Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(16):2895-2909. doi:10.7150/jca.24749 This issue Cite
Research Paper
Clinicopathological and prognostic significance of leucine-rich repeats and immunoglobulin-like domains protein 1 (LRIG1) in malignant tumors: A meta-analysis
Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, Shaanxi 710061, China
Received 2018-1-4; Accepted 2018-6-9; Published 2018-7-30
Abstract
Background: Accumulating studies have demonstrated that the expression of leucine-rich repeats and immunoglobulin-like domains protein1 (LRIG1) is associated with various types of tumors. However, the conclusions of previous studies are not completely consistent. Thus, we conducted this meta-analysis to further explore the authentic value of LRIG1 in cancer outcome and clinical significance.
Methods: We systematically searched electronic databases including PubMed, Web of Science, Embase, Chinese National Knowledge Infrastructure and Wanfang database. The hazard ratios (HRs), odds ratio (OR) and 95 % confidence intervals (CI) were calculated for effect measures.
Results: 16 qualified studies involving 2043 patients with cancer were enrolled. High LRIG1 expression was associated with a good prognosis in malignant tumors (HR: 0.49, 95% CI=0.39-0.59). Furthermore, positive expression rate of LRIG1 was distinctly lower in cancer tissues than that in normal tissues (OR: 0.09, 95% CI=0.05-0.17). Positive LRIG1 expression was definitely related with smaller tumor size (OR: 1.64, 95% CI=1.11-2.42), early tumor stage (OR: 3.67, 95% CI=1.87-7.21), well degree of differentiation (OR: 4.35, 95% CI=2.12-8.93) and negative recurrence (OR: 0.29, 95% CI=0.16-0.53).
Conclusions: LRIG1 expression was associated with a good prognosis in terms of overall survival (OS) and might act as a predictive factor for characteristics of cancer patients.
Keywords: leucine-rich repeats and immunoglobulin-like domains protein1, tumor, meta-analysis
Introduction
Cancer is a leading cause of mortality and a major public health problem worldwide. According to statistical analysis, 1,688,780 new cancer cases and 600,920 cancer deaths are projected to occur in the United States in 2017 [1]. To improve the poor outcome of cancer patients, great efforts have been made on the diagnoses, therapies and prognosis of cancer over the decades. Recently, biomarkers are applied to clinical practice as both predictive and prognostic tools, which play an important role in improving the survival rate of cancer patients [2]. Depending on the facilities and important function of biomarkers for accurate diagnosis, prognostic evaluation and targeting therapy in tumors, identifying novel biomarkers for predicting prognosis is helpful and necessary.
LRIG1, a single-pass transmembrane protein, belongs to leucine-rich and immunoglobulin-like domains family. Emerging data have emphasized the significance of LRIG1 in cell proliferation, apoptosis, and tumorigenesis. Recent studies have shown that LRIG1 is a tumor suppressor by participating in an epidermal growth factor (EGF) driven negative feedback loop [3, 4]. LRIGI acts as a negative regulator of ErbB signaling by promoting the ubiquitination and degradation of EGFR [5]. Genetic ablation of LRIG1 up-regulates the expression of ErbB1-3 and contributes to tumor formation [6]. In addition, LRIG1 has been reported to be an independent prognosis factor and predictive marker of clinicopathology in diverse malignancies. High LRIG1 expression is related to high survival rate and long-term outcome in non-small cell lung cancer [7-10], oropharyngeal cancer [11], cervical adenocarcinoma [12] and hepatocellular carcinoma [13]. However, there is no significant correlation in oesophageal carcinoma [14] and ependymoma [15]. Due to the conflicting findings and small sample sizes of most previous studies evaluating the implications of LRIG1 levels in cancer, this meta-analysis was performed to reveal the authentic value of LRIG1 in cancer outcome and clinicopathological characteristics such as advanced tumor stage, recurrence and lymphatic metastasis.
Material and methods
Search strategy
To identify articles applicable for this meta-analysis, two independent reviewers (Qianqian Zhang and Wenhua Shi) performed a systematic search in PubMed, Web of Science, Embase, Chinese National Knowledge Infrastructure (CNKI) and Wanfang database for studies published before January 2018. The following terms through MeSH headings and keywords were used for searching: (“LRIG1 protein” OR “LRIG1” OR “Leucine-rich repeats and immunoglobulin-like domains protein 1”) AND (“cancer” OR “tumor” OR “neoplasm” OR “malignancies”). We also searched reference lists of primary literatures manually for additional relevant articles. All searches were restricted to English and Chinese publications.
Selection criteria
The inclusion criteria: (1) reporting LRIG1 expression in cancer tissues; (2) case-control, cross-sectional or cohort studies; (3) histologically and pathologically confirmed patients; (4) evaluating the association of LRIG1 expression with clinicopathological features or patient prognosis in malignant tumors; (5) if multiple studies contained overlap or duplicated data, only the most informative and recent study was included.
The exclusion criteria: (1) animal or cell line studies; (2) reviews, case reports, letters or conference abstracts; (3) unqualified or lack of enough data for further quantification analyses. Two independent reviewers evaluated the full articles for the accuracy of the selection process, and any disagreement was resolved by consensus.
Data extraction and methodological quality assessment
The meta-analysis was conducted according to the PRISMA (Preferred Reporting Items for Systemic Reviews and Meta-Analyses) guidelines [16]. The variables collected from each study were: (1) the name of first author, publication year, country, study design and LRIG1 assessment method; (2) age, sex, numbers of patients, case number of different groups, differentiation degree, lymph node metastasis, tumor node metastasis (TNM) classification, recurrence and human papillomavirus (HPV) status; (3) HR and 95% CI of LRIG1 value for OS was collected from the text or Kaplan-Meier curves. The survival information from Kaplan-Meier curves was extracted using Enguage Digitizer 4.1. The HR and 95% CI were determined following the methods described previously [17, 18]. The Newcastle-Ottawa Scale (NOS) [19] was applied to evaluate the quality of each identified studies. We defined studies with scores no less than 6 as qualified to be included in the meta-analysis (Table 1).
Statistical analysis
All statistical analyses were performed using the software Stata SE12.0 (StataCorp, College Station, TX, USA). The pooled OR with 95% CI was calculated to estimate the relationships between LRIG1 expression and clinicopathological parameters of malignant tumors. The HR with 95%CIs were used to assess the role of LRIG1 in predicting prognosis of malignant tumors. Heterogeneity among studies was quantified using the chi-squared and I2 test. I2 >50% or P<0.05 was defined as significant heterogeneity [20]. The fixed-effect model was used to pool the results with no evident heterogeneity existing. Otherwise, the random-effect model was selected. Furthermore, subgroup analysis and meta-regression were performed to explore the potential source of heterogeneity, using covariates such as research area, tumor type and histology. Sensitivity analysis was conducted by omitting individual studies sequentially to assess robustness of included studies. Funnel plots and Egger's test were applied to evaluate the publication bias [21]. P≥ 0.05 means no publication bias. All P values were 2-tailed.
Results
Literature search
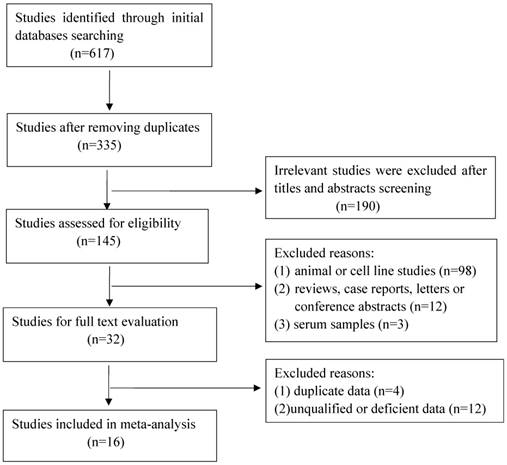
A total of 617 articles were retrieved initially, of which 282 duplicate studies and 190 irrelevant studies were excluded during the initial screening of the titles and abstracts. Subsequently, 113 studies were removed for the following reasons: animal or cell research (n=98); reviews, case reports, letters or conference abstracts (n=12); serum samples (n=3). After this elaborative searching process, we reviewed 32 studies of full text for extracting applicable data, nevertheless, 16 studies were eliminated due to overlapping data, deficient date or low quality. Eventually 16 qualified studies containing 2043 cancer patients were enrolled for further analysis. The flow of literature searching was shown in Figure 1.
Flow chart of the selection for the meta-analysis
Characteristics of enrolled studies
As shown in Table 1, the majority of 16 studies [7-14, 22-29] were carried out in Asia, including 9 studies in China [7-9, 13, 25-29] and 1 study in Japan [24]. The remaining studies were in Sweden [10-12, 14, 22, 23]. All enrolled studies were published from 2005 to 2017 and the sample size of studies ranged from 38 to 347. There are ten different types of cancers including non-small cell lung cancer [7-10], oropharyngeal cancer [11], cervical cancer [12,23,29], primary vaginal carcinoma [22], squamous cell carcinoma of skin [24], oesophageal carcinoma [14], hepatocellular carcinoma [13], colon cancer [25,27], bladder transitional cell carcinoma [26] and gastric cancer [28]. The expression of LRIG1 was detected by immunohistochemistry (IHC) staining in both cancer and normal tissues.
Prognosis value of LRIG1 in malignant tumors
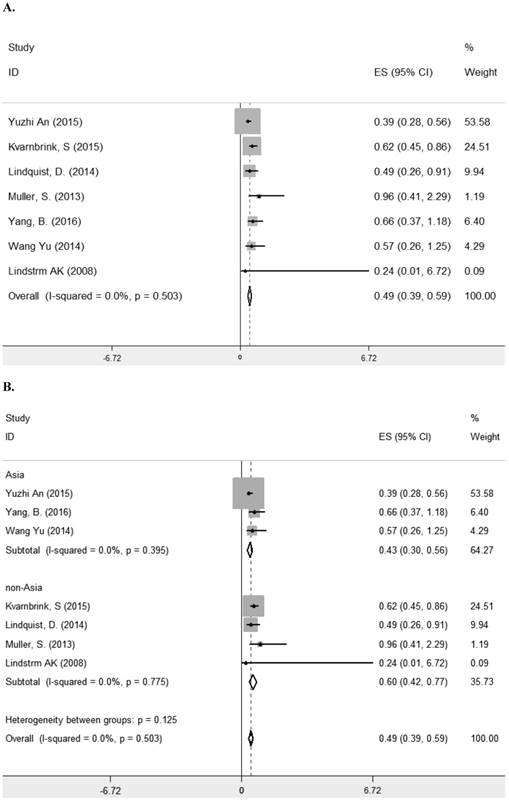
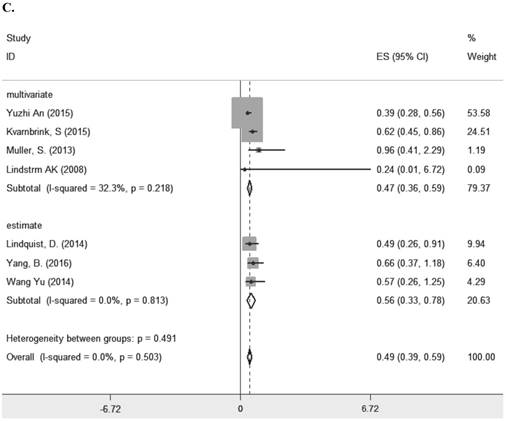
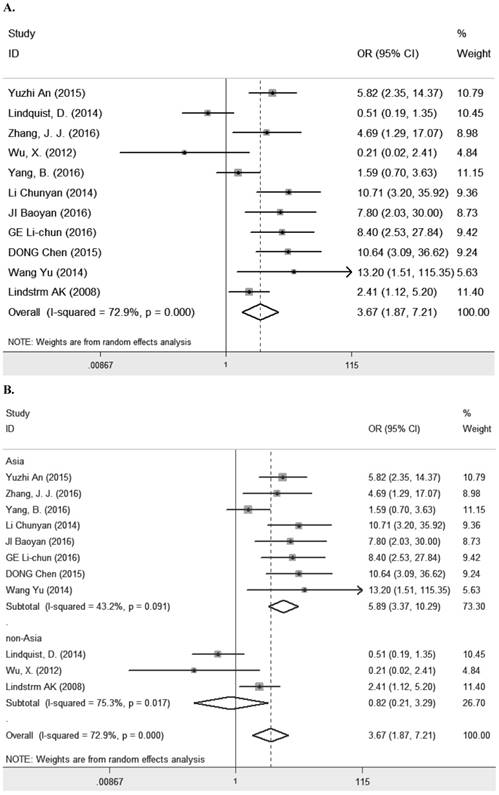
Seven studies [8, 10-13, 23, 29] comprising 1261 patients investigated the relationship between LRIG1 expression and overall survival (OS) in diverse cancer types (Table 1). The combined HR was 0.49 (95% CI=0.39-0.59) (z=9.35, p<0.001), calculating by a fixed model according to the lake of heterogeneity (I2=0.0%) (Figure 2A). The subgroup analysis based on research area revealed that the HR in Asian was 0.43 (95% CI=0.30- 0.56, p<0.001) and in non-Asia region was 0.60 (95% CI=0.42-0.77 p<0.001) (Figure 2B). Considering different methods of extracting HR, the subgroup analysis suggested that the HR in group of multivariate was 0.47 (95% CI=0.36-0.59, p<0.001) and in group of estimate was 0.56 (95% CI=0.33-0.78, p<0.001) (Figure 2C). These results showed that high LRIG1 expression was associated with a good prognosis in malignant tumors, and different methods of extracting HR did not influence these results.
Association between LRIG1 expression and clinicopathological significance
LRIG1 expression: cancer patients VS normal patient
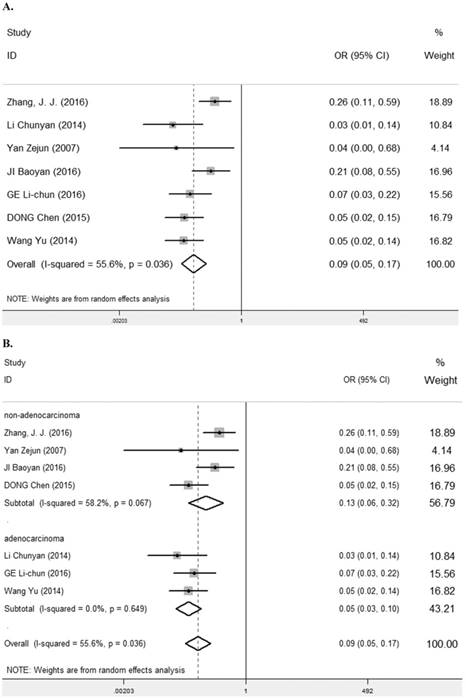
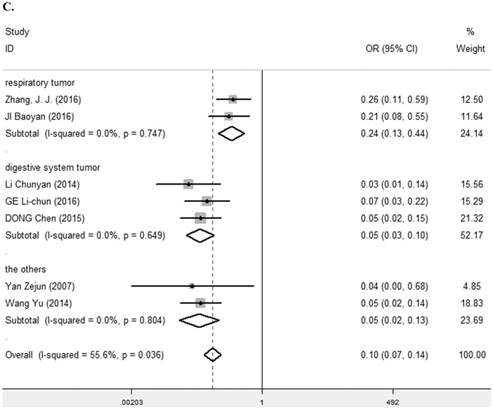
Seven studies [7, 9, 25-29] containing 447 cancer patients and 310 normal patients were assessed for the correlation between LRIG1 expression in cancer and normal tissues (Table 2). A random-effect model was selected due to heterogeneity (I2=55.6%, p=0.036) (Figure 3A) among studies. Positive LRIG1 expression was distinctly lower in cancer tissues than that in normal tissues (pooled OR: 0.09, 95% CI=0.05-0.17) (z=7.38, p<0.001) (Figure 3A). To explore sources of heterogeneity, subgroup analysis stratified by tumor histology and tumor type was conducted. The summary OR in group of adenocarcinoma and non-adenocarcinoma was 0.05 (95% CI=0.03-0.10, p<0.001; I2=0.0%, p=0.649) and 0.13 (95% CI=0.06-0.32, p<0.001; I2=58.2%, p=0.067), respectively (Table 3) (Figure 3B). The summary OR in group of digestive system tumor, respiratory tumor and the others was 0.06 (95% CI=0.03-0.11, p< 0.001; I2=0.0%, p=0.649), 0.24 (95% CI=0.13-0.44, p< 0.001; I2=0.0%, p=0.747) and 0.05 (95% CI=0.02-0.13, p< 0.001; I2=0.0%, p=0.804) (Table 3) (Figure 3C), which demonstrated that tumor type mainly explained the source of heterogeneity.
Baseline characteristics of included studies
| Author | Year | Country | Research design | Method | Cancer type | Age (year) | Total cases | OS HR(95%CI) | score |
|---|---|---|---|---|---|---|---|---|---|
| Kvarnbrink, S | 2015 | Sweden | Retrospective | IHC | Non-small cell lung cancer | 67 | 347 | 0.623(0.449-0.863) | 8 |
| Lindquist, D. | 2014 | Sweden | Retrospective | IHC | Oropharyngeal cancer | - | 278 | 0.49 (0.26-2.91) | 8 |
| Muller, S. | 2013 | Sweden | Retrospective | IHC | Cervical cancer | 48 | 86 | 0.964 (0.407-2.285) | 8 |
| Ranhem, C. | 2017 | Sweden | Retrospective | IHC | Primary vaginal carcinoma | 37-90 | 81 | - | 7 |
| Wu, X. | 2012 | Sweden | Retrospective | IHC | Oesophageal carcinoma | - | 80 | - | 8 |
| Lindström AK | 2008 | Sweden | Retrospective | IHC | Cervical cancer | 59.7 | 128 | 0.24 (0.01-6.72) | 9 |
| Tanemura, A. | 2005 | Japan | Retrospective | IHC | Squamous cell carcinoma of skin | 18-93 | 38 | - | 8 |
| Yuzhi An | 2015 | China | Retrospective | IHC | Non-small cell lung cancer | - | 182 | 0.39 (0.28-0.56) | 8 |
| Zhang, J. J. | 2016 | China | Retrospective | IHC | Non-small cell lung cancer | 60.12±11.4 | 122 | - | 6 |
| Yang, B. | 2016 | China | Retrospective | IHC | Hepatocellular carcinoma | - | 133 | 0.66 (0.37-1.18) | 7 |
| Li Chunyan | 2014 | China | Retrospective | IHC | Colon cancer | 40-76 | 100 | - | 6 |
| Yan Zejun | 2007 | China | Retrospective | IHC | Bladder transitional cell carcinoma | 27-75 | 60 | - | 6 |
| JI Baoyan | 2016 | China | Retrospective | IHC | Non-small cell lung cancer | - | 128 | - | 6 |
| GE Lichun | 2016 | China | Retrospective | IHC | Colon cancer | 36-79 | 106 | - | 6 |
| DONG Chen | 2015 | China | Retrospective | IHC | Gastric cancer | 38-73 | 67 | - | 6 |
| Wang Yu | 2014 | China | Retrospective | IHC | Cervical cancer | 34-78 | 107 | 0.57 0.26-1.25) | 7 |
IHC, immunohistochemistry; OS, overall survival; HR, hazard ratio; CI, confidence interval.
A. Forest plot of HR for the relationship between LRIG1 expression and OS in a fixed effect model. B. Forest plot of subtotal HRs based on different research region. C. Forest plot of subtotal HRs based on different obtainment of HR
LRIG1 expression with clinicopathological parameter
| Author | LRIG1 expression (positive /all) (N) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender(N) | Group(N) | Tumor size (N) | Tumor stage (N) | Tumor differentiation(N) | Lymphatic metastasis (N) | Recurrence(N) | HPV status | |||||||||
| M | F | Control | Cancer | ≤5cm | >5cm | Ⅰ-Ⅱ | Ⅲ-Ⅳ | well | Moderate and poor | yes | No | yes | no | yes | No | |
| Yuzhi An | 62/119 | 40/63 | - | - | - | - | 95/151 | 7/31 | 53/76 | 49/106 | - | - | - | - | - | - |
| Lindquist, D. | 185/20 | 59/72 | - | - | - | - | 24/30 | 220/248 | - | - | - | - | - | - | 194/215 | 50/63 |
| Muller, S. | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 51/56 | 23/29 |
| Ranhem, C. | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 35/37 | 23/34 |
| Tanemura, A | - | - | - | - | - | - | - | - | 19/20 | 4/18 | - | - | 3/7 | 19/31 | - | - |
| Zhang, J. J. | - | - | 50/61 | 33/61 | 25/45 | 8/16 | 29/46 | 4/15 | 14/25 | 19/36 | 17/19 | 16/42 | - | - | - | - |
| Wu, X. | - | - | - | - | - | - | 22/75 | 2/3 | - | - | - | - | - | - | - | - |
| Yang, B. | 34/79 | 31/54 | - | - | 51/83 | 14/50 | 53/103 | 12/30 | - | - | - | - | 19/57 | 46/76 | - | - |
| Li Chunyan | 11/34 | 11/30 | 34/36 | 22/64 | 10/26 | 12/38 | 15/22 | 7/42 | 11/17 | 11/47 | 3/23 | 19/41 | - | - | - | - |
| Yan Zejun | - | - | 8/8 | 20/52 | - | - | - | - | 11/13 | 9/39 | - | - | 2/17 | 18/35 | - | - |
| JI Baoyan | 29/51 | 19/35 | 36/42 | 48/86 | 36/63 | 12/23 | 45/70 | 3/16 | 20/33 | 28/53 | 27/58 | 21/28 | - | - | ||
| GE Li-chun | 12/38 | 10/32 | 31/36 | 22/70 | 11/34 | 11/36 | 12/18 | 10/52 | 6/9 | 16/61 | 4/29 | 18/41 | - | - | - | - |
| DONG Chen | 14/37 | 10/30 | 61/67 | 24/67 | 16/46 | 8/21 | 14/19 | 10/48 | 17/28 | 7/39 | 2/22 | 22/45 | - | - | - | - |
| Wang Yu | - | - | 50/60 | 10/47 | - | - | 9/24 | 1/23 | - | - | 8/20 | 2/27 | - | - | - | - |
| Lindström AK | - | - | - | - | - | - | 47/86 | 14/42 | - | - | - | - | - | - | - | - |
N, cases; M, male; F, female; S, squamous; A, adenocarcinoma; HPV, human papillomavirus.
LRIG1 expression and tumor size
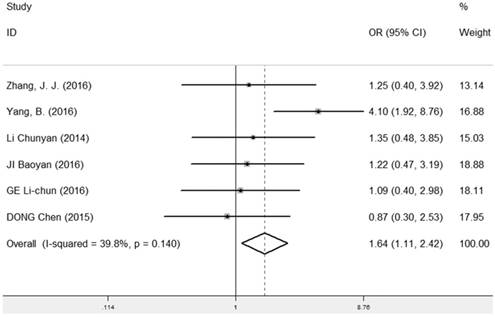
The correlation between LRIG1 expression and different tumor size was evaluated in six studies [7, 9, 13, 25, 27, 28] containing 214 cancer patients and 267 normal patients (Table 2). A fixed-effect model was used and the pooled OR was 1.64 (95% CI=1.11-2.42) (z=2.51, p=0.012) (Figure 4), implying that positive expression rate of LRIG1 was distinctly higher in cancer tissues of tumor size ≤ 5cm than that in cancer tissues of tumor size > 5cm.
LRIG1 expression and tumor stage
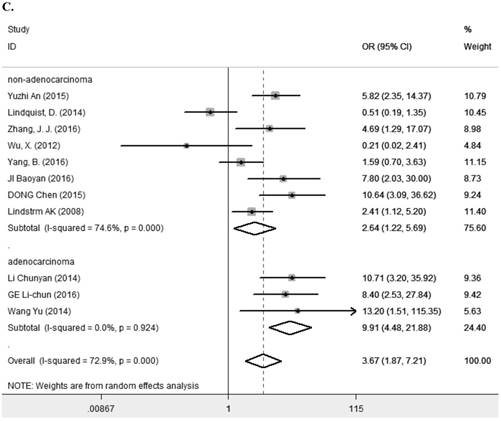
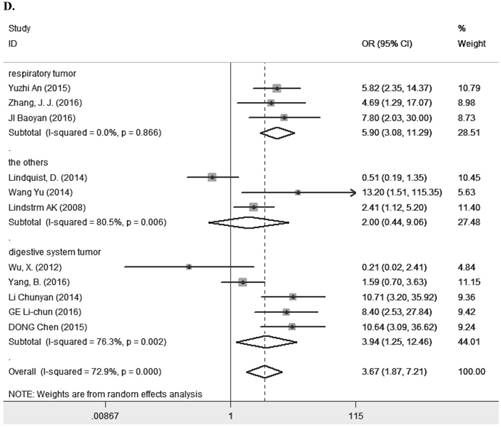
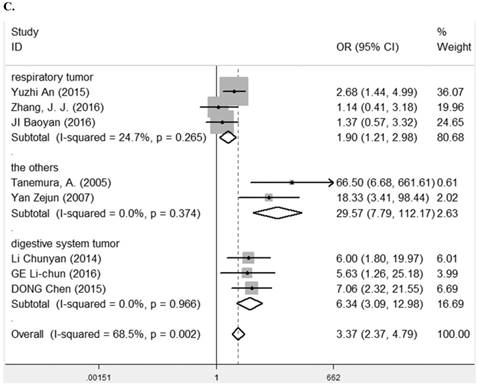
Eleven studies [7-9, 11, 13, 14, 23, 25, 27-29] containing 1194 cancer patients examined the relevance between LRIG1 expression and tumor stage (Table 2). A random-effect model was selected due to heterogeneity (I2=72.9%, p<0.001) (Figure 5A) among studies. The pooled OR was 3.67 (95% CI=1.87-7.21) (z=3.77, p<0.001) (Figure 5A), indicating positive rate of LRIG1 expression was distinctly higher in early stage than that in advanced stage. To explore sources of heterogeneity, univariable meta-regression was carried out, founding that research area partly explained the heterogeneity existing in studies (p=0.013) (Table 4). Furthermore, we applied subgroup analysis stratified by research area, tumor histology and tumor type. The summary OR in group of Asian and non-Asian region was 5.89 (95% CI=3.37-10.29, p<0.001; I2=43.2%, p=0.091) and 3.67 (95% CI=0.21-3.29, p=0.782; I2=75.3%, p=0.017) (Table 3) (Figure 5B). The summary OR in group of adenocarcinoma and non-adenocarcinoma was 9.91 (95% CI=4.48-21.88, p< 0.001; I2=0.0%, p=0.924) and 2.64 (95% CI=1.22-5.69, p=0.013; I2=74.6%, p<0.01) (Table 3) (Figure 5C). The summary OR in group of digestive system tumor, respiratory tumor and the others was 3.94 (95% CI=1.25-12.46, p=0.019; I2=76.3%, p=0.002), 3.12 (95% CI=0.83-11.74, p=0.092; I2=82.5%, p=0.001) and 4.15 (95% CI=0.86-19.97, p=0.076; I2=53.4%, p=0.143) (Table 3) (Figure 5D). These results indicated that research area was an important source of heterogeneity.
LRIG1 expression and tumor differentiation
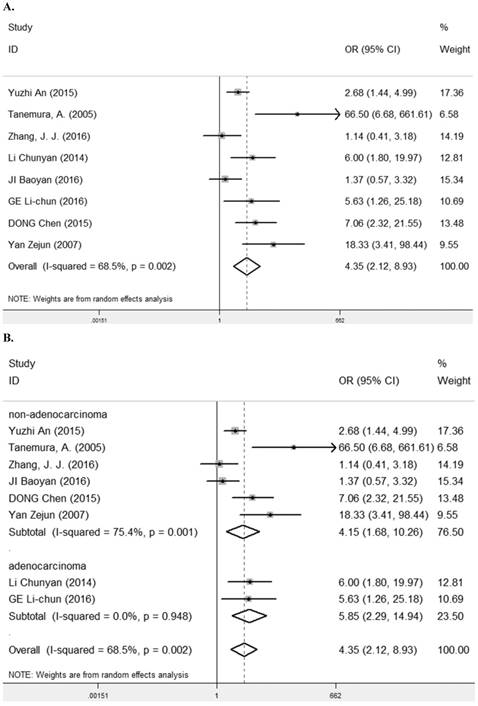
Eight studies [7-9, 24-28] containing 623 cancer patients evaluated the correlation between LRIG1 expression and tumor differentiation (Table 2). A random-effect model was selected due to heterogeneity (I2= 68.5%, p=0.002) (Figure 6A) among studies. The pooled OR was 4.35 (95% CI=2.12-8.93) (z=4.00, p<0.001) (Figure 6A). To explore sources of heterogeneity, subgroup analysis was performed according to tumor histology and tumor type. The summary OR in group of adenocarcinoma and non-adenocarcinoma was 5.85 (95% CI=2.29-14.94, p< 0.001; I2=0.0%, p=0.948) and 4.15 (95% CI=1.68-10.26, p=0.002; I2=75.4%, p=0.001) (Table 3) (Figure 6B). The summary OR in group of digestive system tumor, respiratory tumor and the others was 6.34 (95% CI=3.09-12.98, p< 0.001; I2=0.0%, p=0.966), 1.90 (95% CI=1.21-1.98, p=0.005; I2=24.7%, p=0.265) and 29.57 (95% CI=7.79-112.17, p<0.001; I2=0.0%, p=0.374) (Table 3) (Figure 6C). The results implied that positive rate of LRIG1 expression was distinctly higher in well differentiation than that in moderate and poor differentiation, and subgroup analysis based on tumor type markedly reduced heterogeneity in each group.
A. Forest plot of OR for the relationship between LRIG1 expression in cancer tissues and normal tissues, using a random effect model. B. Forest plot of subtotal ORs for the relationship between LRIG1 expression in cancer tissues and normal tissues based on different tumor histology. C. Forest plot of subtotal ORs for the relationship between LRIG1 expression in cancer tissues and normal tissues based on different tumor type.
Subgroup analysis
| Variable | Subgroup | OR/HR (95% CI) | p-value | z-value | I2 | |
|---|---|---|---|---|---|---|
| Overall survival. | Research area | Asia | 0.43(0.30- 0.56) | < 0.001 | 6.58 | 0.0% |
| non-Asia | 0.60 (0.42-0.77) | < 0.001 | 6.82 | 0.0% | ||
| Obtainment | multivariate | 0.47(0.36-0.59) | < 0.001 | 8.02 | 32.3% | |
| estimate | 0.56(0.33-0.78). | < 0.001 | 4.86 | 0.0% | ||
| Cancer VS normal | histology | adenocarcinoma | 0.05 (0.03-0.10) | < 0.001 | 8.85 | 0.0% |
| non-adenocarcinoma | 0.13 (0.06-0.32) | < 0.001 | 7.63 | 58.2% | ||
| tumor type | digestive system tumor, | 0.06 (0.03-0.11) | < 0.001 | 8.87 | 0.0% | |
| respiratory tumor | 0.24(0.13-0.44) | < 0.001 | 4.53 | 0.0% | ||
| the others | 0.05 (0.02-0.13) | < 0.001 | 6.21 | 0.0% | ||
| Tumor stage | research area | Asia | 5.89 (3.37-10.29) | < 0.001 | 6.23 | 43.2% |
| non-Asia | 3.67(0.21-3.29) | 0.782 | 0.28 | 75.3% | ||
| histology | adenocarcinoma | 9.91 (4.48-21.88) | < 0.001 | 5.67 | 0.0% | |
| non-adenocarcinoma | 2.64(1.22-5.69) | 0.013 | 2.47 | 74.6% | ||
| tumor type | digestive system tumor | 3.94(1.25-12.46) | 0.019 | 2.34 | 76.3% | |
| respiratory tumor | 3.12(0.83-11.74) | 0.092 | 1.69 | 82.5% | ||
| the others | 4.15 (0.86-19.97) | 0.076 | 1.77 | 53.4% | ||
| Tumor differentiation | histology | adenocarcinoma | 5.85 (2.29-14.94) | < 0.001 | 3.69 | 0.0% |
| non-adenocarcinoma | 4.15 (1.68-10.26) | 0.002 | 3.09 | 75.4% | ||
| tumor type | digestive system tumor | 6.34 (3.09-12.98) | < 0.001 | 5.04 | 0.0% | |
| respiratory tumor | 1.90(1.21-1.98) | 0.005 | 2.15 | 24.7% | ||
| the others | 29.57 (7.79-112.17) | <0.001 | 4.85 | 0.0% | ||
OR, odds ratio; HR, hazard ratio; CI, confidence interval.
LRIG1 expression and lymphatic metastasis
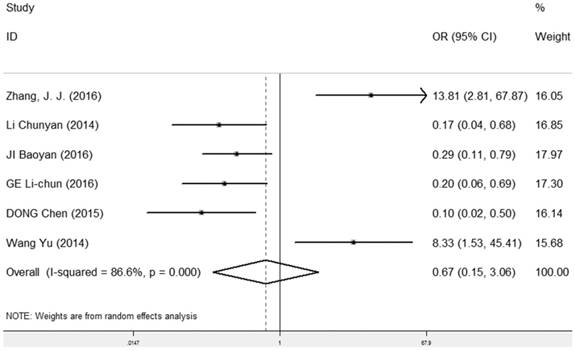
The correlation between LRIG1 expression and lymphatic metastasis was examined in six studies [7, 9, 25, 27-29] containing 395 cancer patients (Table 2). A random-effect model was selected due to heterogeneity (I2=86.6%, p<0.001) (Figure 7) among studies. The OR was 0.67 (95% CI=0.15-3.06) (z=0.52, p=0.604) (Figure 7), indicating that there was no association between positive LRIG1 expression and lymphatic metastasis.
LRIG1 expression and recurrence
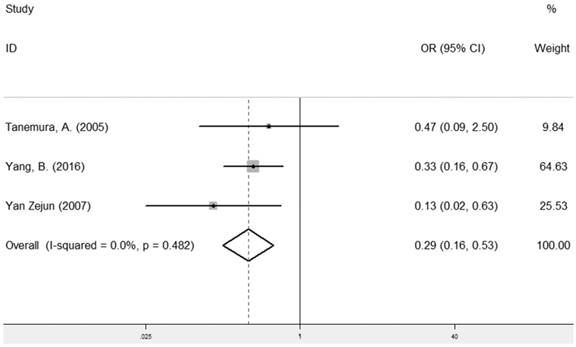
The combined analysis of the correlation between LRIG1 expression and recurrence included three studies [13, 24, 26] containing 223 cancer patients (Table 2). The results showed that positive LRIG1 expression was significantly related with the group of no recurrence (pooled OR: 0.29, 95% CI=0.16-0.53) (z=4.04, p< 0.001) (Figure 8), calculating by a fixed model due to a lack of heterogeneity (I2=0.0%, p=0.482) (Figure 8).
Forest plot of OR for the relationship between LRIG1 expression and tumor size in a fixed effect model.
A. Forest plot of OR for the relationship between LRIG1 expression and tumor stage in a random effect model. B. Forest plot of subtotal ORs for the relationship between LRIG1 expression and tumor stage based on different research region. C. Forest plot of subtotal ORs for the relationship between LRIG1 expression and tumor stage based on different tumor histology. D. Forest plot of subtotal ORs for the relationship between LRIG1 expression and tumor stage based on different tumor type.
Univariable meta-regression
| Variable | Covariate | Level | t-value | p>|t| | 95% CI |
|---|---|---|---|---|---|
| Tumor stage | research area | Asia | Ref | - | - |
| non-Asia | -3.09 | 0.013 | (-3.23 ~ -0.50) | ||
| histology | adenocarcinoma | Ref | - | - | |
| non-adenocarcinoma | -1.73 | 0.117 | (-3.11 ~ 0.41) | ||
| tumor type | digestive system tumor | 0.82 | 0.435 | (-1.38 ~ 2.90) | |
| respiratory tumor | 1.12 | 0.294 | (-1.21 ~ 3.50) | ||
| the others | Ref | - | - |
CI, confidence interval; Ref, reference.
A. Forest plot of OR for the relationship between LRIG1 expression and tumor differentiation in a fixed effect model. B. Forest plot of subtotal ORs for the relationship between LRIG1 expression and tumor differentiation based on different tumor histology. C. Forest plot of subtotal ORs for the relationship between LRIG1 expression and tumor differentiation based on different tumor type.
Forest plot of OR for the relationship between LRIG1 expression and lymphatic metastasis in a random effect model.
Forest plot of OR for the relationship between LRIG1 expression and recurrence in a fixed effect model.
LRIG1 expression and HPV status
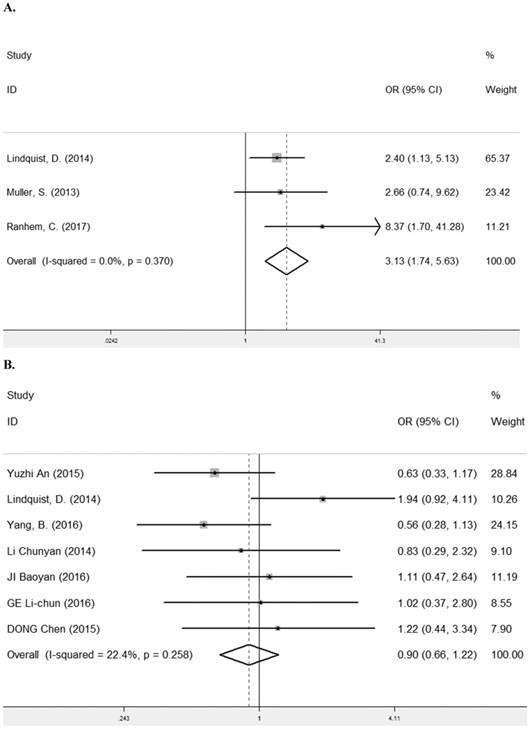
HPV infection is a known risk factor for the carcinomas of female genital tract and the etiological factor of oropharyngeal cancer [30-32]. Three studies [11, 12, 22] containing 434 cancer patients were assessed for the correlation between LRIG1 expression and HPV status (Table 2). The pooled OR was 3.31 (95% CI =1.74-5.63) (z=3.81, p< 0.001) (Figure 9A), calculating by a fixed model (Figure 9A). Due to the majority of cases in three studies were female, we also investigated the relationship between LRIG1 expression and gender in seven studies [7, 8, 11, 13, 25, 27, 28] to eliminate the influence of the gender for results. A fixed-effect model was selected according to low heterogeneity (I2=22.4%, p=0.258) (Figure 9B) among studies. The pooled OR was 0.90 (95% CI =0.66-1.22) (z=0.69, p< 0.488) (Figure 9B). These results indicated that there was no association between positive LRIG1 expression and gender. Besides, positive LRIG1 expression was significantly related with the positive HPV status.
Sensitivity analysis and publication bias
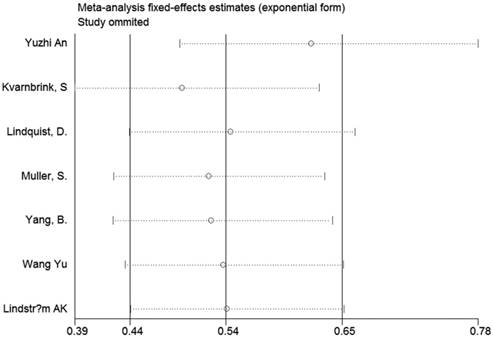
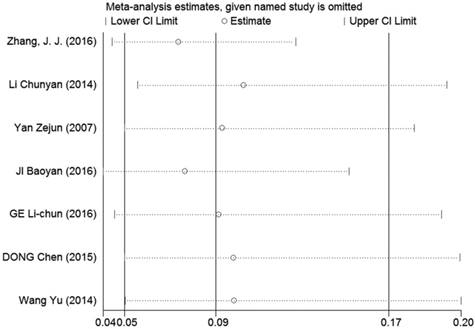
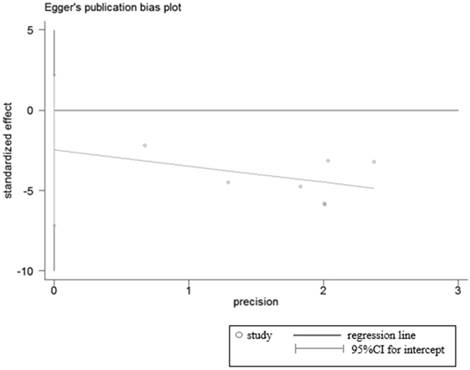
Sensitivity analyses were performed to determine the robustness of summarized HR (Figure 10) and overall OR (Figure 11). As a result, each individual study had no significantly influence on the pooled analysis. Simultaneously, the Egger's test was used to evaluate the publication bias. The result showed that no publication bias existed in this meta-analysis (Figure 12, Figure 13).
A. Forest plot of OR for the relationship between LRIG1 expression and HPV status in a fixed effect model. B. Forest plot of OR for the relationship between LRIG1 expression and gender in a fixed effect model.
Sensitivity analysis of the effect of the individual study on the pooled HRs.
Sensitivity analysis of the effect of the individual study on the pooled OR for the relationship between LRIG1 expression in cancer tissues and normal tissues.
Egger's test for publication bias in OS analysis.
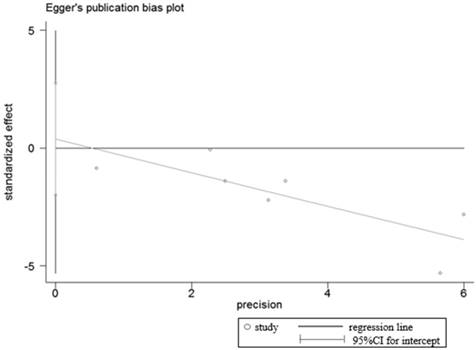
Egger's test for publication bias in studies investigated the relationship between LRIG1 expression in cancer tissues and normal tissues.
Discussion
Cancer is one of the prevalent disease with poor prognosis and there is no effective method to predict and prevent the process of tumorigenesis. Detection of biomarkers is a convenient technique for early diagnosis and prediction of cancer. Thus, new strategies for exploring novel biomarkers have attracted great attention worldwide. LRIG1 is an independent prognosis factor and predictive marker of clinicopathology in solid tumors, which has been proved by accumulating evidence. However, there is no consistent results on the effect of LRIG1 in malignant tumors. To resolve the contradiction. This meta-analysis was performed to estimate the prognostic value of LRIG1 expression and its relationship with clinicopathological significance in various cancers.
In this meta-analysis, high LRIG1 expression was significantly associated with longer overall survival, which was consistent with the conclusion of subgroup analysis. These results suggested that LRIG1 was a good prognostic maker in malignant tumors. Meanwhile, LRIG1 expression was obvious lower in cancer tissues than normal tissues and the same result was found with no heterogeneity in subgroup analysis based on type of tumor. High LRIG1 expression was also related with positive HPV status and tumor progression assessed by its significant association with smaller tumor size, early tumor stage, well degree of differentiation and negative recurrence, indicating the predictive value of LRIG1 in clinical characteristics. However, there was no association between positive LRIG1 expression and lymphatic metastasis. In addition, we conducted meta-regression and subgroup analysis, founding that tumor type and research area may explain the source of heterogeneity among the results from a collection of studies.
Though these useful outcomes were found, there were limitations in our meta-analysis. Firstly, some sample size of enrolled studies was small, which were prone to selection bias. Secondly, several survival data were extracted from the K-M survival curves rather than individual subject data, which could lead to overestimation of the prognostic effect. Finally, statistical heterogeneity existed among studies. However, meta-regression and subgroup analysis were conducted to account the source of heterogeneity.
The present meta-analysis demonstrated the prognostic value of LRIG1 in malignant tumors. Furthermore, recent studies have shown that LRIG1 participates in growth factor receptor signaling through inhibition of oncogenic receptor tyrosine kinases, EGFR, MET and RET to suppress the development of cancer [33-35]. LRIG1 also increases sensitivity of tumor cells to cisplatin, docetaxel and vinorelbine via suppression of EGFR signaling [14]. Estrogen receptor alpha (ERα) up-regulates LRIG1 through direct transcription [36]. In addition, animal experiments have demonstrated that the soluble extracellular part of LRIG1 inhibits glioma growth by interstitial delivery in experimental glioma models and this negative effect of soluble extracellular part of LRIG1 in vivo is largely independent of EGFR status. Based on cell encapsulation technology for in situ delivery and latent anti-tumor effect of soluble extracellular part of LRIG1, it may be a meaningful treatment for glioblastoma in the future [37].
In conclusion, LRIG1 was a potential biomarker for prognosis of tumor and predictive factor in clinical significance, and the anti-tumor effect of LRIG1 may be a potent development direction in oncotherapy.
Abbreviations
OS: overall survival; HR: hazard ratio; U: univariate analysis; LRIG: Leucine-rich repeats and immunoglobulin-like domains protein 1; OR: odds ratio; EGF: epidermal growth factor; EGFR: epidermal growth factor receptor; CI: confidence interval; ERα: estrogen receptor alpha.
Acknowledgements
This work was supported by Chinese National Science Foundation (No. 81670051 and No. 81330002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017Jan;67(1):7-30
2. Rodrigues D, Longatto-Filho A, Martins SF. Predictive Biomarkers in Colorectal Cancer: From the Single Therapeutic Target to a Plethora of Options. BioMed Res Int. 2016;2016:1-12
3. Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R. et al. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun. 2001Jun29;284(5):1155-61
4. Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. 2002;41(4):352-4
5. Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J. et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004Aug18;23(16):3270-81
6. Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK. et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012Mar30;149(1):146-58
7. JI B. Clinical significance of LRIG1 and Fascin-1 expression in patients with non-small cell lung cancer. Hainan Med J. 2016;27(23):3825-7
8. An Y, Zhao Z, Ou P, Wang G. Expression of LRIG1 is Associated With Good Prognosis for Human Non-small Cell Lung Cancer. Medicine (Baltimore). 2015Nov;94(47):e2081
9. Zhang J, Wang X, Zhang Y, Wu J, Zhou N. Leucine-rich repeats and immunoglobulin-like domains protein 1 and fascin actin-bundling protein 1 expression in nonsmall cell lung cancer. J Cancer Res Ther. 2016;12(8):248
10. Kvarnbrink S, Karlsson T, Edlund K, Botling J, Lindquist D, Jirström K. et al. LRIG1 is a prognostic biomarker in non-small cell lung cancer. Acta Oncol. 2015Sep14;54(8):1113-9
11. Lindquist D, Näsman A, Tarján M, Henriksson R, Tot T, Dalianis T. et al. Expression of LRIG1 is associated with good prognosis and human papillomavirus status in oropharyngeal cancer. Br J Cancer. 2014Apr1;110(7):1793-800
12. Muller S, Lindquist D, Kanter L, Flores-Staino C, Henriksson R, Hedman H. et al. Expression of LRIG1 and LRIG3 correlates with human papillomavirus status and patient survival in cervical adenocarcinoma. Int J Oncol. 2013Jan;42(1):247-52
13. Yang B, Dai C, Tan R, Zhang B, Meng X, Ye J. et al. Lrig1 is a positive prognostic marker in hepatocellular carcinoma. OncoTargets Ther. 2016 Nov;Volume 9:7071-9
14. Wu X, Hedman H, Bergqvist M, Bergström S, Henriksson R, Gullbo J. et al. Expression of EGFR and LRIG proteins in oesophageal carcinoma with emphasis on patient survival and cellular chemosensitivity. Acta Oncol. 2012Jan;51(1):69-76
15. Yi W, Haapasalo H, Holmlund C, Järvelä S, Raheem O, Bergenheim AT. et al. Expression of leucine-rich repeats and immunoglobulin-like domains (LRIG) proteins in human ependymoma relates to tumor location, WHO grade, and patient age. Clin Neuropathol. 2009Feb;28(1):21-7
16. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg Lond Engl. 2010;8(5):336-41
17. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998Dec30;17(24):2815-34
18. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007Jun7;8:16
19. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010Sep;25(9):603-5
20. Higgins JPT. Measuring inconsistency in meta-analyses. BMJ. 2003Sep6;327(7414):557-60
21. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997Sep13;315(7109):629-34
22. Ranhem C, Lillsunde Larsson G, Hedman H, Lindquist D, Karlsson MG, Hellström A-C. et al. Expression of LRIG proteins as possible prognostic factors in primary vaginal carcinoma. PLOS ONE. 2017Aug25;12(8):e0183816
23. LindströM AK, Ekman K, Stendahl U, Tot T, Henriksson R, Hedman H. et al. LRIG1 and squamous epithelial uterine cervical cancer: correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer. 2008Mar;18(2):312-7
24. Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases. Dermatol Surg. 2005;31(4):423-30
25. Li C. Down-expression and clinnical significance of lrig1 in colon cancer. Mod Oncol. 2014;22(05):1120-3
26. Yan Z. The expressions and clinical significance of LRIG1 and EGFR in bladder transitional cell carcinoma. J Clin Urol. 2007;22(3):216-8
27. GE L. Expression and significance of Lrig1 in colon cancer tissue and cell lines. Chin J Public Health. 2016;32(10):1003-9996
28. DONG C. Expressions of Lrig1 and EGFR in gastric cancer tissue. Chinese Journal of Public Health. 2015;31(06):819-22
29. Wang Y. Expression of leucine·-rich and immunoglobulin-like domains 1 and epidermal growth factor receptor and their significance in cervical adenocarcinoma. Cancer Res Clin. 2014Dec;26(12):843-6
30. Nowakowski AM, Kotarski J. [Risk factors of cervical cancer and possibilities of primary prevention]. Przegl Epidemiol. 2011;65(1):81-8
31. Chao A, Chen T-C, Hsueh C, Huang C-C, Yang J-E, Hsueh S. et al. Human papillomavirus in vaginal intraepithelial neoplasia. Int J Cancer. 2012Aug1;131(3):E259-268
32. Pytynia KB, Dahlstrom KR, Sturgis EM. Epidemiology of HPV-associated oropharyngeal cancer. Oral Oncol. 2014May;50(5):380-6
33. Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL. et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004Nov5;279(45):47050-6
34. Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL. et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007Mar;27(5):1934-46
35. Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008Jan2;28(1):39-49
36. Krig SR, Frietze S, Simion C, Miller JK, Fry WHD, Rafidi H. et al. Lrig1 is an estrogen-regulated growth suppressor and correlates with longer relapse-free survival in ERα-positive breast cancer. Mol Cancer Res. 2011Oct;9(10):1406-17
37. Johansson M, Oudin A, Tiemann K, Bernard A, Golebiewska A, Keunen O. et al. The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status. Neuro-Oncol. 2013Sep;15(9):1200-11
Author contact
![]() Corresponding author: Dr. Manxiang Li, Department of Respiratory and Critical Care Medicine, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277, West Yanta Road, Xi'an, Shaanxi 710061, China. Telephone: +86-029-85324053; Fax: +86-029-85324053; E-mail address: manxianglicom
Corresponding author: Dr. Manxiang Li, Department of Respiratory and Critical Care Medicine, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277, West Yanta Road, Xi'an, Shaanxi 710061, China. Telephone: +86-029-85324053; Fax: +86-029-85324053; E-mail address: manxianglicom

 Global reach, higher impact
Global reach, higher impact