Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(14):2559-2570. doi:10.7150/jca.24601 This issue Cite
Research Paper
ADAM10 Sheddase Activity is a Potential Lung-Cancer Biomarker
1. Department of Pathology, University of Pittsburgh; UPMC Hillman Cancer Center, Pittsburgh, PA.
2. Department of Immunology, University of Pittsburgh; UPMC Hillman Cancer Center, Pittsburgh, PA.
3. Department of Biostatistics, University of Pittsburgh; UPMC Hillman Cancer Center, Pittsburgh, PA.
4. Department of Medicine, University of Pittsburgh; UPMC Hillman Cancer Center, Pittsburgh, PA.
5. Department of Pharmacology and Chemical Biology, University of Pittsburgh; UPMC Hillman Cancer Center, Pittsburgh, PA.
6. VAPHS, Pittsburgh, PA.
7. BioZyme Inc, Apex, NC.
8. Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA.
9. Center for Systems Biology, Massachusetts General Hospital and Harvard Medical School, Boston, MA.
Received 2017-12-27; Accepted 2018-3-31; Published 2018-6-23
Abstract
Background: Increases in expression of ADAM10 and ADAM17 genes and proteins are inconsistently found in cancer lesions, and are not validated as clinically useful biomarkers. The enzyme-specific proteolytic activities, which are solely mediated by the active mature enzymes, directly reflect enzyme cellular functions and might be superior biomarkers than the enzyme gene or protein expressions, which comprise the inactive proenzymes and active and inactivated mature enzymes.
Methods: Using a recent modification of the proteolytic activity matrix analysis (PrAMA) measuring specific enzyme activities in cell and tissue lysates, we examined the specific sheddase activities of ADAM10 (ADAM10sa) and ADAM17 (ADAM17sa) in human non-small cell lung-carcinoma (NSCLC) cell lines, patient primary tumors and blood exosomes, and the noncancerous counterparts.
Results: NSCLC cell lines and patient tumors and exosomes consistently showed significant increases of ADAM10sa relative to their normal, inflammatory and/or benign-tumor controls. Additionally, stage IA-IIB NSCLC primary tumors of patients who died of the disease exhibited greater increases of ADAM10sa than those of patients who survived 5 years following diagnosis and surgery. In contrast, NSCLC cell lines and patient tumors and exosomes did not display increases of ADAM17sa.
Conclusions: This study is the first to investigate enzyme-specific proteolytic activities as potential cancer biomarkers. It provides a proof-of-concept that ADAM10sa could be a biomarker for NSCLC early detection and outcome prediction. To ascertain that ADAM10sa is a useful cancer biomarker, further robust clinical validation studies are needed.
Keywords: Lung cancer, Tumor tissue, Blood exosomes, Lysate, Fluorogenic peptide substrate, Proteolytic activity matrix analysis, ADAM10, ADAM17, Sheddase activity, Cancer biomarker
Introduction
Lung cancer (LC) is the second most frequent malignancy and the leading cause of cancer-related deaths worldwide. It is a stealth disease that develops without symptoms, and is usually diagnosed in an advanced stage that cannot be successfully treated. Among LC patients, 85% have NSCLC, and among NSCLC patients, only 25% are diagnosed with the localized IA-IIB disease stages. These patients with the localized NSCLC are considered to have favorable prognosis, and are predominately treated with the conventional surgery alone. However, approximately 50% of these patients experience disease recurrence and die within 5 years after diagnosis and surgery [1].
Biomarkers that could detect clinically hidden and discriminate aggressive and non-aggressive LC would enable diagnosis of the early, curable stages of disease in high risk populations (i.e., heavy smokers) and prediction of the disease outcome, respectively. Ultimately, the biomarkers would timely enable personalized effective treatments of the early-stage and aggressive LC. However, although several recent studies have indicated that specific signatures of microRNA expression could distinguish better- and worse-prognosis LC [2], no clinically validated biomarkers of LC are currently available.
Biologically active molecules that are increased in cancer or stroma cells and mediate cancer-cell growth, survival, invasion and metastasis formation could be promising cancer biomarkers and therapy targets [3-7]. ADAM10 (A disintegrin And Metalloproteinase 10) and ADAM17 are plasma-membrane anchored metalloproteinases that have such features [8, 9], being able to hydrolyze proximal to the cell surface, shed and activate ectodomains of transmembrane molecules that mediate epidermis growth, inflammation, and cancer pathogenesis, growth, invasion and metastasis formation [10-28].
Increases in expression of ADAM10 and ADAM17 genes and/or proteins have been inconsistently found in cancer tissues, and have not been validated as clinically applicable biomarkers. [8-10, 28-41]. The enzyme-specific proteolytic activities are solely mediated by the active (uninhibited) mature enzymes, which directly reflect the enzyme cellular functions and might be, therefore, superior biomarkers than the expression of enzyme genes or proteins comprising inactive proenzyme and active (uninhibited) and inactive (inhibited) mature enzyme forms [42, 43]. Because ADAM10sa and ADAM17sa are directly associated with cancer pathogenesis and aggressive behavior, and a particular active form of ADAM10 is selectively displayed in cancer cells [44], the increased specific sheddase activities could be significant biomarkers of cancers [42].
In the present study, using our recent modification of the high-throughput multiplex proteolytic activity matrix analysis (PrAMA) [45] that specifically measures ADAM10sa and ADAM17sa in cell and tissue lysates [46], we determined that ADAM10sa is consistently and selectively upregulated in NSCLC cell lines and patient primary tumors and circulating exosomes, and distinctly more upregulated in the primary tumors of patients having poor prognosis. These findings indicate that ADAM10sa could be a clinically useful early-detection and outcome-prediction biomarker of NSCLC.
Materials and Methods
Reagents
The FRET (Fluorescence-Resonance Energy Transfer) peptide substrates PEPDAB005, PEPDAB008, PEPDAB010, PEPDAB011, PEPDAB013, PEPDAB014 and PEPDAB022, which are based on the cleavage sites in specific proteins known to be processed by different ADAM family members [45, 46], were obtained from BioZyme Inc (Apex, NC). Recombinant ADAM10 and ADAM17 were purchased from R&D Systems (Minneapolis, MN). EDTA-free non-MP protease inhibitor cocktails, tablets and Halt solution, were obtained from Roche Applied Science (Indianapolis, IN) and ThermoFisher Scientific (Pittsburgh, PA), respectively. Trypsin and enzyme-free cell-dissociation solution were obtained from GIBCO-Life Technologies (Grand Island, NY).
Cell lines and peripheral blood cells
NSCLC A549, H520, H596, H441, H358 and H2345, and small-cell lung carcinoma (SCLC) H69 cell lines were obtained from ATCC (Manassas, VA). They were cultured in RPMI-1640 cell-culture medium supplemented with 10% FCS (GIBCO-Life Technologies). Human normal bronchial-epithelium (NHBE) cells, COPD bronchial-epithelium (COPDBE) cells and normal-lung fibroblasts (NLFB) were purchased from Lonza (Allendale, NJ). They were cultured in BEGMTM BulletKitTM medium (Lonza). The cell lines were kept stored in the frozen stocks, and cultured up to 6 passages, when needed for the experiments. Peripheral blood mononuclear leukocytes (PBLs) were obtained using Phycoll-Hypaque (Sigma-Aldrich, St. Louis, Mo) density-gradient centrifugation of healthy-donor peripheral blood provided by the University of Pittsburgh Medical Center (UPMC) Lung Cancer (LC) SPORE Tissue Bank (TB). Natural killer (NK) cells were purified from PBLs using Miltenyi Biotech (San Diego, CA) human NK-cell isolation kit [47]. Dendritic cells (DCs) were generated from PBL monocytes, as previously described [48].
Tissues, plasma and sera
Ten pairs of NSCLC primary-tumor and tumor-free normal-lung tissues and 10 chronic-obstructive-pulmonary-disease (COPD) lung tissues preserved dry-snap-frozen; and 41 pairs of NSCLC primary-tumor and tumor-free normal-lung tissues, 25 NSCLC primary-tumor tissues without control-lung tissues, and 27 COPD-lung tissues, all preserved frozen in the RNA-protection solution RNAlater (Qiagen, Germantown, MD), were obtained surgically. All tumor and paired lung tissues were obtained from stage IA-IIB NSCLC patients. Tissues were verified by histopathology. Most of the tumors were adenocarcinomas (AD) and squamous cell carcinomas (SQ) (Table 1). Plasma specimens (1 mL) were obtained from 5 healthy donors and 4 stage IA-IIB and 1 stage IIIB NSCLC tumor-bearing patients. Serum specimens (1 mL) were obtained from 7 healthy donors, 7 small-pulmonary-nodule (SPN) patients, 7 COPD patients and 6 stage IIIA and 1 stage IIIB NSCLC tumor-bearing patients (Table 1). All the biological specimens were obtained, deidentified, stored frozen at -80oC, and provided by the UPMC-LC-SPORE-TB under the IRB-approved protocol REN16070229/IRB9502100. All tissue and blood donors were either active or former smokers.
Essential demographic, clinical and pathologic information for tumor tissues, plasma and sera
| Plasma | Sera | |||||||
|---|---|---|---|---|---|---|---|---|
| Tumors | NSCLC | Healthy | NSCLC | SPN | COPD | Healthy | ||
| Age (years) | ||||||||
| <60 | 19 | 2 | 2 | 3 | 3 | 3 | 3 | |
| >60 | 47 | 3 | 3 | 4 | 4 | 4 | 4 | |
| Gender | ||||||||
| Male | 35 | 3 | 3 | 5 | 5 | 5 | 5 | |
| Female | 31 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Stage | ||||||||
| IA | 17 | 2 | ||||||
| IIA | 4 | |||||||
| IIIA | 1 | 6 | ||||||
| IB | 26 | 1 | ||||||
| IIB | 18 | 1 | ||||||
| IIIB | 1 | 1 | ||||||
| Histological type | ||||||||
| Squamous | 21 | 3 | 1 | |||||
| Adeno | 37 | 1 | 2 | |||||
| Adeno- Squamous | 3 | 1 | 4 | |||||
| Large cell | 5 | |||||||
| Tumor size | ||||||||
| T1 | 21 | 2 | 3 | |||||
| T2-T3 | 45 | 3 | 4 | |||||
| Lymph node metastases | ||||||||
| Negative | 42 | 3 | 4 | |||||
| Positive | 24 | 2 | 3 | |||||
| Outcome during 6 years | ||||||||
| Died | 17 | 4 | 3 | |||||
| Alive | 18 | 1 | 4 | |||||
Isolation of exosomes
The standard fractionated ultracentrifugation [49] and the newly developed size-defined filtration methods were used to isolate exosomes from peripheral blood plasma and sera, respectively. Plasma fractionated ultracentrifugation was performed by 20-min centrifugation at 2000 g, to remove platelets; filtration through 0.45 μm pore filter, to remove cell debris; and 40-min centrifugation at 100,000 g using 30% sucrose gradient, to pellet exosomes. The exosomes were washed twice in PBS by 60-min centrifugations at 100,000 g.
Size-defined filtration was performed by passing sera through a stack of inter-connected 450, 220 and 20 nm pore-size ExoMir filters (Bioo Scientific Co., Austin, Tx). The first two filters retained cell debris and cell-derived particles larger than exosomes. The third 20 nm pore-size filter retained only exosomes and enabled complete removal of smaller particles and soluble blood proteins and washing of the retained exosomes using PBS. Exosomes are subsequently lysed and their active enzymes extracted in the 20 nm pore-size filter.
Preparation of cell, tissue and exosome lysates
All procedures were performed at 0oC [46]. Single cell suspensions of viable cells were prepared from cultured adherent cells using the non-enzymatic cell-dissociation solution. Detached cells were washed twice in culture medium and three times in PBS. Frozen tissues were thawed on ice and washed three times in PBS. Cell and tissue lysates were prepared using a lysis buffer composed of 50 mM Tris-HCl/150 mM NaCl buffer supplemented with 0.1% SDS, 1% sodium deoxycholate, 1% Nonidet P40, 1% Triton X-100, and 1% EDTA-free protease inhibitor cocktail (Roche). Cells (106) were lysed in 50 μL of the lysis buffer for 1 h. Tissues (10-50 mg) were lysed by homogenization in 500 μl of the lysis buffer using the tissue-grinder system (ThermoFisher Scientific) and 1-h incubation. Exosomes obtained from 1 mL of peripheral blood plasma or sera were lysed in 200 or 500 μL of lysis buffer, respectively. The lysates were centrifuged at 16,000 g for 20 min, their supernatants were collected, and protein concentrations determined using Quick Start Bradford Dye Reagent (Bio-Rad, Hercules, CA).
ELISA
Human ADAM10 and ADAM17 ELISA kits were obtained from Antibodies Online Inc (Atlanta, GA) and R&D Systems, respectively. ELISAs were performed using 10 μg of lysates.
Metalloprotease enzyme activity assay
The assay was performed in 100 μL of assay-buffer solutions of 1 to 10 μg of cell-, tissue- or exosome-lysate proteins, or 10 ng of recombinant ADAM10 (rADAM10) or rADAM17 and supplemented with 10 μM PEPDAB substrates (BioZyme) [46]. The assay buffer consisted 25 mM Tris-HCl buffer pH 8, supplemented with 0.1 mg/μL BSA, 0.0006% Brij-35, 1% glycerol, 0.5% Roche-tablet (Roche) and 0.5% ThermoFisher Scientific Halt (Halt) broad-spectrum protease inhibitor cocktails without EDTA. Totally processed substrate was obtained using 1% trypsin. The enzymatic reaction was induced by incubating the lysate/substrate solutions in 96-well white-wall, flat-bottom LUMITRAC-200 plates (Sigma Aldrich) at 37oC for 1 to 4 h. Fluorescence was measured in a TECAN-infinite-200-pro fluorimeter (Technical Communities, Inc., San Bruno, CA) using 485 nm excitation and 530 nm emission wavelengths. The experiments were performed in duplicates or triplicates. The obtained data were computed using the formulas: 1) Experimental fluorescence-intensity gain (EFIG) = Experimental-sample mean fluorescence intensity (ESMFI) - Background-sample mean fluorescence intensity (BSMFI); 2) Trypsin-induced fluorescence-intensity gain (TIFIG) = Trypsin-sample mean fluorescence intensity (TSMFI) - BSMFI; 3) pM processed substrates = EFIG x 106/TIFIG.
Modified Proteolytic Activity Matrix Analysis (PrAMA)
The assay was performed as described before [45, 46]. Briefly, PrAMA is an integrated mathematical modeling analysis that interprets dynamic signals from panels of moderately specific fluorogenic substrates to deduce a profile of specific MP-proteolytic activities. Deconvolution of enzyme signals from complex mixtures of proteases is accomplished by using individual MP-cleavage signatures for the substrate-panel measured with purified enzymes. The method relies upon the assumption that the total observed substrate cleavage is the linear sum of cleavage from all enzyme activities in a complex biological mixture. As a consequence of this assumption, a vector of specific enzyme activities, E, may be inferred from a signature of substrate cleavages, V0, by solving the following equation:
ET=[S]0-1C-1V0T
where [S]0 is the concentration of substrate, and C comprises a matrix of kinetic parameters that describes the efficiency (kcat/KM) with which particular enzymes cleave the substrates. The modified PrAMA was performed using systematically optimized and validated combinations of sensitivity (Syntherror) and specificity (Sigmathreshold) parameters to achieve maximal (100%) sensitivity and specificity with no cross-reactivity between rADAM10 and rADAM17, and lysates of wild-type and ADAM10- or ADAM17-knockout cells, control and ADAM10-cDNA-transfected ADAM10-knockout cells, control and ADAM10-siRNA- or ADAM17-siRNA-transfected NSCLC cells, and control and GI254023X-inhibited NSCLC-cell or -tissue lysates (46). Ten-thousand bootstrapping repetitions for each data-set were analyzed. The computation was performed using Matlab-2009a MathWorks (Natick, MA).
Statistics
Data were statistically evaluated using the SPSS-10.0 (SPSS Inc., Chicago, IL) and R-3.3.2 (https://www.r-project.org) programs. Data are reported as means + standard deviation (SD) or standard error (STE), and medians, quartiles and ranges. Wilcoxon Sum-Rank tests and Wilcoxon Rank-Sum tests were used to compare two groups of independent and paired data, respectively. Pearson's coefficients were used to measure correlations. Linear-regression analysis with 95% confidence-band of the best fit line was performed to evaluate correlations. The receiver-operating-characteristic (ROC) curve analysis was conducted to assess the ability of biomarkers to discriminate groups. The ROC curve, area under ROC (AUC), cutoff, sensitivity (SE), specificity (SP) and accuracy (AC) were evaluated. Optimal cutoffs were chosen using the maximized Youden Index (SE+SP-1). To avoid over-fitting of the data, a 10-fold cross-validation was used to estimate the predictive values of models. Most of the tests were two-sided. The one-sided tests were indicated in the figure legends. p values <0.05 were considered significant.
Results
ADAM10sa is upregulated in cultured lung cancer cells
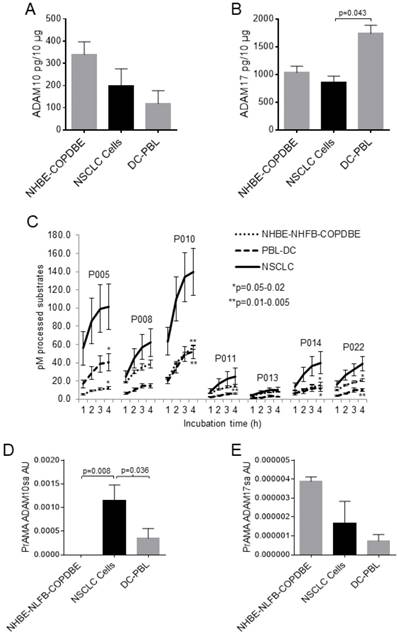
A cancer biomarker candidate should be significantly different in cancerous and non-cancerous cells. Therefore, we initially tested the levels of ADAM10 and ADAM17 proteins and sheddase activities in NSCLC, normal lung, COPD lung and inflammatory cells. All the tested cells, including NSCLC, NHBE, COPDBE (NHBE-COPDBE), DCs and PBLs (DC-PBL), contained substantial amounts of ADAM10 and ADAM17 proteins as measured by ELISA (Figs. 1A, 1B, Supplementary Figs. 1A, 1B). ADAM10 and ADAM17 protein levels were marginally decreased in NSCLC cells relative to NHBE-COPDBE cells and DC-PBL, respectively. In contrast, the processing of PEPDAB005, 010, 011, 014 and 022 substrates was significantly increased in NSCLC-cell lysates relative to the non-cancerous NHBE-NHFB-COPDBE-cell and DC-PBL lysates (Fig. 1C). Subsequently, PrAMA inferred from these findings that ADAM10sa was also significantly increased in NSCLC-cell lysates relative to NHBE-NLFB-COPDBE-cell (p=0.008) and DC-PBL (p=0.036) lysates (Fig. 1D, Supplementary Fig. 1C). Contrarily, ADAM17sa was marginally decreased in NSCLC-cell and DC-PBL lysates relative to NHBE-NLFB-COPDBE cell lysates (Fig. 1E, Supplementary Fig. 1D). These findings show that ADAM10sa, but not ADAM10 and ADAM17 protein expression or ADAM17sa, is increased in NSCLC cells. They also indicate that the protein expression and sheddase activity of either ADAM10 or ADAM17 do not correlate in the examined cells.
ADAM10sa levels are high in lung cancer cells, but undetectable or low in non-cancerous cells. Lysates of cancerous NSCLC A549, H520, H2347, H596, H441 and H358, and SCLC H69 cells; and non-cancerous NHBE, NLFB, COPDBE, PBL (n=5), DCs (n=8) and/or NK cells (n=3) were tested for the presence of (A) ADAM10 and (B) ADAM17 proteins using ELISAs, (C) enzymatic activities by hourly measuring processing of PEPDAB substrates from 1 h to 4 h of incubation, and PrAMA (D) ADAM10sa and (E) ADAM17sa inferred from the 3-h incubation enzymatic activities presented in (C) using the high-sensitivity/high-specificity Syntherror/Sigmathreshold 0.5/1.55 parameters. (A, B) Means + STE of ADAM10 and ADAM17 pg/10 μg of cell lysates are presented. (C) Means + STE of pM processed substrates with 10 μg of the cell lysates are shown (*p=0.02-0.04, **p=0.003-0.007, significance of differences between NSCLC and control cells). (D, E) Means + STE of PrAMA ADAM10sa and PrAMA ADAM17sa arbitrary units (AU) are presented. In (B) and (D), one-sided significance test was used.
ADAM10sa is upregulated in NSCLC tumors
In order to determine whether ADAM10sa and/or ADAM17sa are upregulated in tumors and are potential NSCLC biomarkers, we comparatively examined these enzymes in paired stage IA-IIB NSCLC primary tumors and cancer-free lung tissues. Such tissue specimens were available cryopreserved either dry or in RNAlater. Using smaller tissue cohorts (n=10), we first determined that tissues stored either way consistently displayed significant amounts of ADAM10sa and ADAM17sa. The obtained data also indicated that ADAM10sa might be upregulated in NSCLC tumors (Supplementary Fig. 2). More detailed PrAMA of multiple enzymes showed that MMP2 activity (MMP2a) and MMP14a, but not MMP9a (Supplementary Figs. 3A-3C), MMP1a or ADAM9sa (data not shown), might be also upregulated in NSCLC primary tumors relative to COPD and normal lungs. However, among the examined specific enzyme activities, ADAM10sa appeared to be highly superior, being more than 100-, 40- and 30-fold higher than the MMP and ADAM17 activities in NSCLC tumors, COPD lungs and normal lungs, respectively (Supplementary Fig. 3D-3F).
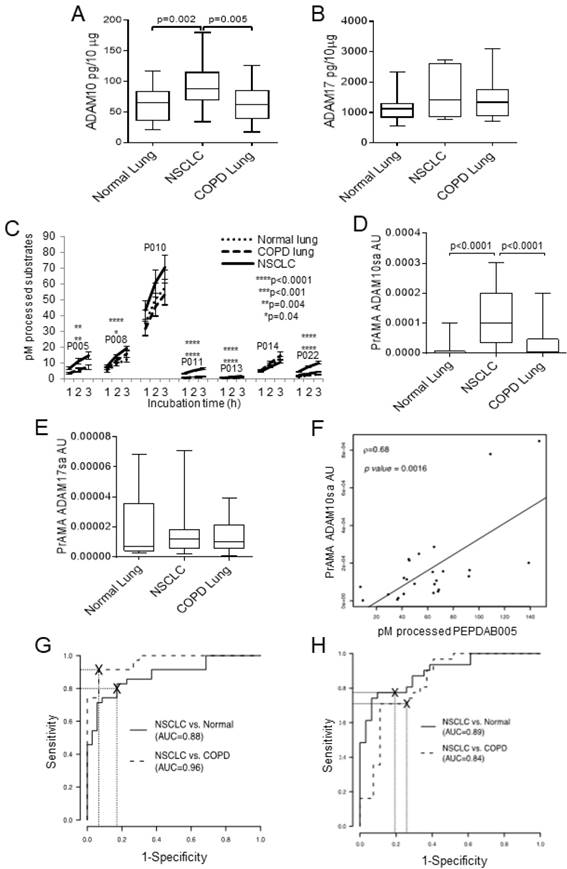
Next, we investigated ADAM10 and ADAM17 protein expression and sheddase activity in lysates of larger cohorts of paired NSCLC tumor and cancer-free normal lung (n=31) and COPD lung (n=27) specimens cryopreserved in RNAlater (Fig. 2). We found that ADAM10 protein levels were inconsistently and moderately increased in NSCLC primary tumors relative to normal or COPD lungs (p=0.002-0.005), while ADAM17 protein expression levels were similar in these tissues (Figs. 2A, 2B). In contrast, the NSCLC tumor lysates processed significantly better PEPDAB005, 008, 011, 013 and 022 than normal or COPD lung lysates (Fig. 2C). Notably, PEPDAB005 processing was especially increased in tumor lysates (Figs. 2C, Supplementary Fig. 4). Consequently, PrAMA inferred from these findings that ADAM10sa was highly significantly increased in tumors relative to the low enzyme activity in normal and COPD lungs (p<0.0001, Fig. 2D). In contrast, ADAM17sa was similar in the tumors and non-cancerous lungs (Fig. 2E). Importantly, PEPDAB005 processing and the most dominant enzyme activity, ADAM10sa, significantly correlated in NSCLC tumor lysates (Fig. 2F), indicating that the PEPDAB005 processing was mostly mediated by ADAM10sa. Furthermore, ROC-curve analyses showed that both PEPDAB005 processing (Fig. 2G) and ADAM10sa (Fig. 2H) could distinguish NSCLC tumors from non-cancerous normal or COPD lungs with significant and similar sensitivity (SE), specificity (SP) and accuracy (AC) [(PEPDAB005 processing: SE=0.83, 0.91; SP=0.83, 0.93; AC=0.83, 0.92), (ADAM10sa: SE=0.75, 0.71; SP=0.94, 0.89; AC=0.84, 0.79), respectively]. These findings show that ADAM10sa is significantly upregulated in NSCLC tumors, and is, therefore, a potential biomarker that could discriminate cancerous and non-cancerous lung tissues. They also indicate that PEPDAB005 processing could be an ADAM10sa-surrogate marker in NSCLC tumors.
While ADAM10 protein expression is moderately increased, ADAM10sa is highly increased in NSCLC tumors. Lysates of 31 pairs of primary-tumor and tumor-free normal lung tissue specimens obtained from stage IA-IIB NSCLC patients, and of 27 lung tissue specimens obtained from COPD patients, were cryopreserved in RNAlater for 1 to 7 years before testing. The lysates were examined for the presence of ADAM10 and ADAM17 proteins and sheddase activities. (A) ADAM10 and (B) ADAM17 proteins were assessed using ELISAs. Data are box plots with medians, quartiles and ranges of ADAM10 and ADAM17 pg/10 μg lysates. (C) The enzymatic activities were measured by processing PEPDAB substrates. Data are presented as pM means + STE of processed substrates with 10 μg lysates (*p=0.041, **p=0.0029-0.0044, ****p<0.0001). (D) ADAM10sa and (E) ADAM17sa were determined by analyzing PEPDAB-processing data obtained at 3 h of incubation using the PrAMA high-sensitivity/high-specificity Syntherror/Sigmathreshold 0.5/1.33 parameters. Data are box plots with medians, quartiles and ranges of ADAM10sa and ADAM17sa AU per 10 μg of lysates. (F) Correlation of PEPDAB005 processing and ADAM10sa in tumor lysates was determined by assessing the Pearson correlation coefficient (ρ=0.64, p=0.0016). ROC curve analyses were performed to assess the potentials of (G) PEPDAB005 processing and (H) ADAM10sa to distinguish NSCLC tumor lysates from non-cancerous normal or COPD lung-tissue lysates. (G) and (H) figures present areas under ROC (AUC) for NSCLC tumor vs normal lung (AUC=0.88, 0.89, respectively), and NSCLC tumor vs COPD lung (AUC=0.96, 0.84, respectively).
NSCLC primary-tumor lysates of patients who died better process PEPDAB005 than those of patients who survived 5 years after diagnosis and surgery. Lysates of stage IA-IIB NSCLC primary tumors of patients who survived (n=18) or died (n=17) of the disease during 5 years after diagnosis and surgical therapy were examined following their cryopreservation in RNAlater for 1 to 5 years. The lysates were examined for the presence of (A) ADAM10 protein by ELISA and (B) enzymatic activities by measuring processing of PEPDAB005 at 3 h of incubation. Data are presented as box plots with medians, quartiles and ranges of ADAM10 protein pg/10 μg, and pM of processed PEPDAB005 with 10 μg of tumor lysates, respectively. (C) ROC curve analysis was performed to determine the potential of PEPDAB005 processing to distinguish NSCLC primary tumors of patients having poor and good prognosis (AUC=0.88).
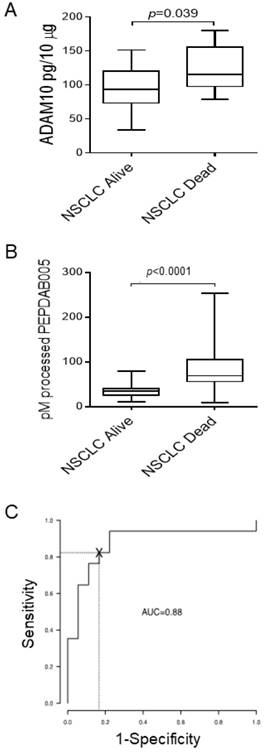
ADAM10sa may be more upregulated in NSCLC tumors of poor-prognosis patients
About 50% of patients diagnosed with the localized NSCLC (stage IA-IIB) die within 5 years after initial diagnosis and surgical therapy. Since ADAM10 plays key roles in cancer pathogenesis, invasion and metastases formation [10, 28], we examined whether the levels of ADAM10 protein and ADAM10sa-surrogate marker PEPDAB005 processing are more increased in primary tumors of NSCLC patients who died (n=17) from the disease relative to those who survived (n=18) 5 years after diagnosis and therapy (Fig. 3). ADAM10 protein levels were inconsistently and marginally increased (p=0.039) in tumor lysates of NSCLC patients who died relative to those who survived (Fig. 3A). Importantly, PEPDAB005 processing was highly significantly increased (p<0.0001) in tumor lysates of patients who died relative to those who survived (Fig. 3B). ROC curve analysis showed that the PEPDAB005 processing could distinguish with significant sensitivity, specificity and accuracy (SE=0.94, SP=0.78, AC=0.86) NSCLC tumors of patients having poor prognosis from those having good prognosis (Fig. 3C). These findings strongly indicate that ADAM10sa is more elevated in primary tumors of NSCLC patients having poor prognosis than in those of patients having good prognosis.
ADAM10sa is increased on blood exosomes of tumor-bearing NSCLC patients
Cancer cells produce in tumor microenvironment and release into peripheral blood large amounts of microvesicles (30 to 100 nm in diameter), named exosomes [50]. Exosomes are derivatives of and structurally related to endosomes. They are morphologically defined by a limiting membrane expressing on its external surface plasma-membrane-bound molecules, and could be a good source of cell-surface biomarkers [50-54]. Since ADAM10 and ADAM17 are plasma-membrane integral molecules, they are likely expressed on exosomes too.
Initially, we determined that unfractionated human sera of tumor-bearing patients and control individuals similarly processed PEPDAB substrates masking the specific enzymatic activity of exosomes that were greatly diluted in their natural environment (data not shown). Therefore, we performed the investigation using purified peripheral-blood plasma and serum exosomes. First, we assessed the presence of ADAM10sa and ADAM17sa in plasma exosomes obtained by the standard fractionated ultra-centrifugation (Figs. 4A, 4B, Supplementary Fig. 5). Plasmas were acquired from tumor-bearing stage IA-IIB NSCLC patients and healthy individuals. Exosome lysates were simultaneously tested with five PEPDAB substrates. We found that the exosome lysates of NSCLC patients significantly better processed only PEPDAB005 than those of healthy individuals (p=0.024, Fig. 4A). Consequently, PrAMA inferred that NSCLC-patient exosomes contained increased ADAM10sa (Fig. 4B), and unchanged ADAM17sa (Supplementary Fig. 5), relative to healthy-individual exosomes. Remarkably, PEPDAB005 processing and ADAM10sa significantly correlated in NSCLC-patient exosome lysates (Fig. 4C). These findings indicated that the enzyme activity measured in NSCLC-patient exosomes by PEPDAB005 processing was mostly ADAM10sa. Furthermore, ROC-curve analysis suggested that both PEPDAB005 processing and ADAM10sa could distinguish with significant and equal sensitivity, specificity and accuracy NSCLC tumor-bearing patients from healthy individuals (SE=0.80, SP=1.00, AC=0.90) (Fig. 4D). These data indicated that ADAM10sa, but not ADAM17sa, was selectively increased in exosomes of NSCLC tumor-bearing patients. They also suggested that ADAM10sa could be effectively quantified in NSCLC-patient blood exosomes by testing only PEPDAB005 processing.
ADAM10sa is increased in peripheral-blood exosomes of tumor-bearing NSCLC patients. Plasma of NSCLC tumor bearing patients (stage IA-IIB, n=4; and IIIB, n=1) and healthy individuals (n=5) were isolated by fractionated centrifugations and lysed. (A) The exosome lysates were examined using the enzymatic activity assays measuring PEPDAB substrate processing at 3 h of incubation. Presented data are medians, quartiles and ranges of pM of processed substrates with 60 μg of exosome lysates (*p=0.024, significance of differences between healthy-individual and NSCLC-patient exosomes). These data were analyzed by PrAMA, and PrAMA data were presented as boxplots with medians, quartiles and ranges of arbitrary units (AU) of (B) ADAM10sa (*p=0.061) and (Supplementary Fig. 4) ADAM17sa. In (A) and (B), one-sided test was implemented. (C) Correlation of PEPDAB005 processing and ADAM10sa in exosome lysates was determined by assessing the Pearson correlation coefficient (ρ=0.88, p=0.043). (D) ROC curve analyses were performed to assess the potentials of PEPDAB005 processing and ADAM10sa to distinguish blood exosomes of NSCLC tumor-bearing patients and healthy individuals. ROC curves and their analyses were identical for two biomarkers (AUC=0.80). (E) Serum exosomes of healthy individuals (n=7), and SPN (n=7), COPD (n=7) and NSCLC tumor-bearing patients (stage IIIA, n=6; and IIIB, n=1) were isolated by the size-defined filtration, lysed and assessed using PEPDAB005 processing. Data are presented as box plots with medians, quartiles and ranges of pM of processed PEPDAB005 with 5 μg of exosome lysates. (F) ROC curve analyses were performed to assess the potential of PEPDAB005 processing to discriminate blood exosomes of NSCLC tumor-bearing patients from those of healthy individuals and SPN or COPD patients (AUC=1, 1, 0.93, respectively).
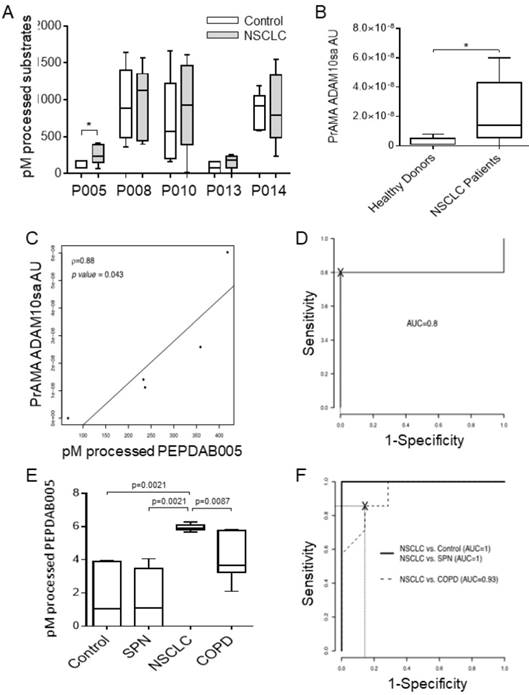
The exosome purification with the fractionated ultra-centrifugation is complex, labor-intensive and time-consuming, and could limit the potential routine clinical investigation of the exosome biomarkers in numerous blood specimens. Therefore, we introduced a simple and rapid size-defined filtration method to isolate exosomes. This method enabled consistent isolation of 500 to 1300 μg of microvesicles from 1 mL serum. In addition, as PEPDAB005 processing closely correlated with ADAM10sa both in NSCLC tumors and patient exosomes (Figs. 2F, 4C), it was applied as ADAM10sa-surrogate marker in the further studies of exosomes. Using the simplified method, we could simultaneously assess 28 specimens of serum exosomes obtained from NSCLC tumor-bearing patients (n=7), healthy individuals (n=7), and COPD (n=7) and SPN (n=7) patients (Fig. 4E). We found that exosome lysates of NSCLC-patient processed significantly better PEPDAB005 than those of healthy individuals (p=0.0021), or SPN (p=0.0021) and COPD (p=0.0087) patients. ROC-curve analysis showed that the ADAM10sa-surrogate assay could distinguish NSCLC tumor-bearing patients from healthy individuals, or SPN and COPD patients with significant sensitivity, specificity and accuracy (SE: 1.0, 1.0, 1.0; SP: 1.0, 1.0, 0.72; AC: 1.0, 1.0, 0.86, respectively). These findings strongly indicated that ADAM10sa was selectively increased on exosomes of NSCLC tumor-bearing patients and, therefore, could represent a significant NSCLC-detection biomarker capable of accurately distinguishing NSCLC-tumor-bearing patients not only from healthy individuals but also from patients having inflammatory or benign-tumor pulmonary diseases. The findings also suggested that the PEPDAB005 processing could be an ADAM10sa-surrogate marker of NSCLC-patient exosomes.
Discussion
The expressed genes and proteins of cancer pathogenesis and aggressiveness contributing enzymes ADAM10 and ADAM17 have been explored as potential cancer biomarkers. The enzyme markers were frequently found increased in malignant tumors or patient blood, but the increases were not consistent or robust, and were not clinically validated as cancer biomarkers [3-9, 29, 32-36, 38-40, 51-54].
We provide herein the evidences that ADAM10sa is moderately increased in NSCLC cells, highly increased in NSCLC primary tumors, further increased in NSCLC aggressive tumors, and notably increased in blood exosomes of tumor-bearing NSCLC patients. The enzyme activity was consistently high in NSCLC primary tumors and consistently low in the control tissues, indicating that it may be a significant NSCLC biomarker. In contrast, the ADAM10-protein expression was inconsistently, moderately or marginally increased in NSCLC primary tumors and largely overlapped with the enzyme expression levels in the control tissues, confirming that ADAM10-protein expression may be a poor NSCLC biomarker.
ADAM10 sheds and activates ErbB, Eph and Notch, which subsequently trigger key signaling pathways of cancer initiation, progression, invasion and metastases formation [8, 10, 12-18, 55-57]. In solid tumors, hypoxia develops and enhances ADAM10 expression and activity of cancer cells via induction and accumulation of hypoxia-inducible transcription factor 1α (HIF-1α) [58]. These findings concur with ours and explain why ADAM10 protein expression is not increased in normoxic cultured NSCLC cells, but is increased in hypoxic NSCLC tumor tissues. The hypoxia-upregulated ADAM10 renders cancer cells resistant to innate immunity by increasing shedding of the NK-cell activating ligand MICA from cancer-cell surface. The hypoxia induction of ADAM10 could be partially inhibited by the anti-hypoxia treatment with nitric oxide mimetics, indicating a potentially new anti-cancer therapy [58].
Contrasting ADAM10 protein expression, ADAM10sa is moderately increased in cultured NSCLC cells and highly increased in NSCLC primary tumors. These findings could not be simply explained by the hypoxia-induced enhancement of ADAM10 expression [58]. Recent studies have shown that cancer cells both in culture and in tumor, but not non-transformed cells, express a large, unprocessed, active form of ADAM10 (having occupied catalytic-domain active site with prodomain). This tumor form of ADAM10 is activated by disulfide isomerization in its membrane-proximal cysteine-reach substrate-recognition domain, which is mediated by reactive oxygen radicals highly produced in tumor microenvironment [44]. Thus, it is conceivable that the tumor form of ADAM10 is selectively expressed by NSCLC cells and both upregulated by hypoxia and activated by oxygen radicals in NSCLC-tumor microenvironment thus resulting in the excessive ADAM10sa found in NSCLC primary tumors and especially in their aggressive species obtained from poor-prognosis patients.
Using the specific enzyme activity assay that employs both processing of multiple fluorogenic substrates and PrAMA, we detected significant ADAM10sa increases in peripheral blood exosomes of NSCLC patients bearing relatively small, T1 or T2 primary tumors, indicating that the increased ADAM10sa in peripheral blood exosomes could be an early detection biomarker of NSCLC. As PrAMA-ADAM10sa is the most dominant MP activity in NSCLC tumors that closely correlates with PEPDAB005 processing in both NSCLC tumors and exosomes of NSCLC tumor-bearing patients, the single substrate processing could be alone an ADAM10sa-surrogate marker in NSCLC tumors and, especially, in NSCLC-patient blood exosomes having a less complex enzyme composition. Indeed, the one-substrate assay detected in exosomes of tumor-bearing NSCLC patients the elevation of enzyme activity with the same accuracy as the five-substrate PrAMA-ADAM10sa. These findings indicate that PEPDAB005 processing alone could be a simple, sensitive and inexpensive blood test, which could be applied in early detection and follow-up studies of NSCLC, when large cohorts of blood exosome specimens are simultaneously tested or the quantities of isolated exosomes are insufficient for the full-scale, multiple-substrate PrAMA-ADAM10sa examination.
Having the key roles in all phases of cancer development [8, 10, 12-18, 55-57], ADAM10 has been considered not only a potentially significant cancer biomarker but also a highly promising therapy target [10, 59]. However, numerous clinical trials with broad-spectrum small-molecule inhibitors of MP catalytic-domain active sites implemented in patients with advanced cancers have so far failed, showing major musculosclerotic toxicity and no clinical benefits [60, 61]. Subsequent studies have indicated that the causes of the promising therapy failure could be the non-selective inhibition of both cancer-promoting (i.e., MMP2, MMP9, MMP14, ADAM10, ADAM17) and cancer-suppressing (i.e., MMP3, MMP8, MMP12) MP activities, lack of inhibition of the proteolytic-activity independent cancer-promoting functions of MPs mediated by the beyond catalytic domain exosites, and dose-limiting toxicity [61-63]. Therefore, more selective small-molecule inhibitors targeting catalytic-domain active sites, mutated TIMP variants with improved selectivity and affinity, and specific antibodies targeting both catalytic and non-catalytic functions of pro-tumorigenic MMPs and ADAM10 have been developed [61-63]. Preclinical studies of the new-generation inhibitors provide highly promising proofs-of-principle that anti-cancer therapies with more specific MP inhibitors could be clinically beneficial [44, 61-66]. This was exemplified by the findings that the small-molecule MP inhibitor INCB3619, selectively targeting ADAM10, ADAM17, MMP2 and MMP12, effectively reduced shedding of ErbB ligands and retained growth of human cancer xenografts in nude mice without inducing fibroplasia toxicity [66]. More importantly, the specific targeting of the cancer-prevalent active form of ADAM10 with a monoclonal antibody led to inhibition of Notch signaling, tumor growth and tumor regrowth after chemotherapy in murine models [44]. The potential future anti-ADAM10 therapy as well as other current and future therapies of NSCLC could be personalized and made more beneficial by the ADAM10sa-biomarker-based early detection, outcome prediction and follow-up.
In conclusion, our study provides a proof-of-concept that the increased ADAM10sa in primary tumor lesions and blood exosomes could be a significant prognostic, early detection and follow-up biomarker of NSCLC and perhaps some other cancers. ADAM10sa could be also a companion marker to enroll patients into future therapies that target ADAM10. To confirm our findings and validate ADAM10sa as a clinically useful NSCLC biomarker, additional retrospective and prospective studies of large cohorts of tissue and blood specimens are required.
Abbreviations
LC, Lung cancer; NSCLC, Non-small cell lung carcinoma; ADAM, A Disintegrin And Metalloproteinase; ADAM10sa, ADAM10 sheddase activity; ADAM17sa, ADAM17 sheddase activity; MP, metalloproteinases; MMP, Matrix metalloproteinases, MMPa, MMP activity; FRET, Fluorescence-Resonance Energy Transfer; PrAMA, Proteolytic Activity Matrix Analysis; ROC, receiver operating characteristic; AUC, area under ROC; SE, Sensitivity; SP, Specificity; AC, Accuracy; COPD, Chronic obstructive pulmonary disease; SPN, Small pulmonary nodule; NHBE, Normal human lung bronchial epithelium; COPDBE, human COPD lung bronchial epithelium; NLFB, human normal lung fibroblasts; PBL, peripheral blood leukocytes; NK, natural killer; DC, dendritic cells.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank Dr. Jill Siegfried, Dr. William Bigbee and the UPCI Lung Cancer SPORE for providing NSCLC tumor specimens.
Grant Support
The authors are grateful to the supporting fund from VA Merit Award Grant 1-I01-BX000993-01/ VUJ-ONCA-053-12S (NL Vujanovic) and NIH R01 CA96504 (DA Lauffenburger).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fry W, Philips JL, Menck HR. Ten-year survey of lung cancer treatment and survival in hospitals in the United States. Cancer. 1999;86:1867-76
2. Inamura K, Ishikawa Y. MicroRNA in lung cancer: Novel biomarkers and potential tools for treatment. J Clin Med. 2016;3:pii E36. doi: 10.3390/jcm5030036
3. Lu C, Soria JC, Tang X, Xu XC, Wang L, Mao L. et al. Prognostic factors in resected stage I non-small-cell lung cancer: A multivariate analysis of six molecular markers. J Clin Oncol. 2004;22:4575-83
4. Milas I, Komaki R, Hachiya T, Bubb RS, Ro JY, Langford L. et al. Epidermal growth factor receptor, cyclooxygenase-2, and BAX expression in the primary non-small cell lung cancer and brain metastases. Clin Cancer Res. 2003;9:1070-6
5. Selvaggi G, Novello S, Torri V, Leonardo E, De Giuli P, Borasio P. et al. Epidermal growth factor receptor over expression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncology. 2004;15:28-32
6. Brabender J, Park J, Metzger R, Schneider PM, Lord RV, Hölscher AH. et al. Prognostic significance of cyclooxygenase 2 mRNA expression in non-small cell lung cancer. Ann Surgery. 2002;235:440-3
7. Sienel W, Hellers J, Morresi-Hauf A, Lichtinghagen R, Mutschler W, Jochum M. et al. Prognostic impact of matrix metalloproteinase-9 in operable non-small cell lung cancer. Int J Cancer. 2003;103:647-51
8. Guo J, He L, Yuan P, Wang P, Lu Y, Tong F. et al. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncol Rep. 2012;28:1709-18
9. Ni SS, Zhang J, Zhao WL, Dong XC, Wang JL. ADAM17 is overexpressed in non-small cell lung cancer and its expression correlates with poor patient survival. Tumour Biol. 2013;34:1813-8
10. Saftig P, Reiss K. The “a disintegrin and metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? European J Cell Biol. 2011;90:527-35
11. Miller MA, Oudin MJ, Sullivan RJ, Wang SJ, Meyer AS, Im H. et al. Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov. 2016;6:382-99
12. Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W. et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615-24
13. Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact mediated axon repellent. Science. 2000;289:1360-5
14. Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E. et al. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123:291-304
15. Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J. et al. Distinct role for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769-79
16. Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D. et al. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and b-catenin nuclear signaling. EMBO J. 2005;24:742-52
17. Ranganathan P, Weaver KL, Capobianco AJ. Notch signaling in solid tumors a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338-51
18. Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Deu. 2007;21:2511-24
19. Black RA. Tumor necrosis factor-alpha converting enzyme. Int J Biochem Cell Biol. 2002;34:1-5
20. Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF. et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729-33
21. Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE. et al. TACE/ADAM17 Processing of EGFR ligands indicates a role as a physiological convertase. Ann N Y Acad Sci. 2003;995:22-38
22. Horiuchi K, Miyamoto T, Takaishi H, Hakozaki A, Kosaki N, Miyauchi Y. et al. Cell surface colony-stimulating factor 1 can be cleaved by TNF-alpha converting enzyme or endocytosed in a clathrin-dependent manner. J Immunol. 2007;179:6715-24
23. Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC. et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281-4
24. Rovida E, Paccagnini A, Del Rosso M, Peschon J, Dello Sbarba P. TNF-alpha-converting enzyme cleaves the macrophage colony-stimulating factor receptor in macrophages undergoing activation. J Immunol. 2001;166:1583-9
25. Sahin U, Blobel CP. Ectodomain shedding of the EGF-receptor ligand epigen is mediated by ADAM17. FEBS Lett. 2007;581:41-4
26. Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ. et al. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem. 2002;277:12838-45
27. Black RA, Doedens JR, Mahimkar R, Johnson R, Guo L, Wallace A. et al. Substrate specificity and inducibility of TACE (tumour necrosis factor alpha-converting enzyme) revisited: the Ala-Val preference, and induced intrinsic activity. Biochem Soc Symp. 2003;70:39-52
28. Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929-41
29. Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donald H, Rocken C. Increased expression of ADAM family members in human breast and breast cancer lines. J Cancer Res Clin Oncol. 2005;131:41-8
30. Zheng X, Jiang F, Katakowski M, Kalkanis SN, Hong X, Zhang X. et al. Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer Sci. 2007;98:674-84
31. Blanchot-Jossic F, Jarry A, Masson D, Bach -Ngohou K, Paineau J, Denis MG. et al. Up-regulated expression of ADAM17 in human colon carcinoma: co-expression with EGFR in neoplastic and endothelial cells. J Pathol. 2005;207:156-63
32. Roemer A, Schwettmann L, Jung M, Roigas J, Kristiansen G, Schnorr D. et al. Increased mRNA expression of ADAMs in renal cell carcinoma and their association with clinical outcome. Oncol Rep. 2004;11:529-36
33. Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E. et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39-50
34. Tanaka Y, Miyamoto S, Suzuki SO, Oki E, Yagi H, Sonoda K. et al. Clinical significance of heparin-binding epidermal growth factor-like growth factor and a disintegrin and metalloprotease 17 expression in human ovarian cancer. Clin Cancer Res. 2005;11:4783-92
35. Karan D, Lin FC, Bryan M, Ringel J, Moniaux N, Lin MF. et al. Expression of ADAM (a disintegrin and metalloproteases) and TIMP-3 (tissue inhibitor of metalloproteinase-3) in human prostate adenocarcinomas. Int J Oncol. 2003;23:1365-71
36. McGowan PM, McKiernan E, Bolster F, Ryan BM, Hill AD, McDermott EW. et al. ADAM-17 predict adverse outcome in patients with breast cancer. Ann Oncol. 2008;19:1075-81
37. Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T. et al. L1, a novel target of betta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol. 2005;168:633-42
38. Yoshimura T, Tomita T, Dixon MF, Axon AT, Robinson PA, Crabtree JE. ADAMs (A Disintegrin And Metalloprotease) messenger RNA expression in Helicobacter pylori-infected, normal and neoplastic gastric mucosa. J Infect Dis. 2002;185:332-40
39. Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of hematological malignancies. Biochem Biophys Res Commun. 1997;235:437-42
40. McCulloch DR, Aki P, Samaratunga H, Herington AC, Odorico DM. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor 1, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10:314-23
41. Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A. et al. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003;362:869-75
42. Ge L, Baskic D, Basse P, Vujanovic L, Unlu S, Yoneyama T. et al. Sheddase activity of tumor necrosis factor-alpha converting enzyme is increased and prognostically valuable in head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2913-22
43. Bose AK, Janes KA. A high-throughput assay for phosphoprotein-specific phosphatase activity in cellular extracts. Mol Cell Proteomics. 2013;12:797-806
44. Atapattu L, Saha N, Chheang C, Eissman MF, Xu K, Vail ME. et al. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J Exp Med. 2016;213:1741-57
45. Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG. et al. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb). 2011;3:422-38
46. Yoneyama T, Gorry M, Miller MA, Gaither-Davis A, Lin Y, Moss ML. et al. Modification of proteolytic activity matrix analysis (PrAMA) to measure ADAM10 and ADAM17 sheddase activities in cell and tissue lysates. J Cancer. 2017;8:3916-32
47. Magee M, Vujanovic LN, Butterfield LH, Vujanovic NL. Isolation, culture and propagation of natural killer (NK) cells. Lotze MT, Thomson AW Ed. Natural Killer Cells - Basic Science and Clinical Application, Elsevier Academic Press. 2010:125-35
48. Janjic BM, Lu G, Pimenov A, Whiteside TL, Storkus WJ, Vujanovic NL. Innate direct anticancer effector function of human immature dendritic cells. I. Involvement of a potent apoptosis-inducing pathway. J Immunol. 2002;168:1823-30
49. Munich S, Sobo-Vujanovic A, Buchser WJ, Beer-Stolz D, Vujanovic NL. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology. 2012;1:1074-83
50. Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247-54
51. Hakulinen J, Sankkila L, Sugiyama N, Lehti K, Keski-Oja J. Secretion of active membrane type 1 matrix metalloproteases (MMP-14) into extracellular space in microvesicular exosomes. J Cell Biochem. 2008;105:1211-8
52. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603-7
53. Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42-6
54. Roy R, Zurakowski D, Pories S, Moss ML, Moses MA. Potential of fluorescent metalloprotease substrates for cancer detection. Clin Biochem. 2011;44:1434-9
55. Ranganathan P, Weaver KL, Capobianco AJ. Notch signaling in solid tumors: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338-51
56. Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511-24
57. Espinoza I, Pochampally R, Xing E, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013;6:1249-59
58. Barsoum IB, Hamilton TK, Li X, Cotechini T, Miles EA, Siemens DR. et al. Hypoxia Induces Escape from Innate Immunity in Cancer Cells via Increased Expression of ADAM10: Role of Nitric Oxide. Cancer Res. 2011;71:7433-41
59. Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechol. 2008;9:2-8
60. Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387-92
61. Radisky ES, Raeeszadeh-Sarmazdeh M, Radisky DC. Therapeutic potential of matrix metalloproteinase inhibition in breast cancer. J Cell Biochem. 2017;118:3531-48
62. Shay G, Lynch CC, Fingleton B. Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2015;44-46:200-6
63. Radisky ES, Radisky DC. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Front Biosci (Landmark Ed). 2015;20:1144-63
64. Zhang W, Liu S, Liu K, Ji B, Wang Y, Liu Y. Knockout of ADAM10 enhances sorafenib antitumor activity of hepatocellular carcinoma in vitro and in vivo. Oncol Rep. 2014;32:1913-22
65. Woods N, Trevino J, Coppola D, Chellappan S, Yang S, Padmanabhan J. Fendiline inhibits proliferation and invasion of pancreatic cancer cells by interfering with ADAM10 activation and β-catenin signaling. Oncotarget. 2015;6:35931-48
66. Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q. et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007;13:1892-902
Author contact
![]() Corresponding author: Nikola L. Vujanovic, UPMC Hillman Cancer Center, Research Pavilion, 5117 Centre Avenue, Pittsburgh PA, 15213; phone number, 412-623-3211; fax number, 412-623-1119; e-mail address, vujanovicnledu
Corresponding author: Nikola L. Vujanovic, UPMC Hillman Cancer Center, Research Pavilion, 5117 Centre Avenue, Pittsburgh PA, 15213; phone number, 412-623-3211; fax number, 412-623-1119; e-mail address, vujanovicnledu

 Global reach, higher impact
Global reach, higher impact