinical and experimental pathology. 2015;8:11871-6
10. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z. et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575-84
11. Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. American journal of translational research. 2016;8:4095-105
12. Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin CZ. et al. A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. International journal of molecular sciences. 2016:17
13. Gong H, Wen H, Zhu X, Lian Y, Yang X, Qian Z. et al. High expression of long non-coding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39:1010428317705336
14. Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z. et al. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surgical oncology. 2017;26:527-34
15. Lin J, Zhan Y, Liu Y, Chen Z, Liang J, Li W. et al. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8:24202-12
16. Su W, Xu M, Chen X, Chen N, Gong J, Nie L. et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Molecular cancer. 2017;16:142
17. Wang Q, Du X, Yang M, Xiao S, Cao J, Song J. et al. LncRNA ZEB1-AS1 contributes to STAT3 activation by associating with IL-11 in B-lymphoblastic leukemia. Biotechnology letters. 2017;39:1801-10
18. Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by Long Noncoding RNA ZEB1-AS1 in Gastric Cancer Promotes Tumor Proliferation and Invasion. DNA and cell biology. 2017
19. Fu J, Cui Y. Long noncoding RNA ZEB1-AS1 expression predicts progression and poor prognosis of colorectal cancer. The International journal of biological markers. 2017;32:e428-e33
20. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
21. DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemporary clinical trials. 2015;45:139-45
22. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017
Author contact
![]() Corresponding authors: Shihe Shao, shaoshihe2006com, +86-511-85038449; Chen Shao, shaochen84com, +86-511-8503058
Corresponding authors: Shihe Shao, shaoshihe2006com, +86-511-85038449; Chen Shao, shaochen84com, +86-511-8503058

 Global reach, higher impact
Global reach, higher impact