Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(16):2834-2843. doi:10.7150/jca.26046 This issue Cite
Research Paper
Upregulated Expression of Long Non-Coding RNA, LINC00460, Suppresses Proliferation of Colorectal Cancer.
1. Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China.
2. Key Laboratory of Nanobiological Technology of National Health and Family Planning Commission, Xiangya Hospital , Central South University, Changsha, China.
3. Institute of Reproductive and Stem Cell Engineering, School of Basic Medicine, Central South University, Changsha, China.
4. Xiangya Stomatological Hospital & School of Stomatology, Central South University, Changsha, China.
Received 2018-3-14; Accepted 2018-5-30; Published 2018-7-16
Abstract
Through bioinformatics analysis, a novel lncRNA, LINC00460, was implicated in the development of multiple cancers. However, the precise expression pattern, clinical significance and biological function of LINC00460 in colorectal cancer (CRC) remain unknown. Network databases were used to investigate the correlation between LINC00460 and CRC. In situ hybridization was performed to verify the precise expression pattern and clinical significance of LINC00460 in a CRC tissue microarray, which included 92 pairs of CRC and adjacent normal tissues. The effect of LINC00460 on proliferation was evaluated by MTT, colony formation assays and flow cytometry employing SW620 and HCT116 cell lines. Cell migration and matrigel invasion assays were performed to investigate whether LINC00460 is involved in the metastasis of CRC. The expression of LINC00460 was significantly upregulated in CRC tissues and cells, associated with early stage CRC and low disease-free survival. The downregulated of LINC00460 expression increased cell proliferation by regulating the cell cycles of SW620 and HCT116 cells. LINC00460 knockdown did not affect cell migration or invasion in vitro. These findings suggest that LINC00460 may be an interesting target for the development of CRC.
Keywords: long non-coding RNA, LINC00460, colorectal cancer, proliferation
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy worldwide and the second leading cause of cancer-related death in the United States[1]. Although the treatment of CRC has gradually improved, the 5-year survival rate of patients with CRC remains relatively low because the prognosis of the newly diagnosed colorectal cancer patient is determined by the clinicopathological stage of the disease[2]. Most patients are diagnosed at an advanced stage, and unfortunately, malignant proliferation, extensive invasion and lymphatic metastasis has already occurred in advanced stage CRC[3]. Colorectal carcinogenesis is a multifactorial and multistep process that may involve several biological pathways and an accumulation of genetic alterations[4]. Therefore, as an effective therapeutic strategy, early diagnostic approach or new prognostic prediction, the identification of aberrantly expressed biomarkers that can affect the development and progression of CRC is a critical need.
Long non-coding RNA (lncRNA) molecules, which are more than 200 nucleotides in length without protein-coding capacity, have shown lower conservation and crucial function in multiple biological processes, such as modulating gene expression at the epigenetic, transcriptional and posttranscriptional levels[5, 6]. More importantly, accumulative evidence has demonstrated that aberrant lncRNA expression may contribute to different disease states, particularly cancers.[7]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was identified as a prognostic parameter to promote cell survival and metastasis in non-small cell lung cancer and even other cancers[8]. HOX transcript antisense RNA (HOTAIR) was also significantly overexpressed in multiple tumors types and was able to induce the proliferation and metastasis of these tumors[9]. Therefore, it is important to identify CRC-related lncRNAs and investigate their potential roles and prominent effects in colorectal carcinogenesis[10].
With the development and popularity of high-throughput sequencing and gene microarray technologies, a increasing number of gene expression analyses have been successfully shared on the web[11]. A significant amount of information on lncRNA is now available through databases, including TCGA, GEO, lncRNAdb, NONCODE, lncRNAtor, LncRNADisease, ncFANs, InCeDB, LNCipedia, and psiDR[12-14]. These databases are powerful tools for preliminarily predicting the valuable lncRNA expression between normal and cancerous samples, even in all stages of cancer. In the present study, we identified a new human lncRNA in CRC, named LINC00460, which is an intergenic lncRNA, that is located at chromosome 13q33.2 and is transcribed into a 935 bp transcript. To date, the function of LINC00460 has been poorly annotated. Studies have shown that LINC00460 is significantly up-regulated in non-small cell lung cancer (NSCLC) tissues, and that this molecule promotes the migration and invasion of lung cancer cells by inducing the epithelial-mesenchymal transition[15, 16]. Cao et al.[17] reported that patients with head and neck squamous cell carcinoma (HNSCC) with higher levels of LINC00460 had a relatively poor prognosis. LINC00460 might be a novel biomarker for accurate prognostic predictions of patients with HNSCC. Wang et al.[18] demonstrated an opposite outcome, showing that LINC00460 expression was downregulated in CRC tissues and serum samples compared to controls via microarray analysis and quantitative polymerase chain reaction (qRT‑PCR). However, the expression of LINC00460 in many cancers, especially in CRC, has not yet been well characterized and needs to be further explored.
Thus, the present study aimed to investigate the precise expression pattern, clinical significance and biological function of LINC00460 in CRC development.
Materials and Methods
Patient data and tissue samples
Paired CRC and adjacent non-tumor tissues were collected from 92 patients who underwent resection of primary CRC between 2014 and 2016 in the Department of Gastrointestinal Surgery at Xiangya Hospital, Central South University (Changsha, China). All specimens were confirmed by histopathological examination. Patient clinicopathological characteristics and information were obtained from medical records. None of the patients had received preoperative treatment, including radiotherapy or chemotherapy. Tumor staging was performed according to the tumor-node-metastasis (TNM) staging system of Union for International Cancer Control (UICC). All patients with CRC have been followed upon confirmation of the diagnosis, at intervals of 1-2 months until February 2018, and the median follow-up period was 24 months (range 17-39 months). Follow-up studies included physical examination, laboratory analysis, and computed tomography if necessary. Overall survival (OS) was defined as the interval between the dates of surgery and death. Disease-free survival (DFS) was defined as the interval between the dates of surgery and recurrence. If there was no recurrence, then the data were collected based on the date of death or the last follow-up.
Tissue samples were fixed in 5% DEPC paraformaldehyde (Servicebio, Wuhan, China), and stored at 4°C right after taken during operation. The tissue samples were dehydrated and embedded in paraffin for a week. All the paraffin blocks were made into tissue microarrays for in-situ hybridization (ISH). The adjacent non-tumor samples obtained from the same patients were located at least 5 cm away from the tumor edge.
The present study was conducted according to the principles expressed in the Declaration of Helsinki. All patients understood the procedure and provided written informed consent before enrolling in the study. The present study was approved by the Research Ethics Committee of Xiangya Hospital, China.
Cell culture
Human colon adenocarcinoma (COAD) cell lines SW620, HCT116, CX-1, HT29 and normal colonic epithelial cell lines NCM460 were obtained from the Cell Center of Xiangya School of Medical, Central South University (Changsha, China), and the cell lines were identified by short tandem repeat (STR) sequencing (Genesky Biotechnology Company, Shanghai, China). All cell lines were cultured in RPMI-1640 media (HyClone, Waltham, USA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, Beit Haemek, Israel) and 1% penicillin-streptomycin (100 U/ml penicillin and 100 mg/ml streptomycin) (Biological Industries, Beit Haemek, Israel) in humidified air at 37°C with 5% CO2. The medium was changed every 2 days. The cells were collected for all experiments at the logarithmic growth phase.
Transfection
To overexpress LINC00460, the coding sequence of LINC00460 was amplified and subcloned into the pcDNA3.1(+/-) vector. Plasmid vectors (pcDNA-LINC00460 and empty vector) were purchased from Aokesw Biological Company (Chengdu, China). SW620 and HCT116 cells were seeded onto six-well plates. When the cell densities were approximately 70%, LINC00460-expressing plasmid or a negative control vector were transfected into cells by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA) according to the manufacturer's protocol. The same strategy was used to silence LINC00460. Three specific siRNA oligonucleotides for the human LINC00460 and nonspecific siRNA (si-NC), as a negative control, were purchased from Ribobio biological Company (Guangzhou, China). The sequences of the siRNAs targeting LINC00460 were si-h-LINC00460_1 5'-AGACC TAATA GCCAA TAAG-3'; si-h-LINC00460_2 5'-CCATG TGAAG TGTAG AACA-3'; and si-h-LINC00460_3 5'-TGGTG ACAAT AACAC TGTG-3'.
RNA extraction and qRT-PCR analysis
According to the manufacturer's protocol, total RNA was extracted from tissues and cells with TRIzol Reagent (Invitrogen, Carlsbad, USA) and resuspended in RNase-free water. Then, the RNA samples were quantified for 500 ng/μl by using spectrophotometer (BioTek Instrument, Winooski, USA). Only samples with an A260:A280 ratio between 1.8 and 2.1 were analyzed further. The Reverse Transcriptase kit was provided by TaKaRa (Otsu, Japan). The PCR amplifications were performed in three duplicates for each sample. β-actin was used as an internal reference gene for normalization. Relative expression levels of genes were calculated by using the comparative cycle threshold (Ct) (2-ΔΔCt) method and then converted to fold-changes. The following primer sequences were used: LINC00460-QF: 5'-GGATG AACCA CCATT GCC-3'; LINC00460-QR: 5'-CCCAC GCTCA GTCTT TCT-3'; β-actin-F: 5'-CAGCC GCTTC ACCTA CAGC-3'; β-actin-R: 5'-TTTTG TATTC AATCA CTGTC TTGCC-3'.
In situ hybridization
According to ISH of LncRNA kit provided by Servicebio (Wuhan, China), after deparaffinization and rehydration, the tissue microarray was digested with 20 μg/ml proteinase K at 37°C for 30 min, incubated with prehybridization solution at 65°C for 30 min and then hybridized overnight at 37°C, with probes at 8 ng/μl. After washing with a downgraded concentration of SSC (2xSSC, 10 min at 37°C, 1 SSC, 2x5 min at 37°C, 0.5 SSC, 10 min at room temperature), the sections were treated with blocking reagent at room temperature for 30 min and Alkaline Phosphatase Anti-Digoxin (anti-DIG-AP) at 37°C for 50 min. Finally, the signal was revealed with BCIP/NBT chromogen, and the cell nuclei were counterstained with nuclear fast red solution. Subsequently, the expression of LINC00460 was observed by microscopy. A semi-quantitative scoring criterion was used as previously described. The staining intensity and number of positive cells were independently scored by two experienced pathologists. Each sample was assessed from five randomly selected fields under a microscope at ×200 magnification. The sequences of the probes were 5'-GGCTG AGGCA TTTCT AACAG GGCTG GAGGA-3' (labeled at the 5' end with DIG-dUTP).
Cell migration and Invasion assay
Invasion capacity of transfected tumor cell was assessed by using Transwell Cell Culture Inserts (BD Biosciences, New Jersey, USA) in 24-well plates. A total of 60 μl of 1mg/ml Matrigel (BD Biosciences, New Jersey, USA) were placed into the upper chamber at 37°C and dried for 4-5 h. A total of 2 x 105 cells in 100 µl of serum-free medium were added to the upper chamber. The bottom well contained 600 μl of RPMI-1640 medium supplemented with 10% FBS. After incubating for 24 h at 37°C in 5% CO2, the cells on the upper membrane surface were wiped by using a cotton swab, and the cells that had crossed the filter pores were fixed with 800 μl of methanol, and subsequently stained with 800 μl of 0.1% crystal violet solution. Under a microscope at ×200 magnification, the number of invasive cells in each chamber were counted from five randomly selected fields for three independent experiments and averaged. The migration assay was conducted in a similar manner without the Matrigel coating.
Cell proliferation and colony formation assays
Approximately 2×103 SW460 and HCT116 cells were seeded into each well of 96-well plates after transfection with si-LINC00460/si-NC or pcDNA-LINC00460/empty vector for 48 h. Cell viability was assessed at 0, 1, 2, 3, 4 and 5 days. Then, 10 µl of 1 mg/ml MTT reagent (Vazyme, Nanjing, China) was added to each well, followed by incubation at 37°C, 5%CO2 for 4 h, and subsequently 100 μl of dimethylsulfoxide (DMSO, Vazyme, Nanjing, China) was added to dissolve the crystals. The absorbance at 490 nm in each well was detected by the enzyme marker (BioTek Instrument, Winooski, USA). All experiments were performed in quintuplicate and repeated once. For the colony formation assay, approximately 500 transfected cells were placed into each well of a fresh six-well plate and maintained in 1640 media containing 10% FBS for 2 weeks replacing the medium every 4 days. After 14 days, the colonies were fixed with methanol and stained with 0.1% Giemsa stain. Visible colonies were manually counted. Each sample was assessed in triplicate.
Flow cytometry for cell cycle analysis and apoptosis
Approximately 2×106 SW620 or HCT116 cells for cell cycle distribution were collected at 48 h after transfection with si-Linc00460 or si-NC. The cells were fixed in chilled 75% ethanol at 4°C overnight, and subsequently resuspended in 800 μl of phosphate-buffered saline (PBS) (added 1% BSA). The fixed cells were stained with 100 μl of propidium iodide (PI) (Sigma, San Antonio, USA) (500 μg/ml) and 100 μl RNase A (Sigma, San Antonio, USA) (100 μg/ml) at 37°C for 30 min. For apoptosis analysis, after washing three times with chilled PBS, SW620 or HCT116 cells were double stained with 5 μl of Annexin V-FITC (Sigma, San Antonio, USA) and 5 μl of PI. The cells were analyzed by flow cytometry (Beckman Coulter, South Kraemer, USA) using cell Modifit software. The percentage of the cells in G0/G1, S, and G2/M phases was counted and compared. The cells were discriminated into viable, dead, early apoptotic, and late apoptotic cells, and then the relative ratio of apoptotic cells was compared with that of the control. Each assay was performed in triplicate.
Statistical analysis
All statistical analyses were performed with the SPSS 17.0 statistical software package (IBM, Chicago, USA). Two-sided P values were calculated, and a threshold of P<0.05 was considered statistically significant. The significance of the differences between groups was estimated by Student's t test, χ2 test, or Wilcoxon test, as appropriate. The association between lncRNAs levels and clinical pathology features were analyzed with Kruskal-Wallis H test. Overall survival (OS) or Disease-free survival (DFS) rates were calculated by using the Kaplan-Meier method, and the log-rank test was used to estimate the statistical differences between survival curves.
Results
LINC00460 expression is upregulated in human colorectal cancer
The information for LINC00460 expression was obtained from the Cancer RNA-Seq Nexus (CRN) database and RNA-Seq datasets GEPIA[19]. As shown in Fig 1A, 1B, LINC00460 expression levels were significantly upregulated in colon cancer tissues compared with those in adjacent normal tissues (P < 0.01). To validate the findings from the bioinformatics analysis, we further used ISH to reveal LINC00460 expression in tissue microarrays with 92 paired colon/rectal cancer and adjacent normal tissue samples. According to the semi-quantitative scoring, LINC00460 expression was classified into two groups: moderately and strongly positive staining (scores: 2 and 3), which were regarded as high LINC00460 expression, and negative and weakly positive staining (scores: 0 and 1), which were regarded as low LINC00460 expression. LINC00460 expression was 60.9% (56/92) in CRC tissues and 24.7% (22/89) in adjacent normal tissues (Figs. 1C, 1D). Statistical analysis by using a χ2 test revealed that the LINC00460 signal was much stronger in CRC samples than in adjacent tissues samples (60.9% (56/92) vs. 24.7% (22/89), P < 0.001). The results of ISH were consistent with the database analysis.
LINC00460 expression correlates with the invasion depth and TNM stages of colorectal cancer
Subsequently, we analyzed the association between different expression levels of LINC00460 and the clinicopathological parameters of the patients (Table 1). Only 70 patients with complete information and two patients with partial data were included in the analysis of clinicopathological factors. The data indicated that a stronger LINC00460 signal was significantly associated with a shallow depth of invasion (P = 0.01) and earlier pathological stages (P = 0.046). However, LINC00460 expression was irrelevant to other parameters, such as age (P = 0.292), sex (P = 0.469), tumor location (P = 0.697), tumor size (P = 0.442), histologic differentiation (P = 0.591), lymph node metastasis (P = 0.194), distant metastasis (P = 0.805) and CEA level (P = 0.623), in CRC.
LINC00460 expression is upregulated in human colorectal cancer and associated with poor prognosis. A. RNA-Seq data from TCGA for LINC00460 in COAD were revealed using CRN (http://syslab4.nchu.edu.tw/). B. LINC00460 expression in COAD tissues (n=275) compared with that in non-cancerous tissues (n=349) was analyzed by using the GEPIA database. C. ISH revealed LINC00460 expressions in tissue microarray with 92-paired CRC and adjacent normal tissue samples. A semi-quantitative scoring criterion was used to calculate both the intensity of the positive staining (scores: negative = 0, weak = 1, moderate = 2, or strong = 3) and the proportion of positive cells of interest (scores: 0∼5% = 0, 6%∼25% = 1, 26%∼50% = 2, 51%∼75% = 3, >75%= 4). The product of their scores can be divided into the following categories: negative (scores: 0), weakly positive (scores: 1~4), moderately positive (scores: 5~8), strong positive (scores: 9~12). D. LINC00460 signal was much stronger in CRC samples than that in adjacent tissues samples (P < 0.001). N: Normal C: Cancer. E. Kaplan-Meier survival plots were shown in GEPIA. F. Kaplan-Meier OS and DFS curves were plotted according to the LINC00460 expression level.
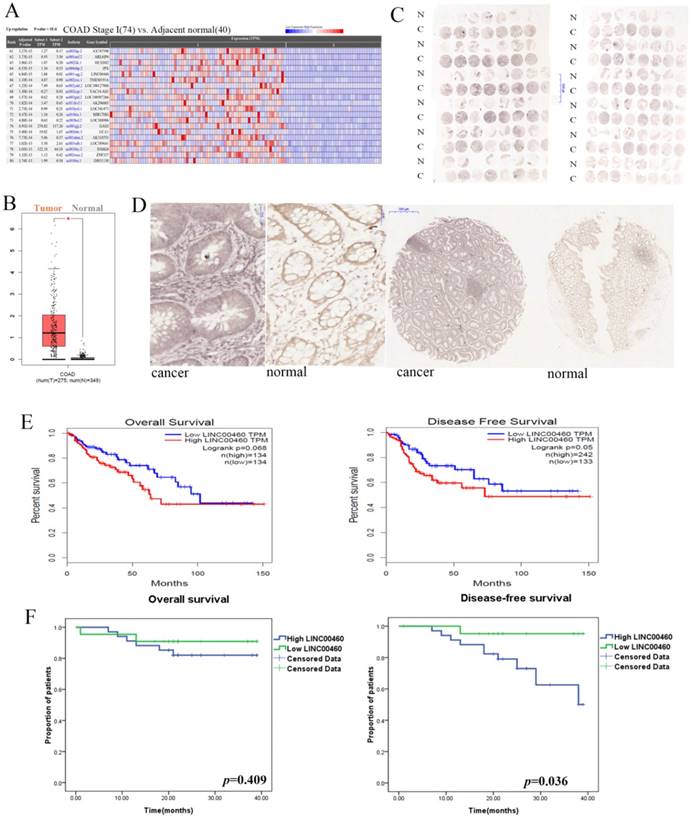
High LINC00460 expression was associated with poor disease-free survival rate
Next, we integrated several public datasets of CRC in GEPIA. High LINC00460 expression was associated with poor DFS (P =0.05), and did not affect OS (P =0.068). (Figure 2E). The CRC patients in the present study were followed up, and the correlation between LINC00460 expression level and their survival outcomes was evaluated. Fifty-six patients were included in this analysis, and only 16 out of 72 cases were excluded from the prognostic analysis due to loss or incomplete follow-up. OS and DFS curves were plotted according to the LINC00460 expression level. As shown in Figure 2F, high LINC00460 expression seemed to be associated with poorer DFS of CRC (P = 0.036). However, Kaplan-Meier analysis and log-rank tests showed no statistical evidence of an association between LINC00460 expression and the OS of CRC (P = 0.409).
Correlation between LINC00460 expression and clinicopathological characteristics in colorectal cancer.
| Clinical parameter | LINC00460 | Kruskal-Wallis H | |||
|---|---|---|---|---|---|
| Negative | Weak | Moderate | Strong | test ( P value) | |
| Age (years) | |||||
| ≤50 | 1 | 8 | 5 | 4 | 0.292 |
| >50 | 1 | 17 | 22 | 14 | |
| Gender | |||||
| Male | 1 | 16 | 17 | 9 | 0.469 |
| Female | 1 | 9 | 10 | 9 | |
| Location | |||||
| Right | 0 | 9 | 7 | 6 | 0.697 |
| Transverse | 0 | 1 | 4 | 2 | |
| Left | 1 | 12 | 14 | 8 | |
| Rectal | 1 | 3 | 2 | 2 | |
| Size | |||||
| ≤5 cm | 2 | 15 | 13 | 10 | 0.442 |
| >5 cm | 0 | 10 | 14 | 8 | |
| Histologic differentiation | |||||
| Well | 1 | 4 | 7 | 6 | 0.591 |
| Moderately | 1 | 15 | 16 | 10 | |
| Poorly | 0 | 5 | 4 | 2 | |
| Invasion depth | |||||
| T1 | 0 | 0 | 3 | 3 | 0.01* |
| T2 | 0 | 0 | 0 | 3 | |
| T3 | 2 | 5 | 10 | 4 | |
| T4 | 0 | 19 | 14 | 7 | |
| Lymph node metastasis | |||||
| N0 | 2 | 14 | 20 | 12 | 0.194 |
| N1 | 0 | 4 | 4 | 4 | |
| N2 | 0 | 7 | 3 | 1 | |
| Distant metastasis | |||||
| Yes | 0 | 3 | 0 | 2 | 0.805 |
| No | 2 | 22 | 27 | 16 | |
| TNM stages | |||||
| I | 0 | 0 | 3 | 5 | 0.046* |
| II | 2 | 14 | 17 | 7 | |
| III | 0 | 8 | 7 | 4 | |
| IV | 0 | 3 | 0 | 2 | |
| CEA | |||||
| ≤5 ng/ml | 1 | 17 | 20 | 14 | 0.623 |
| >5 ng/ml | 1 | 6 | 7 | 4 | |
*p<0.05
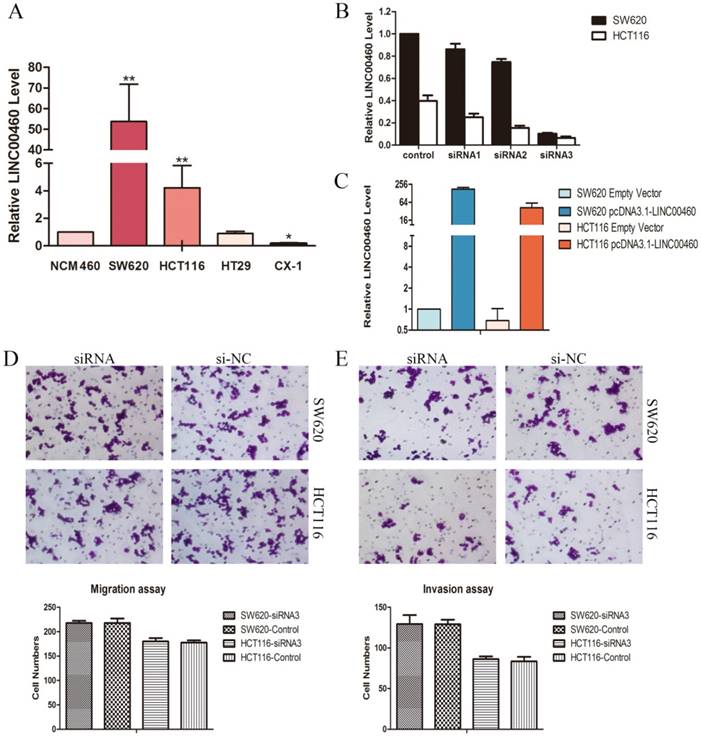
High LINC00460 expression was detected in the human colon cancer cell lines: SW620 and HCT116
QRT-PCR was applied to quantify the LINC00460 expression levels in diverse CRC cell lines including SW620, HCT116, HT29, and CX-1, compared with those in a normal human colon epithelial cell line (NCM460). Markedly high LINC00460 expression was found in the SW620 and HCT116 cells lines compared to other cells (Figure. 2A). Thus, the SW620 and HCT116 cell lines were selected for in-vitro study.
To explore the biological functions of LINC00460 in CRC, we silenced endogenous LINC00460 by using three different short interfering RNAs (siRNAs) targeting LINC00460 in SW620 and HCT116 cells. The effect of knockdown in these cells was confirmed by qRT-PCR. SiRNA-3 achieved 89.8% and 83.2% silencing efficiencies in SW620 and HCT116 cells, respectively (Figure. 2B), and thus, siRNA-3 was selected for further study. LINC00460 was cloned into the pcDNA3.1- vector (pcDNA3.1-LINC00460) and overexpressed in the SW620 and HCT116 cell lines by transient transfection. QRT-PCR analysis was performed at 72 h post-transfection, revealing that LINC00460 expression was increased 176.5-fold in SW620 cells and 77.4-fold in HCT116 cells compared with control cells (Figure. 2C).
LINC00460 knockdown did not affect cell migration and invasion in vitro
Migration and invasion are the mechanisms of cancer cell metastasis. To investigate whether LINC00460 is involved in the metastasis of CRC, cell migration assays and matrigel invasion assays were performed. The results showed that LINC00460 knockdown in SW620 and HCT116 cells did not affect the migration and invasive capabilities of these cell lines compared with those of the control cells (Figure 2D, 2E).
LINC00460 significantly inhibited the proliferation and colony formation of colorectal cancer cells
We evaluated cancer cell proliferation with MTT and colony formation assays. MTT assays demonstrated that the knockdown of LINC00460 expression significantly promoted the cell viability of the SW620 and HCT116 cells lines. Similarly, LINC00460 overexpression inhibited the growth and proliferation of SW620 and HCT116 cells (Figure. 3A). Colony formation assay revealed that the colony-forming ability was decreased following the overexpression of LINC00460 in SW620 and HCT116 cells (Figure. 3B).
Silencing LINC00460 significantly suppressed colorectal cancer cell progression at the G1 phase of the cell cycle and did not affect the rate of apoptosis
To further characterize the anti-proliferation effect of LINC00460 on CRC, flow cytometry was performed to explore whether LINC00460 silencing promoted the proliferation of CRC cells by altering cell cycle progression or apoptosis. The results revealed that the cell cycle progression of both SW620 and HCT116 cells was significantly decreased at the G1 phase; additionally, the percentage of cells at the S or G2 phase was increased when LINC00460 was knocked down (Figure. 3C). This result suggested that LINC00460 expression could attenuate DNA synthesis and promote cell cycle G1 phase arrest. Additionally, in apoptosis assays, LINC00460 knockdown did not increase the rate of early or late apoptosis in SW620 and HCT116 cells (Figure. 3D).
LINC00460 knockdown did not affect cell migration and invasion in vitro. A. Relative expression levels of LINC00460 in diverse CRC cell lines compared with that in a normal human colon epithelial cell line (NCM460) by using qRT-PCR. B. Relative expression levels of LINC00460 in SW620 and HCT116 cells transfected with si-NC or three si-LINC00460 molecules were measured by using qRT-PCR. C. Relative expression levels of LINC00460 in SW620 and HCT116 cells transfected with empty vector or pcDNA3.1-LINC00460 were measured by using qRT-PCR. D. The representative pictures of cell migration assays in SW620 and HCT116 cells with LINC00460 depletion. The average number of migrating cells was counted. E. The representative pictures of Matrigel invasion assays in SW620 and HCT116 cells with LINC00460 depletion. The average number of migrating cells was counted. All data were presented as the means±SD (n=3). *, P<0.05, and **, P<0.01.
Discussion
Zhang et al.[20] selected LINC00460 to identify worthy lncRNA molecules for research by using network analysis. They predicted that LINC00460 might play an important role in tumorigenesis and metastasis through the processes of cell cycle and death. However, experimental validation was not performed. Thus, we were deeply interested in comprehensively investigating the role of LINC00460 in CRC.
Silencing LINC00460 significantly promoted cell proliferation via decreasing colorectal cancer cell at the G1 phase of the cell cycle and did not affect the rate of apoptosis. A. MTT assays were used to determine the cell viability with LINC00460 knockdown and overexpression in SW620 and HCT116 cells. B. The representative picture of colony formation with LINC00460 overexpression in SW620 and HCT116 cells. The average number of colonies was counted. C. Flow cytometry was used to analyze the cell cycle progression. The bar chart represents the percentages of cells in the G1/G0, S, or G2/M phase. D. Flow cytometry was used to analyze the apoptosis. The bar chart represents the percentages of apoptotic cells. E. MethHC database (http://methhc.mbc.nctu.edu.tw/php/search.php?opt=gene) revealed DNA methylation level of LINC00460 in COAD. All data are presented as the means ± SD. *, P < 0.05, and **, P < 0.01.
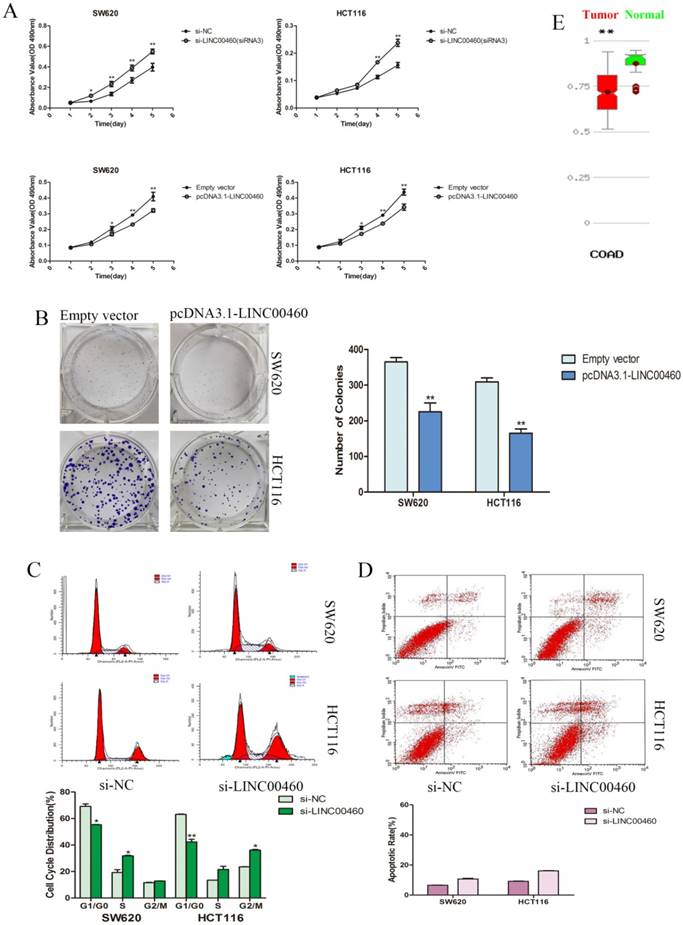
In the present study, through the CRN and GEPIA database, we found that LINC00460 was remarkably upregulated in COAD tissues (Figure. 1A, 1B). Next, we performed ISH to verify the results in the database. Tissue microarray analyses showed that LINC00460 was significantly upregulated in CRC compared with paired non-tumor tissues (Figure. 1C, 1D). This finding is consistent with the prediction in bioinformatics, which contradicts the result presented by Wang et al[18]. Next, we investigated the biological function of LINC00460 in the SW620 and HCT116 cell lines. The results showed that LINC00460 knockdown could promote cell proliferation by facilitating the S or G2 phase transition of cell cycle progression (Figure. 3A, 3C). Conversely, LINC00460 overexpression contributed to the opposite effect (Figure. 3A, 3B), suggesting that LINC00460 may act as a novel tumor suppressor to affect DNA synthesis in CRC. We further analyzed the association between LINC00460 expression and clinicopathological parameters in CRC tissues. The results revealed that high LINC00460 expression was more frequently detected in tumors with shallow invasion depth and an early TNM stage (Table. 1). Therefore, we proposed that the high level of LINC00460 expression might play important roles in CRC development. However, altered LINC00460 expression had no effect on CRC cell migration or invasion (Figure. 2D, 2E). Since cell migration and invasion analyses are used to evaluate cancer progression[21], LINC00460 might not participate in the advanced or metastatic stages of CRC, which is consistent with the results of the clinicopathological parameters. Surprisingly, we found that patients with higher LINC00460 expression showed shorter DFS. Additionally, OS was not correlated with LINC00460 expression (Figure 1F). Thus, we propose that LINC00460 expression inhibits CRC, and can be used to follow and reflect development.
In the present study, a series of in-vitro assays demonstrated that LINC00460 acts as a tumor suppressor gene in CRC cells. However, surprisingly, LINC00460 expression in CRC was upregulated in the present study. The underlying mechanism remains elusive. One study regarded it as an analog of negative feedback regulation[22]. We speculate that the expression of LINC00460 might only impact the early stages of CRC, and with tumor progression, LINC00460 function would be converted by some regulator or manipulated by epigenetic means. The database for the methylation of human cancer genes, MethHC, revealed that methylation might play a critical role in the regulation of LINC00460 expression in COAD (Figure 3E). Notably, there is another explanation. In the present study, in situ hybridization revealed positive staining not only in the cancerous epithelial layer of the colorectum, but also in the lamina propria, which contains stem cells, endocrine cells and immune-related cells (Figure 1)[23]. Thus, the increased LINC00460 expression in colorectal cancer tissues may result from tumor cell secretion or activation by a kind of immunocompetent cells. When carcinogenesis occurs in the local epithelium of the colorectum, an aberrant increase in LINC00460 release into the microenvironment from cancer-associated cells modulates the growth of colorectal cancer cells, however, the function of oncogenes overwhelmingly drowns out the effect of LINC00460 on colorectal cancer cells. These hypotheses suggest that LINC00460 may provide a novel direction for future research on CRC.
Conclusions
In summary, we demonstrated and confirmed that the expression of LINC00460 was significantly upregulated in CRC tissues and cells, associated with early stage CRC and low disease-free survival. Downregulated LINC00460 expression could increase cell proliferation by regulating the cell cycles of the SW620 and HCT116 cell lines. These findings suggest that LINC00460 may be an interesting target for the development of CRC. Further investigation is needed to explore the precise mechanism of LINC00460 in the CRC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 81472692]. We would like to thank the staff of the Key Laboratory of Nanobiological Technology of National Health and Family Planning Commission.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008 CD005390
3. Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288-96
4. Wang X, Yang HH, Liu Y, Zhou Q, Chen ZH. Lycopene Consumption and Risk of Colorectal Cancer: A Meta-Analysis of Observational Studies. Nutr Cancer. 2016;68:1083-96
5. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
6. Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun. 2017;8:14422
7. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-19
8. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-41
9. Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219
10. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P. et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65:1494-504
11. Lu X, Huang C, He X, Liu X, Ji J, Zhang E. et al. A Novel Long Non-Coding RNA, SOX21-AS1, Indicates a Poor Prognosis and Promotes Lung Adenocarcinoma Proliferation. Cell Physiol Biochem. 2017;42:1857-69
12. Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y. et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890-902
13. Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol Cell. 2015;59:698-711
14. Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
15. Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:1003-10
16. Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y. et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80-90
17. Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T. et al. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94-101
18. Wang R, Du L, Yang X, Jiang X, Duan W, Yan S. et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142:2291-301
19. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
20. Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2017
21. Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P. et al. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250
22. Gao S, Lin Z, Li C, Wang Y, Yang L, Zou B. et al. TFPI2AS1, a novel lncRNA that inhibits cell proliferation and migration in lung cancer. Cell Cycle. 2017;16:2249-58
23. Park SH, Moon Y. Integrated stress response-altered pro-inflammatory signals in mucosal immune-related cells. Immunopharmacol Immunotoxicol. 2013;35:205-14
Author contact
![]() Corresponding author: Zi-Hua Chen, Hepatobiliary and Enteric Surgery Center, Xiangya Hospital, Central South University, Changsha, China; Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China. Xiangya Road No. 87, Kaifu District, Changsha, Hunan Province, China. Tel: +8613808413012; E-mail:zihuaccom. Postcode: 410008
Corresponding author: Zi-Hua Chen, Hepatobiliary and Enteric Surgery Center, Xiangya Hospital, Central South University, Changsha, China; Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China. Xiangya Road No. 87, Kaifu District, Changsha, Hunan Province, China. Tel: +8613808413012; E-mail:zihuaccom. Postcode: 410008

 Global reach, higher impact
Global reach, higher impact