Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(14):2580-2588. doi:10.7150/jca.24896 This issue Cite
Research Paper
Long non-coding RNA CRYBG3 regulates glycolysis of lung cancer cells by interacting with lactate dehydrogenase A
1. School of Radiation Medicine and Protection, Medical College of Soochow University, Suzhou 215123, China
2. Collaborative Innovation Center of Radiological Medicine of Jiangsu Higher Education Institutions, Suzhou 215123, China
3. Medical College of Soochow University, Suzhou 215123, China
4. The Second Affiliated Hospital of Soochow University, Suzhou 215123, China
5. Center for Radiological Research, College of Physician and Surgeons, Columbia University, New York, NY 10032, USA
*These authors contributed equally.
Received 2018-1-12; Accepted 2018-5-5; Published 2018-6-23
Abstract
Cancer cells usually utilize glucose as a carbon source for aerobic glycolysis, a phenomenon known as the Warburg effect. And a high rate of glycolysis has been observed in lung cancer cells. The growing evidence indicates that long non-coding RNAs (lncRNAs) are important players in lung cancer initiation and progression. However, the correlation between lncRNAs and glycolysis remains unclear. In this study, we recognized a lncRNA, LNC CRYBG3, which can interact with lactate dehydrogenase A (LDHA), a vital enzyme of glycolysis, is highly upregulated in both clinical lung cancer tissues and in vitro cultured lung cancer cell lines. A positive correlation between the expression level of LNC CRYBG3 and LDHA expression levels is observed. In another hand, LNC CRYBG3 is a regulator of glycolysis and its overexpression promoted the uptake of glucose and the production of lactate whereas the knockdown of LNC CRYBG3 led to opposite results and suppressed cell proliferation. These results indicated that LNC CRYBG3 might be a novel target for lung cancer treatment.
Keywords: LNC CRYBG3, LDHA, Glycolysis, Pyruvate, Lactate, Lung cancer
Introduction
Lung cancer is the leading cause of cancer mortality in the world [1]. As the major subtype, non-small cell lung cancer (NSCLC) comprises about 85 percent of lung cancer cases [2]. In spite of advancements and efforts for treatment, the five-year survival rate of NSCLC is still poor with only approximately 15% [3, 4]. Although considerable progress on oncogenes has been developed, the molecular basis underlying NSCLC progression remains to be clarified.
Cancer cells take up glucose and transform it into lactate under aerobic conditions, which is known as the Warburg effect. Sufficient amounts of nucleotides, proteins, and lipids derived from this type of glucose metabolism are required for cancer cell rapid growth and division [5]. Critical to this highly glycolytic phenotype is lactate dehydrogenase A (LDHA), which catalyzes the last step of aerobic glycolysis. Abnormal expression of LDHA has been observed in many human cancers, such as pancreatic cancer [6], hepatocellular carcinoma [7], and breast cancer [8]. Inhibition of LDHA reduces cell malignant transformation and remarkably delays tumor formation, indicating the underlying role of LDHA in tumor initiation or maintenance [9]. Related mechanisms of tumor progression inhibited by LDHA suppression have been revealed [10, 11]. In lymphoma, the reduction of LDHA induces oxidative stress and alters cellular energy metabolism, which ultimately contributes to cell death [10]. In breast cancer, LDHA knockdown suppresses tumorigenicity through the induction of oxidative stress-mediated mitochondrial apoptosis [11].
Long non-coding RNA (lncRNA) is a set of RNAs without the function of encoding proteins, but regulating gene expression at chromatin modification, transcriptional or posttranscriptional levels [12]. Reports have demonstrated the biological functions of lncRNAs in several types of cancers, including ovarian cancer [13], renal cell carcinoma [14], gastric cancer [15], prostate cancer [16], hepatocellular carcinoma [17] as well as in lung cancer [18-20]. Moreover, the dysregulation of lncRNAs is harmful to human health, which is found to participate in tumorigenesis and progression [21]. It becomes well-known during the last decade that noncoding RNAs act as key regulators of cancer metabolism [22-24]. Cancer cells display increased metabolic autonomy by taking up nutrients avidly and regulating major metabolic pathways to support fast growth and proliferation [25]. However, the correlation between lncRNAs and glycolysis remains unclear.
In the present study, the relationship between a novel lncRNA, (named LNC CRYBG3) and LDHA and the influence of LNC CRYBG3 on glucose metabolism in lung cancer cells were revealed for the first time. Moreover, the cellular functions of LNC CRYBG3 were also explored.
Materials and Methods
Cell culture
A549 and H1299 cells were maintained in RPMI-1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 1% penicillin sodium and 100 μg/mL streptomycin, at 37ºC in 5% CO2 in a humidified incubator (Thermo Scientific, NC, USA). Beas-2B cells were maintained in DMEM medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 1% penicillin sodium and 100 μg/mL streptomycin, at 37ºC in 5% CO2 in a humidified incubator (Thermo Scientific, NC, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were harvested at indicated time-points by using TRIzol reagent (Invitrogen, CA, USA). Total RNAs were reverse-transcribed using PrimeScript RT Reagent Kit (Takara, Kusatsu, Shiga, Japan). A PCR analysis was performed using PowerUp™ SYBR® Green Master Mix (Life Technologies, Grand Island, NY, USA), and amplified PCR products were quantified and normalized with GAPDH/18S rRNA. The PCR program was carried out with a Life Technologies system (Vii7A, NY, USA) and initiated by 2 min at 95°C before 40 thermal cycles, 30s each at 95°C and 40s at 62°C with a final extension of 10 min at 72 °C. Data were analyzed by C(t) value comparison method and normalized by control expression in each sample. The primer set for LNC CRYBG3 were: (Forward: GAAGAGTGGAAGTGCGAAGGA; Reverse: GGCAATGACCCCATTAGCTC). The primer set for GAPDH were: (Forward: GCACCGTCAAGGCTGAGAAC; Reverse: TGGTGAAGACGCCAGTGGA).
Gene silencing and overexpression
1×105 A549 cells were transfected with 1×106 LNC CRYBG3 shRNA lenti-virus particles (Sangon, Shanghai, China) for 24 h (cell line short as A549-sh cells). Control shRNA (cell line short as A549-LV-NC) and GFP lentivirus particles were also operated according to the manufacturer's instruction. Cells were subsequently screened with MEM medium containing 2 μM puromycin (Invitrogen) and double-checked with RT-PCR. Adenovirus particles were used for LNC CRYBG3 over-expression (Sangon, Shanghai, China).
Lactate dehydrogenase (LDH) assay
Cell death was evaluated by the quantification of plasma membrane damage which resulted in the release of lactate dehydrogenase (LDH). The level of LDH released in the cell culture supernatant was detected by LDH cytotoxicity assay detection kit (Beyotime, shanghai, China) following the manufacturer's instructions.
Sample preparation for GC-MS analysis
For GC-MS analysis, 2×106 cells were suspended in 200 μL of 100% methanol and stored at -80°C. After being thawed at room temperature, the suspensions were centrifuged at 14,000×g at 4°C for 5 min. Then, the supernatant was transferred into a new Eppendorf tube and dried by vacuum. The dried sample was re-dissolved in 50 μL of methoxyamine solution (15 mg/mL). Afterwards, the sample was placed in a water bath of 70°C for 1h for the oximation reaction, which was followed by a silylation reaction with 50 μL of N-Methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) (Sigma Aldrich, USA), also in a water bath of 70°C for 1h. Finally, the derivatized sample was centrifuged at 13,000 rpm for 15 min and the supernatant was used for the GC-MS analysis according to the reference [26].
RNA pull down
LNC CRYBG3, and antisense LNC CRYBG3 transcript were synthesized and constructed into 18T-pMD plasmid. LNC CRYBG3 RNAs were obtained with a RNA in vitro transcription system containing biotin-labeled dUTP. Cells were harvested and resuspended in 1 mL ice-cold RIP buffer containing RNase and protease inhibitors (Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit, Merck). The cell pellet shear was performed on ice using a Dounce homogenizer with 20 strokes and centrifuge at 15,000×g for 15 min at 4°C to clear the cell lysate. And then, the supernatant was transferred into a new tube. Folded RNA was then mixed with cell lysate and incubated at room temperature for 2 h. 60 μL washed Streptavid in agarose beads were added to each binding reaction and further incubated at RT for 1 h. Beads were boiled in Laemmli loading buffer. Samples were used to perform SDS-PAGE and Western Blot assay.
RNA immunoprecipitation analysis
A549 cells were harvested by scraping cells in IP lysis buffer and mechanically sheared using a homogenizer. LDHA anti-body was added to the cell extract and incubated overnight at 4 °C. Streptavidin-coated magnetic beads were then added and incubated for 2 h at 16 °C. As followed, magnetic beads were resuspended in 1 mL TRIzol. The isolated RNA was reverse transcribed to cDNA and then analyzed by qRT-PCR. The PCR cycle parameters for LNC CRYBG3 enrichment and the primer pairs were seen as above.
Western blot
Cells were harvested and lysed using RIPA buffer. Samples were sonicated and centrifuged at 12,000 g for 15 min at 4ºC. The concentration of total protein was determined by using DC Protein Assay Kit I (Bio-Rad, Richmond, CA, USA). Then, the samples were denatured at 100ºC for 5 min. Total proteins were separated by 12% SDS-PAGE and transferred to hybond nitrocellulose membrane (Amersham, NJ, USA). The membrane was blocked with 5% nonfat milk powder in Tris-buffered saline (pH 7.5) and hybridized overnight with primary antibodies β-tubulin, LDHA (Abcam, UK), which were then detected with horseradish peroxidase- conjugated anti-IgG for 2 h at room temperature and visualized with an ECL kit (Millipore, Billerica, MA, USA). Proteins expression levels were normalized to the loading controls basing on their intensity analyzed with Image J (National Institutes of Health, USA).
Immunohistochemistry
Histological sections of formalin-fixed paraffinembedded samples were analyzed for the presence of LDHA. After deparaffinization and antigen retrieval using an autoclave oven technique, sections (5 μm) were incubated at 4°C overnight and incubated with LDHA Rabbit Polyclonal Antibody (Abcam, UK) at 4°C. Antigen-antibody complexes were detected by the cobalt-3, 3'-diaminobenzidine reaction. There were 10 mice in each group and the transplanted tumors from at least 3 mice were sliced for immunohistochemical staining. LDHA expression was shown in brown, the brown area and intensity were quantified by image J (National Institutes of Health, USA).
Statistics
All experiments were independently repeated at least three times and all data were presented as the mean ± standard error. Student's t-tests were used for statistical analysis. Probability (p) values less than 0.05 were considered to be statistically significant.
Results
LNC CRYBG3 is up-regulated in lung cancer
Lung cancer has been reported as the leading cause of cancer-related deaths worldwide. Based on the TCGA database, lung cancer patients with a high LDHA expression level showed a lower survival percentage than those with a low LDHA expression (Fig.1A). The “low” and “high” LDHA expression stand for lower or higher than 90% percent NSCLC samples (both 90% percentile). In our previous work, we identified a radiation-inducible lncRNA, LNC CRYBG3, and its potential target LDHA protein with Pull-down assay and Mass Spectrum (Supporting Figure S1), implying that LNC CRYBG3 might interact with LDHA protein in lung cancer. Therefore, we detected the expression of LNC CRYBG3 and LDHA in clinic lung cancer tissues and discovered the positive correlation between the expression level of LNC CRYBG3 and LDHA in lung carcinoma tissues or lung cancer cells. As shown in Fig.1B, the expression level of LNC CRYBG3 in NSCLC tissues was higher than that of normal tissues (p<0.05). Besides, the level of LNC CRYBG3 further increased in lung tissues after metastasis, which suggested the higher malignant tumors were associated with the higher expression level of LNC CRYBG3. Similarly, the expression level of LDHA in NSCLC tissues was also increased in lung tissues after metastasis (p<0.05, Fig.1C), which suggested the higher malignant tumors were associated with the higher expression level of LDHA. Considering the similar alternation trends of LNC CRYBG3 and LDHA expression in lung tissue after metastasis, the relationship between them was explored. We observed a positive correlation between the expression level of LNC CRYBG3 and LDHA mRNA levels in the clinical lung cancer tissues (R2=0.8799, Fig. 1D). Moreover, a positive correlation between the expression level of LNC CRYBG3 and LDHA mRNA levels was observed in in vitro cultured cell lines (R2=0.8419, Fig. 1E). In order to further study about the function of LNC CRYBG3, two lung cancer cell lines and one epithelial lung cell line were chosen. Compared to the immortalized human embryonic lung cell line Beas-2B, the expression of both LNC CRYBG3 and LDHA mRNA was more prominent in the two human lung cancer cell lines, H1299 and A549 (Fig.1F and 1G). The expression of both LNC CRYBG3 and LDHA mRNA in H1299 cells was higher than A549 cells, respectively. These findings suggest that LNC CRYBG3 and LDHA may function as lung tumor promoters.
LNC CRYBG3 is up-regulated in lung cancer. A, TCGA database shows a low patients survive percentage when LDHA mRNA expression level is high, whereas a high patients survive percentage when LDHA mRNA expression level is low. B, The absolute expression levels of LNC CRYBG3 were determined in 23 clinical lung cancer tissues and four normal lung tissues by using quantitative RT-PCR. C, The absolute expression levels of LDHA were determined in 23 clinical lung cancer tissues and four normal lung tissues by using quantitative RT-PCR. D, A significant positive correlation between LNC CRYBG3 mRNA expression and LDHA mRNA expression was observed in the clinical lung cancer tissues. E, A significant positive correlation between LNC CRYBG3 mRNA expression and LDHA mRNA expression was observed in the cells. F, The mRNA levels of LDHA in various cell lines were determined via qRT-PCR. β-tubulin was used as an internal control. Data are presented as the mean ± standard error (n = 3). G, The expression of LNC CRYBG3 in vitro cultured one immortalized human epithelial lung cell line Beas-2B and two human lung carcinoma cell lines, A549 and H1299, was also quantified with quantitative RT-PCR. All the experiments were independently repeated three times.
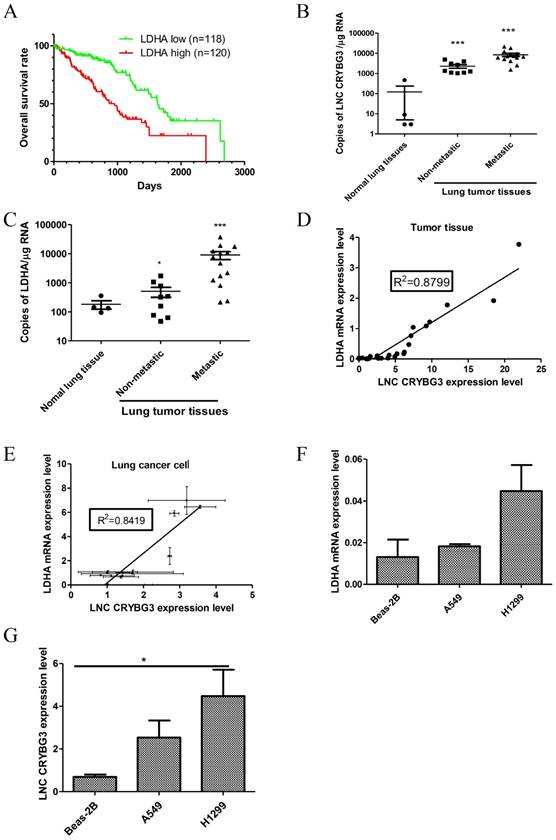
LNC CRYBG3 promotes glycolysis in lung cancer cells
Apart from being oxidized by aerobic respiration by tricarboxylic acid cycle (TCA), glucose can be degenerated to pyruvic acid and then transformed to lactate via glycolysis under aerobic conditions. To determine how LNC CRYBG3 regulate the glucose metabolism, glucose uptake, glycolysis, and TCA intermediates were assessed as well in both A549 cells and H1299 cells, in which the expression levels of LNC CRYBG3 were artificially modified with the adenovirus transfection for overexpression or small hairpin RNAs to knock down LNC CRYBG3 (Supporting Figure S2). The results showed that over-expressed LNC CRYBG3 promoted glucose and pyruvate consumption, while knock-down of LNC CRYBG3 with shRNA reduced their consumption in both two cell lines (Fig. 2A, 2B & 2E). Moreover, lactate levels in the H1299 cell line displayed consistent alternation trends with the expression of LNC CRYBG3 (Fig. 2D). Which further confirming the correlation between LNC CRYBG3 and LDHA. Meanwhile, simultaneous transfection of both adenovirus and shRNA neutralized the pyruvate uptake and lactate production at least partially in H1299 cell lines (Fig. 2B, 2D & 2F). It is noteworthy that there were no statistically significant changes in the levels of TCA intermediates including citrate, α-ketoglutarate, succinate, fumarate and malate, after LNC CRYBG3 overexpression or knock-down (Supporting Figure S3), which indicates that LNC CRYBG3 mainly regulates glycolysis in lung cancer and has little effect on the glucose oxidation metabolism through TCA.
LNC CRYBG3 promotes glycolysis in lung cancer cells. A&B, Pyruvate uptake levels were measured by mass spectrum in the two lung cancer cell lines (A549, H1299) in which LNC CRYBG3 was expressed or knocked down, as described in the Methods. C&D Lactate production levels were measured in the stable cell lines in which LNC CRYBG3 was expressed or knocked down. E&F, Glucose uptake levels were measured in the stable cell lines in which LNC CRYBG3 was expressed or knocked down. The representative meanings of the symbols for each group were as below: Ctrl was control group, NC was negative control group, LNC CRYBG3 was LNC CRYBG3 overexpression group, sh1 was LNC CRYBG3 knocking down group, sh1+ LNC CRYBG3 was LNC CRYBG3 overexpressed after LNC CRYBG3 knocked down group. The experiments were independently repeated three time and the P value was calculated using the Student's t-test.*, p< 0.05.
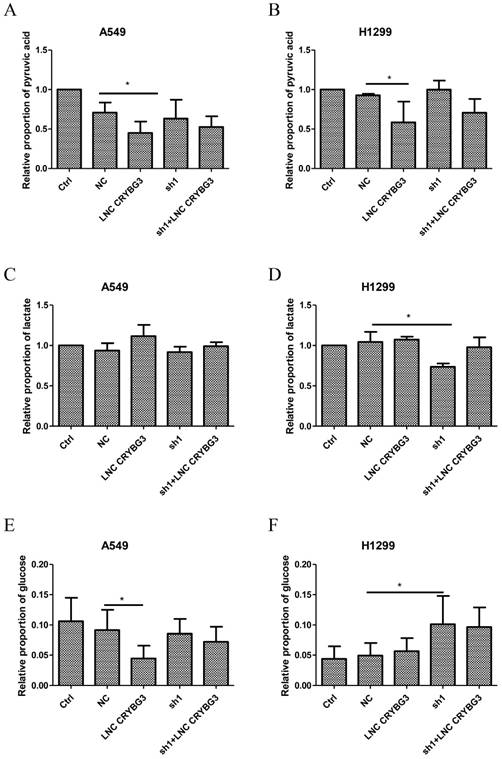
LNC CRYBG3 interacts with LDHA
Since LDHA is one of the key enzymes of glycolysis and also the potential target of LNC CRYBG3 as revealed by pull-down assay, we analyzed the impact of LNC CRYBG3 expression on the catalytic activity of LDHA. As depicted in Fig. 3A, the LDHA activity in A549 cells remarkably increased when LNC CRYBG3 was overexpressed, whereas it was significantly inhibited when LNC CRYBG3 was knocked-down. Overexpression of LNC CRYBG3 rescued the suppression of LDHA activity by its shRNAs. Similar change trends were observed in H1299 cell line (Fig. 3B). Considering that the expression of LNC CRYBG3 is higher in H1299 cell line than that of A549 cell line (Fig.1G), the effect of overexpressed LNC CRYBG3 on LDHA activity was compromised. We further examined the expression of LDHA protein with Western blotting. Transfection of LNC CRYBG3 shRNA resulted in reduced expression of LDHA protein whereas the overexpression of LNC CRYBG3 induced the expression of LDHA protein (Fig. 3C & 3D). Basing on the LDHA protein expression in the lung cancer cells, the effect of LNC CRYBG3 on the LDHA proteins expression in subcutaneous xenotransplanted tumor was examined in nude mice. LNC CRYBG3 shRNA lenti-virus or LNC CRYBG3 overexpression lenti-virus were transfected in subcutaneous xenotransplanted tumor in nude mice (Supporting Figure S2C). As shown in Fig. 3E and 3F, the knockdown of LNC CRYBG3 dramatically suppressed the expression of LDHA protein. When LNC CRYBG3 was overexpressed, the LDHA protein levels were up-regulated, even though the changes were not significant. RNA pull-down assay combined with mass spectrometry showed that LDHA is one of the candidate targets of LNC CRYBG3. Therefore, RNA pull-down assay and RNA immunoprecipitation assay were performed, which displayed that LDHA was pulled down by LNC CRYBG3 (Fig. 3G) and LNC CRYBG3 was precipitated by LDHA (Fig. 3H).These results demonstrated the interaction of LNC CRYBG3 with LDHA.
LNC CRYBG3 interacts with LDHA. A&B, The activity of LDH dehydrogenase were analysed by LDH Cytotoxicity Assay Kit. C&D, The protein levels of LDHA were determined by Western blotting. The intensity of each band was quantified with ImageJ. The experiments were independently repeated three times. E, Detection of LDHA immunoreactivity in transplanted tumor. The experiments were independently repeated three times. F, Brown indicates the amount of LDHA expression, the brown area and depth were statistics by image J. G, A demonstration of the results of RNA pull-down analysis. LNC CRYBG3, and antisense LNC CRYBG3 transcript were synthesized and constructed into 18T-pMD plasmid. LNC CRYBG3 RNAs were obtained with a RNA in vitro transcription system containing biotin-labeled dUTP. Cells were harvested and resuspended in RIP buffer containing RNase and protease inhibitors. Folded RNA was then mixed with cell lysate and incubated at room temperature. Streptavid in agarose beads were added to each binding reaction and incubated at RT. Samples were used to perform SDS-PAGE and Western Blot assay. H, Histogram of LNC CRYBG33 enrichment after RNA immunoprecipitation assays (RIP) with LDHA antibodies. A549 cells were lysed in RIP lysis buffer. Cell extracts were incubated with magnetic beads conjugated with LDHA antibody. Samples were incubated with Proteinase K to isolate immunoprecipitated RNA. The purified RNA was used to qRT-PCR analysis. The P value was calculated using the Student's t-test.*, p < 0.05; ***, p < 0.01.
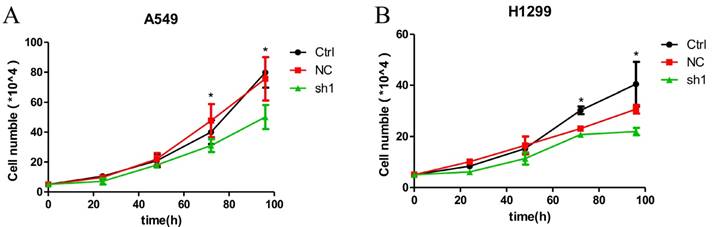
Effects of LNC CRYBG3 in lung cancer cell proliferation. A&B, Growth cureves. The amount of the indicated stable cells cultured in vitro was counted at different time points. Data are presented as mean ± standard error. The P value was calculated using the Student's t-test.*, p < 0.05.
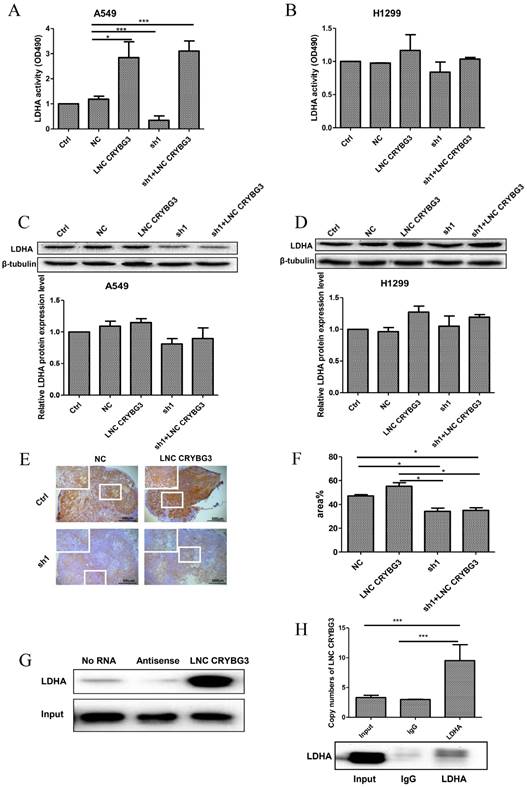
Knockdown of LNC CRYBG3 suppresses lung cancer cell proliferation
Given that LNC CRYBG3 expression was up-regulated in lung cancer, we wondered whether it is involved in regulating the biological behaviors of lung cancer cells. Basing on the high expression of LNC CRYBG3 in the lung cancer cells (Fig. 1), we constructed stable cell lines expressing small hairpin RNAs of LNC CRYBG3 in A549 and H1299 cells. The results indicated that the knockdown of LNC CRYBG3 led to suppressed cell proliferation (Fig. 4A and 4B). Taken together, these findings showed that LNC CRYBG3 is positively correlated with lung cancer cell proliferation.
Discussion
In recent years, accumulating evidence indicates that dysregulation of lncRNA expression causes complex human diseases including human malignancies [27-29]. The abnormal expression of lncRNAs is also involved in cancer cell growth or metastasis as well as lung cancer [19, 30, 31]. LncRNAs can participate in various biological process, such as cell differentiation, transcriptional control, invasion and reprogramming stem cell [32, 33]. However, the underlying mechanism of lncRNA in lung cancer is still unclear. Our previous work has identified a radiation-inducible lncRNA, named LNC CRYBG3 [42]. In this report, we found that the average level of LNC CRYBG3 in lung cancer tissues was significantly higher than that of normal lung tissues and positively correlated with their malignancy. In recent studies, multiple lncRNAs have been explained in lung cancer. For example, MALAT1 exerts oncogenic functions in lung cancer [29], lncRNA DB327252 has an oncogene-like function in lung cancer [34], and lncRNA PVT1 promotes cell proliferation, migration, and invasion in NSCLC [18]. To our best knowledge, it is the first time to report the role of LNC CRYBG3 in lung cancer. The proliferation assay in our study showed that the overexpression of LNC CRYBG3 significantly promoted lung cancer cell proliferation, while the downregulation of endogenous LNC CRYBG3 inhibited cell proliferation. These findings suggested that LNC CRYBG3 plays an important role in regulating in lung cancer progression and might be a potential target for lung cancer treatment.
Cancer metabolism is characterized as aerobic glycolysis or Warburg effect, which affects TCA metabolism and causes the unusual utilization of glucose for producing lactate even in sufficient oxygen [35]. However, whether lncRNAs are involved in the Warburg effects remain unknown. It has been reported that lncRNAs CRNDE could promotes the metabolic changes by which cancer cells switch to aerobic glycolysis (Warburg effect) [36]. In our study, the levels of main metabolites involved in TCA cycle including citrate, α-ketoglutarate, succinate, fumarate and malate didn't show significant alternations (Supporting Figure S3) after either overexpression or knockdown of LNC CRYBG3 in A549 and H1299 lung cancer cell lines. Nevertheless, the pyruvate and lactate in aerobic glycolysis displayed evidently opposite change trends when LNC CRYBG3 level was modified. These results indicated that LNC CRYBG3 could regulate metabolism in lung cancer mainly by affecting glycolysis rather than TCA cycle.
LDHA, which catalyzes the last step of aerobic glycolysis, is a major subunit of LDH. It is involved in early carcinogenesis and tumor progression [37]. Abnormal LDHA expression is universal in many human cancers, such as pancreatic cancer[6], hepatocellular carcinoma [7], and breast cancer [8], etc. According to our research, there is a high LDHA mRNA level in lung cancer cell lines A549 and H1299, as well as the high LDHA protein level in tumor tissues. However, the response of LDHA activity in H1299 cells with altered CRYBG3 seemed to be insignificant. The possible reason is that LNC CRYBG3 background expression in the H1299 cells is higher than that in A549 cells, then overexpression of the LNC CRYBG3 hardly resulted in any additive effect on the H1299 cell line (Fig.3B). In addition, LDHA is also reported to be an adverse independent prognostic factor for lung cancer [38]. As far as we know, LDHA is regulated by some factors, including HIF hydroxylase, H2S and MiR449a [39-41]. However, the relationship between lncRNAs and LDHA hasn't been revealed yet. In our report, we discovered that LDHA mRNA levels positively correlated with the expression levels of LNC CRYBG3 and the LDHA activity is specifically upregulated by LNC CRYBG3. Pull down assay and RIP assay also identified LNC CRYBG3 interacts with LDHA. All these results demonstrated that LNC CRYBG3 is a regulator in lung cancer by interacting with LDHA and promoting the aerobic glycolysis. To the best of our knowledge, this is the first paper to report the interaction between lncRNA and LDHA in lung cancer.
In summary, LNC CRYBG3 could regulate lung cancer metabolism through interacting with LDHA. Our findings suggested that, LNC CRYBG3 plays an important role in lung cancer progression by promoting the LDHA-mediated Warburg effect and it may be served as a novel target for the treatment of lung cancer. And, the underlying mechanisms of LNC CRYBG3 in interacting with LDHA remained to be clarified in our following studies.
Supplementary Material
Supplementary figures.
Acknowledgements
We would like to thank Mr. Chao Xu for technical help with GC-MS analysis. This work was supported by the National Natural Science Foundations of China (No. 11335011, 11405235, 81602794, U1432117 and U1432124), China Postdoctoral Science Foundation (2017T100399), Jiangsu Postdoctoral Science Foundation (1701176B), Jiangsu Science Foundation (BK20160334), Collaborative Innovation Center of Radiological Medicine of Jiangsu Higher Education Institutions and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692-697
3. Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y. et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528-538
4. Lawrence RE, Salgia R. MET molecular mechanisms and therapies in lung cancer. Cell Adh Migr. 2010;4:146-152
5. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Reviews Cancer. 2004;4:891-899
6. Shi M, Cui JJ, Du JW, Wei DY, Jia ZL, Zhang J, Zhu ZG, Gao Y, Xie KP. A Novel KLF4/LDHA Signaling Pathway Regulates Aerobic Glycolysis in and Progression of Pancreatic Cancer. Clinical Cancer Research. 2014;20:4370-4380
7. Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP, Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. Febs Journal. 2012;279:3898-3910
8. Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, Fodstad O, Tan M. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene. 2009;28:3689-3701
9. Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance (vol 9, pg 425, 2006). Cancer Cell. 2006;10:172-172
10. Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Jagt DLV, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2037-2042
11. Wang ZY, Loo TY, Shen JG, Wang N, Wang DM, Yang DP, Mo SL, Guan XY, Chen JP. LDH-A silencing suppresses breast cancer tumorigenicity through induction of oxidative stress mediated mitochondrial pathway apoptosis. Breast Cancer Research and Treatment. 2012;131:791-800
12. Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crino L, Chiari R, Metro G. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33:18
13. Zhong Y, Gao D, He S, Shuai C, Peng S. Dysregulated Expression of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2016;26:1564-1570
14. Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947-2955
15. Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-2329
16. Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M. et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383
17. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666-681
18. Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929-6935
19. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464
20. Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309
21. Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357-366
22. Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301-308
23. Zhao XY, Lin JD. Long Noncoding RNAs: A New Regulatory Code in Metabolic Control. Trends Biochem Sci. 2015;40:586-596
24. Pulito C, Donzelli S, Muti P, Puzzo L, Strano S, Blandino G. microRNAs and cancer metabolism reprogramming: the paradigm of metformin. Ann Transl Med. 2014;2:58
25. Chen B, Li H, Zeng X, Yang P, Liu X, Zhao X, Liang S. Roles of microRNA on cancer cell metabolism. J Transl Med. 2012;10:228
26. Zhao M, Lau KK, Zhou X, Wu J, Yang J, Wang C. Urinary metabolic signatures and early triage of acute radiation exposure in rat model. Mol Biosyst. 2017;13:756-766
27. Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, Yu Q, Li YY, Kang Y, Li H. et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016;7:12693-12703
28. Lalevee S, Feil R. Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7:877-879
29. Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099-1107
30. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243
31. Chen J, Hu L, Wang J, Zhang F, Chen J, Xu G, Wang Y, Pan Q. Low Expression LncRNA TUBA4B is a Poor Predictor of Prognosis and Regulates Cell Proliferation in Non-Small Cell Lung Cancer. Pathol Oncol Res. 2017;23:265-270
32. Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2017;484:586-591
33. Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093
34. Xu E, Yu X, Zeng Q, Qiao G, Xuan Y, Tang Y, Zhu Q, He J. Functional role of lncRNA DB327252 in lung cancer. J Thorac Dis. 2016;8:2793-2802
35. Vaitheesvaran B, Xu J, Yee J, Q-Y L, Go VL, Xiao GG, Lee WN. The Warburg effect: a balance of flux analysis. Metabolomics. 2015;11:787-796
36. Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372-386
37. Ooi AT, Gomperts BN. Molecular Pathways: Targeting Cellular Energy Metabolism in Cancer via Inhibition of SLC2A1 and LDHA. Clin Cancer Res. 2015;21:2440-2444
38. Li AC, Xiao WW, Wang L, Shen GZ, Xu AA, Cao YQ, Huang SM, Lin CG, Han F, Deng XW, Zhao C. Risk factors and prediction-score model for distant metastasis in nasopharyngeal carcinoma treated with intensity-modulated radiotherapy. Tumour Biol. 2015;36:8349-8357
39. Untereiner AA, Olah G, Modis K, Hellmich MR, Szabo C. H2S-induced S-sulfhydration of lactate dehydrogenase a (LDHA) stimulates cellular bioenergetics in HCT116 colon cancer cells. Biochem Pharmacol. 2017;136:86-98
40. Li L, Liu H, Du L, Xi P, Wang Q, Li Y, Liu D. MiR-449a Suppresses LDHA-Mediated Glycolysis to Enhance the Sensitivity of Non-Small Cell Lung Cancer Cells to Ionizing Radiation. Oncol Res. 2017
41. Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393-402
42. Li PF, Hu WT, Pei HL, Ding N, Zhou GM. Expression Profile of Long Non-coding RNAs in Response to Ionizing Radiation. Nuclear Physics Review. 2015;04:484-489
Author contact
![]() Corresponding authors: Guangming Zhou, 199 Renai Road, Suzhou 215123, China. Phone: +86 512 65884829; E-mail: gmzhouedu.cn and Chang Wang, 199 Renai Road, Suzhou 215123, China. Phone: +86 512 65880067, E-mail: wangchangedu.cn
Corresponding authors: Guangming Zhou, 199 Renai Road, Suzhou 215123, China. Phone: +86 512 65884829; E-mail: gmzhouedu.cn and Chang Wang, 199 Renai Road, Suzhou 215123, China. Phone: +86 512 65880067, E-mail: wangchangedu.cn

 Global reach, higher impact
Global reach, higher impact