Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(16):2953-2962. doi:10.7150/jca.25542 This issue Cite
Research Paper
The inhibitory effects of COL1A2 on colorectal cancer cell proliferation, migration, and invasion
1. Department of General Surgery, Shengjing Hospital of China Medical University, Shenyang, China
2. Department of Ear-nose-throat department, Shengjing Hospital of China Medical University, Shenyang, China
3. Department of Graduate School, China Medical University, Shenyang, China
Received 2018-2-13; Accepted 2018-6-23; Published 2018-7-30
Abstract
Purpose: Collagen type I alpha 2 chain (COL1A2) has been shown to participate in the development of various human malignancies. However, the role of COL1A2 in human colorectal cancer (CRC) remains unknown. This study investigated the expression pattern of COL1A2 in primary CRC tissues as well as the correlation of COL1A2 expression with clinicopathological features and prognosis of CRC. The function of COL1A2 in CRC cell proliferation, migration, and invasion as well as the possible mechanisms were also examined.
Methods: Real-time PCR and immunohistochemical analysis were performed to determine the expression of COL1A2 in primary cancer tissues and adjacent normal tissues from CRC patients. A COL1A2-expressing lentiviral vector was transfected into CRC cells, and cell counting kit-8 and Transwell assays were used to explore the effects of COL1A2 on CRC cell proliferation, migration, and invasion. Microarray-based mRNA expression profile screening was performed to reveal the possible signaling pathways involved in COL1A2-regulated cell behaviors.
Results: COL1A2 was significantly downregulated in primary CRC tissues. The mRNA levels of COL1A2 in CRC tissues were correlated with tumor differentiation, invasion, and lymph node metastasis. Overexpression of COL1A2 inhibited proliferation, migration, and invasion of CRC cell lines (SW480 and SW620). The microarray analysis showed that COL1A2 overexpression regulated numerous oncogenes and cancer-related signaling pathways. Among them, altered expression of ten representative cancer-related genes in these pathways were further confirmed by western blotting.
Conclusions: Our study identified COL1A2 as a novel tumor suppressor in CRC and provided a potential therapeutic approach to treat CRC.
Keywords: Collagen type I alpha 2 chain, colorectal cancer, proliferation, migration, invasion, microarray
Introduction
Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide, with over one million new cases and 700,000 deaths each year [1]. CRC progression is a multifactorial and multistage process. A variety of genes are significantly dysregulated in CRC carcinogenesis, which facilitates diagnosis, treatment, and prevention of CRC. Therefore, identifying important genes involved in CRC carcinogenesis and elucidating the underlying biological mechanisms will contribute to the development of more effective diagnostic and treatment options for this disease.
There are a variety of members in the collagen family, among which type I collagen is the most abundant. Type I collagen is a heterotrimeric protein consisting of two α1 chains (COL1A1) and one α2 chain (COL1A2). Type 1 collagen has distinctive biochemical and biomechanical properties, and accounts for the major fibrillary component of extracellular matrix. It is one of the proximal structures that tumor cells of epithelial origin must destroy to permit cell invasion and migration. Growing evidence has shown that COL1A2 is downregulated in many malignancies, including head and neck cancer, bladder cancer, and malignant melanoma [2-4]. However, in other cases such as hepatoma, ovarian cancer, and pancreatic cancer, COL1A2 is upregulated and may serve as a useful diagnostic and prognostic biomarker of these tumors [5-7]. The differential expression of COL1A2 may give rise to distinct collagen-mediated effects in human malignancies. Recently, researchers have found that the COL1A2 gene is hypermethylated and its mRNA expression is markedly decreased in CRC samples [8]. In contrast, upregulation of COL1A2 has also been reported in tumor tissues and blood of CRC patients compared with those of healthy individuals [9, 10]. However, the precise role of COL1A2 in CRC progression remains unclear.
In the present study, we examined the expression pattern of COL1A2 in clinical samples from patients with CRC, and investigated the effects of COL1A2 on CRC cell proliferation, migration, and invasion in vitro. Furthermore, mRNA expression profile screening results showed that COL1A2 overexpression modulated multiple signaling pathways, which are critically important for the development and progression of CRC, through the regulation of these signaling pathway-related genes. These novel findings may provide valuable information to explore the molecular mechanisms underlying COL1A2 function in CRC development, which could help develop effective therapeutic strategies to treat CRC.
Materials and Methods
Clinical tissue samples and cell culture
CRC tissues and paired adjacent normal tissues were obtained from 88 patients diagnosed with CRC in Shengjing Hospital of China Medical University, between 2014 and 2016. All CRC cases were pathologically confirmed. Patients did not receive chemotherapy or radiotherapy before surgery. The patients were followed up, and their complete clinical data were collected. Overall survival (OS) was defined as the interval between the dates of surgery and death. If disease recurrence was not diagnosed, patients were censored at the date of death or last follow-up. In addition, a total of 20 paraffin-embedded human colorectal specimens were collected after surgical resection from January 2014 to December 2014 at the Shengjing Hospital of China Medical University. This study was approved by the Ethical Committee of Shengjing Hospital of China Medical University. All patients signed a written informed consent form.
Human CRC cell lines (HT29, HCT116, SW480, SW620, Caco-2, and COLO-205) were purchased from the Cell Center of Shanghai Institutes for Biological Sciences (Shanghai, China) and were maintained in DMEM, RPMI 1640, or Ham's F12K medium (Hyclone, Logan, UT, USA) containing 10% FBS and 100 U/mL penicillin in a humidified atmosphere of 5% CO2 at 37°C.
RNA extraction and quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIzol (Takara, Dalian, China) following the manufacturer's instructions, and cDNA was synthesized using PrimeScript reverse transcription reagent (Takara). The mRNA levels of COL1A2 were determined using SYBR Premix Ex TaqTM II kit (Takara) according to the manufacturer's protocol. The primers used in this study are indicated in Table 1. Gene expression levels were normalized to the internal control, glyceraldehyde-3-phostphate dehydrogenase (GAPDH).
Primer details
| Name | Primer Sequence | Distribution | AT (ºC) | Product Size (bp) | ET (s) |
|---|---|---|---|---|---|
| COL1A2 | F:5'-GTGGCAGTGATGGAAGTGTG | NM_000089.31172 | 60.16 | 125 | 40 |
| R:5'-AGGACCAGCGTTACCAACAG | 60.17 | 40 | |||
| GAPDH | F:5'-GCACCGTCAAGGCTGAGAAC | 60 | 138 | 40 | |
| R:5'-TGGTGAAGACGCCAGTGGA | 60 | 40 |
Abbreviations: COL1A2: collagen type 1 alpha 2 chain; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; AT:annealing temperature; ET: extension time; bp: base pair.
Immunohistochemical (IHC) staining
The paraffin-embedded sections obtained from paired CRC tissues and non-tumor tissues from 20 CRC patients with different genders, ages, sizes of tumors, differentiation stages, invasion depths, and lymphatic metastasis types were deparaffinized in xylene and hydrated in a series of descending percentages of ethanol. Heated citrate buffer was used for antigen retrieval. The slides were blocked with 3% hydrogen peroxide and 10% goat serum, followed by incubation with an anti-COL1A2 antibody (14695-1-AP, Proteintech, Chicago, IL, USA) at 4°C overnight. After a sequential incubation with secondary antibody, streptavidin-horseradish peroxidase complex, and diaminobenzidine, the slides were counterstained with hematoxylin, dehydrated, and mounted. The staining was observed under a light microscope and imaged with an Olympus camera (Olympus, Japan). Three fields were randomly selected in each slide and observed under high-power magnification. The mean optical densities of the positive areas were measured using Image Pro Plus 6.0 (Media Cybernetics, USA). The results were expressed as the exact value of the relative optical density units.
Lentivirus production and transduction
The lentiviral vectors containing COL1A2 cDNA or a negative control (NC) sequence were purchased from Shanghai GeneChem Company (Shanghai, China). SW480 and SW620 cells were plated in 12-well plates and transduced with lentiviral vectors expressing COL1A2 or the NC sequence. The cells were observed under a fluorescence microscope (Olympus) at 72 h post-transduction and were collected to determine overexpression efficiency at 120 h post-transduction by Western blotting.
Western blot assay
The cells were harvested and lysed with lysis buffer supplemented with protease inhibitor cocktail (1 mM) and phenylmethylsulfonyl fluoride (1 mM) for 0.5 h at 4°C. Lysates were loaded in a polyacrylamide gel for electrophoresis and transferred to a PVDF membrane (Millipore, Bedford, MA, USA) using a BG-blotMiMi transfer system (Baygene, Beijing, China). The membrane was then blocked in 5% skim milk for 1 h followed by overnight incubation with primary antibodies against COL1A2 (Lifespan, Seattle, WA, USA), epidermal growth factor receptor (EGFR), baculoviral IAP Repeat Containing 3 (BIRC3), TNF alpha induced protein 3 (TNFAIP3), insulin receptor substrate 1 (IRS1), ETS proto-oncogene 1 (ETS1), insulin like growth factor 1 receptor (IGF1R), C-X-C motif chemokine lgand 1 (CXCL1) (Abcam, Cambridge, UK), cyclin dependent kinase 6 (CDK6), cyclin D1 (CCND1), snail family transcriptional repressor 2 (SNAI2) (Cell Signaling Technology, Danvers, MA, USA) or GAPDH (Santa Cruz, Dallas, TX, USA) at 4°C. Following an additional incubation with secondary antibodies at room temperature for 1 h, the protein bands were detected using an electrochemiluminescence detection kit (Thermol Biotech, Rockford, IL, USA). GAPDH was used as an internal control.
Cell counting kit-8 (CCK8) assay
SW480 and SW620 Cells (2 × 103) were seeded in each well of a 96-well plate, and cell proliferation was evaluated using the CCK8 assay following the manufacturer's instructions. Briefly, the cells were incubated with CCK8 solution (10 µL) in each well for 3 h at 37°C. The optical density (OD) was measured at 450 nm using a microplate reader. The results were representative of at least three independent experiments.
Transwell assay
For the migration assay, serum-starved SW480 and SW620 cells (5 × 104) were added to the upper chamber of a Transwell insert (3422; Corning COSTAR, Corning, NY, USA). For the invasion assay, cells (8 × 104) were added to the upper chamber of a Matrigel (BD, USA)-coated insert. Media containing 10 % FBS was added to the lower chamber. Cells were allowed to migrate or invade for 24 h. Cells that had not migrated or invaded and remained on the top of the membrane were removed with a cotton swab. Cells that had migrated or invaded to the bottom of the membrane were fixed with methanol and stained with crystal violet (Solarbio, Beijing, China). The stained cells were counted in five randomly selected fields (magnification 200×) using a light microscope. The results were representative of at least three independent experiments.
Microarray assay
Total RNA was isolated from SW480 cells transduced with lentiviral vectors expressing COL1A2 or NC using Trizol. RNA was quantified and assessed using a NanoDrop 2000 (Thermo, USA) and 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). High-quality RNA samples were used for the microarray assay. Reverse transcription, hybridization, labeling, and staining were conducted using a GeneChip 3′ IVT expression kit and a GeneChip hybridization wash and stain kit (Affymetrix, Santa Clara, CA, USA). The GeneChips were then scanned using Gene Array Scanner 3000 to generate raw data. The gene expression data were log2-transformed and median-centered for further analyses. The differentially expressed genes (DEGs) were screened using a Limma package (Smyth, 2004) in R language. The genes with a fold change (FC) > 3 and P value < 0.05 were considered differentially expressed.
Ingenuity pathway analysis (IPA)
All DEGs were analyzed using IPA software (Ingenuity Systems, Mountain View, CA, USA), and their involvement in various canonical signaling pathways, cellular processes, molecular regulatory networks, and disease development was assessed.
Statistical analysis
Data were analyzed using GraphPad Prism software V5.0 (Graphpad, San Diego, CA, USA) or SPSS 22.0 software (Chicago, IL, USA). Survival analysis was performed using the Kaplan-Meier analysis. A log rank test was used to compare different survival curves. The association between the clinicopathological characteristics of patients and the expression of COL1A2 or OS rates was analyzed by univariate or multivariate Cox regression models. Statistical significance was assessed using Student's t-test or an analysis of variance when appropriate. A P value < 0.05 was considered statistically significant.
Correlation between COL1A2 expression and clinicopathological characteristics in CRC. A. Real-time qPCR analysis of COL1A2 mRNA levels in CRC tissues and adjacent noncancerous tissues. B. CRC tissues with various degrees of tumor differentiation. C. CRC tissues with different invasion depths. D. CRC tissues with or without lymphatic metastasis. Abbreviations: COL1A2, collagen type 1 alpha 2 chain; CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction.
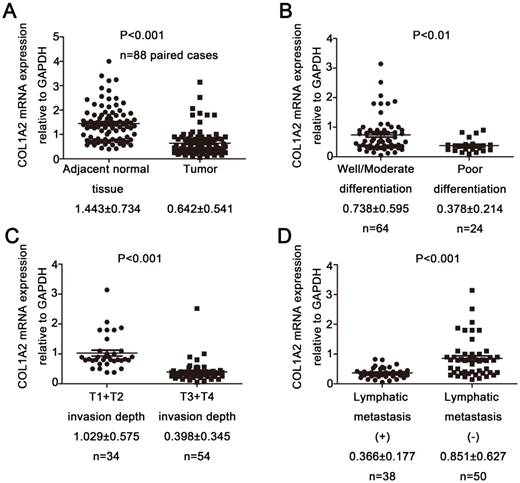
Results
COL1A2 was downregulated and correlated with clinicopathological characteristics in CRC
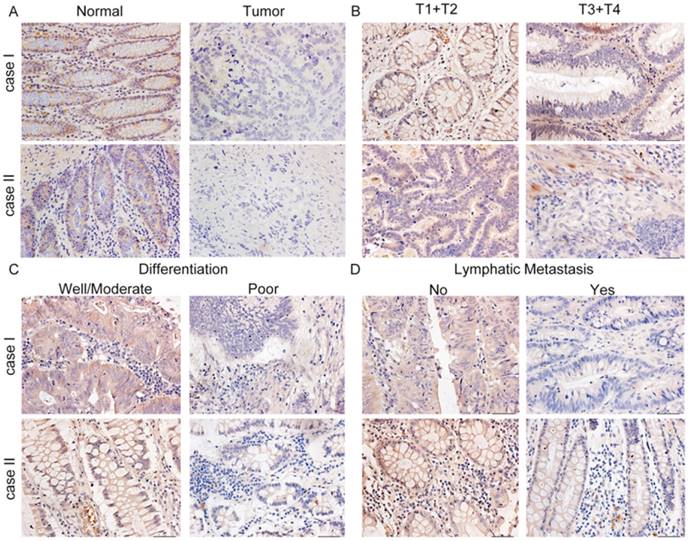
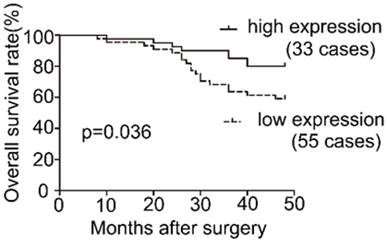
To examine the expression pattern of COL1A2 in CRC, qPCR was performed. The mRNA level of COL1A2 was found to be significantly lower in CRC tissues compared with that in adjacent non-tumor tissues (0.642 ± 0.541 vs. 1.443 ± 0.734, P < 0.001, Fig. 1A). Similar results were also observed between CRC tissues with poor differentiation and well/moderate differentiation (0.378 ± 0.214 vs. 0.738 ± 0.595, P < 0.01, Fig. 1B), T3+T4 and T1+T2 invasion depths (0.398 ± 0.345 vs. 1.029 ± 0.575, P < 0.001, Fig 1C), as well as patients with and without lymphatic metastasis (0.366 ± 0.177 vs. 0.851 ± 0.627, P < 0.001, Fig 1D). The correlation between the clinicopathological characteristics and COL1A2 mRNA levels in CRC patients was shown in Table 2. These results suggest that mRNA expression of COL1A2 may be inversely correlated with CRC development and progression. We next investigated whether protein expression of COL1A2 was correlated with these clinicopathological characteristics. As shown in Fig. 2A-2D, the IHC staining of COL1A2 in CRC tissues, CRC tissues with poor differentiation, greater tumor invasion depth, and lymphatic metastases was significantly weaker compared to adjacent normal tissues (0.084 ± 0.024 vs. 0.132 ± 0.030, P < 0.001), CRC tissues with well/moderate differentiation (0.071 ± 0.027 vs. 0.089 ± 0.022, P = 0.001), less invasion depth (0.076 ± 0.020 vs. 0.095 ± 0.026, P = 0.015), and without lymphatic metastases (0.070 ± 0.027 vs. 0.090 ± 0.020, P = 0.016), respectively. These data collectively suggest that COL1A2 may have an adverse effect on CRC development, serving as a potential biomarker of CRC severity. To further evaluate the correlation between COL1A2 expression and overall survival (OS) in patients with CRC, we performed a Kaplan-Meier analysis. Eighty-eight patients were available for follow-up at 6-48 months. As shown in Fig. 3, CRC patients with high COL1A2 expression had a significantly higher OS rate than those with low COL1A2 expression (P = 0.036). The univariate Cox proportional hazards analysis showed that COL1A2 expression (P = 0.034), lymphatic invasion (P = 0.000), or differentiation (P = 0.003, Table 3) was strongly associated with OS rates. The multivariate Cox analysis also demonstrated that COL1A2 expression (P = 0.025) and lymphatic invasion (P = 0.027) were independent prognostic factors for OS rates (Table 3). Taken together, these data suggest a critical role of COL1A2 in CRC development.
Representative immunohistochemical staining for COL1A2 expression in CRC tissues. A. Strong staining of COL1A2 in adjacent noncancerous tissues (×400). B. CRC tissues of T1+T2 and T3+T4 with relatively weak COL1A2 staining (×400). C. Weak staining of COL1A2 in CRC tissues with well/moderate and poor differentiation (×400). D. Weak staining of COL1A2 in CRC tissues without lymphatic metastasis and with lymphatic metastasis (×400). Abbreviations: COL1A2, collagen type 1 alpha 2 chain; CRC, colorectal cancer.
Correlation between the clinicopathological characteristics and COL1A2 mRNA levels in CRC patients
| COL1A2 mRNA expression relative to GAPDH | P value | ||
|---|---|---|---|
| Adjacent normal tissue (n) | 88 | 1.443 ± 0.734 | <0.01 |
| Tumor samples (n) | 88 | 0.642 ± 0.541 | |
| Smoking | |||
| Yes | 40 | 0.614 ± 0.503 | 0.801 |
| No | 48 | 0.644 ± 0.591 | |
| Alcohol drinking | |||
| Yes | 32 | 0.652 ± 0.471 | 0.854 |
| No | 56 | 0.629 ± 0.587 | |
| Gender | |||
| Male | 50 | 0.653 ± 0.596 | 0.828 |
| Female | 38 | 0.627 ± 0.468 | |
| Age(years) | |||
| ≤65 | 36 | 0.705 ± 0.474 | 0.334 |
| >65 | 52 | 0.590 ± 0.588 | |
| Tumor size (cm) | |||
| <5 | 43 | 0.751 ± 0.649 | 0.053 |
| ≥5 | 45 | 0.527 ± 0.374 | |
| Differentiation | |||
| Well/Moderate | 64 | 0.738 ± 0.595 | <0.01 |
| Poor | 24 | 0.378 ± 0.214 | |
| Invasion depth | |||
| T1+T2 | 34 | 1.029 ± 0.575 | <0.01 |
| T3+T4 | 54 | 0.398 ± 0.345 | |
| Lymphatic metastasis | |||
| Yes | 38 | 0.366 ± 0.177 | <0.01 |
| No | 50 | 0.851 ± 0.627 |
Values are presented as mean ± standard deviation. Abbreviations: COL1A2: collagen type 1 alpha 2 chain; CRC: colorectal cancer; GAPDH: glyceraldehyde-3-phosphate dehydrogenase
Survival analysis of COL1A2 expression in patients with CRC.
Univariate and multivariate analyses of individual parameters for correlation with OS rate
| Variable | Classification | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | ||
| Gender | Male/Female | 0.725 | 0.336-1.565 | 0.413 | |||
| Age | >65/≤65 | 0.659 | 0.305-1.421 | 0.287 | |||
| Tumor size | ≥5/<5 cm | 2.097 | 0.934-4.706 | 0.073 | |||
| Invasion depth | T1+T2/ T3+T4 | 1.578 | 0.686-3.630 | 0.283 | |||
| Lymphatic metastasis | Yes/No | 5.890 | 2.312-15.004 | 0.000* | 3.136 | 1.140-8.623 | 0.027* |
| Differentiation | Well-Moderate/Poor | 6.776 | 1.880-24.419 | 0.003* | 3.248 | 0.979-10.771 | 0.054 |
| COL1A2 | Low/High | 2.904 | 0.987-4.978 | 0.034* | 3.274 | 0.988-8.851 | 0.025* |
HR: hazard ratio; CI :confdence interval* P < 0.05
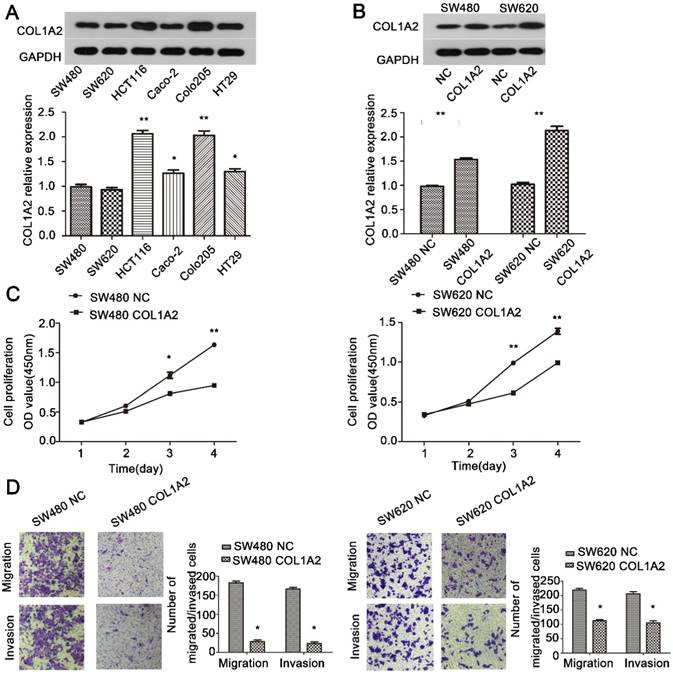
The effects of COL1A2 overexpression on CRC cell behaviors. A. Western blotting for COL1A2 protein expression in CRC cell lines. B. Western blotting for COL1A2 protein expression in COL1A2-overexpressing SW480 and SW620 cells. C. Effects of COL1A2 overexpression on SW480 and SW620 cell proliferation. D. Effects of COL1A2 overexpression on migration and invasion of SW480 and SW620 cells. Data were expressed as mean ± standard deviation. *P < 0.05, **P < 0.01, compared to control cells (n = 3). Abbreviations: COL1A2, collagen type 1 alpha 2 chain; CRC, colorectal cancer.
Overexpression of COL1A2 inhibits CRC cell proliferation, migration, and invasion
To further explore the role of COL1A2 in CRC carcinogenesis, we detected the expression of COL1A2 in SW480, SW620, HCT116, Caco-2, COLO-205, and HT29 cell lines by western blotting. As shown in Fig. 4A, SW480 and SW620 cells showed relatively low expression of COL1A2, and thus were selected to generate stable cell lines with COL1A2 overexpression. We transduced these two cell lines with lentiviral vectors expressing COL1A2, and confirmed that COL1A2 was dramatically upregulated in both cell lines, compared with the cells transduced with lentiviral vectors containing the NC sequence (Fig. 4B). Importantly, COL1A2-overexpressing cells exhibited a much lower proliferation rate compared to control cells (Fig. 4C), indicating that COL1A2 plays a suppressive role in CRC cell growth. Consistent with this finding, COL1A2 overexpression significantly inhibited the migratory and invasive capacities of both SW480 and SW620 cells (Fig. 4D). Taken together, these data suggest a protective role of COL1A2 in CRC growth and metastasis.
Microarray and IPA results on the effects of COL1A2 overexpression in SW480 cells. A. Heat map representation of genes between COL1A2-overexpressing and control SW480 cells. In the microarray heatmap, red and green represent upregulated and downregulated genes, respectively, compared with control SW480 cells. B. The signaling pathways of COL1A2 overexpression-related diseases were shown in descending order by inverse log of the P-value in IPA. C. The COL1A2 overexpression-regulated signaling pathways were shown in descending order by inverse log of the P-value, based on IPA databases. Orange represents activated pathways and blue represents inhibited pathways. D. Western blotting for the protein expression of the downstream target genes of COL1A2. The consistency of protein and mRNA expression of each gene were compared and summarized in Table 4. *P < 0.05, compared to the negative control group (n = 3). Abbreviations: COL1A2, collagen type 1 alpha 2 chain; CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction; IPA, ingenuity pathway analysis.
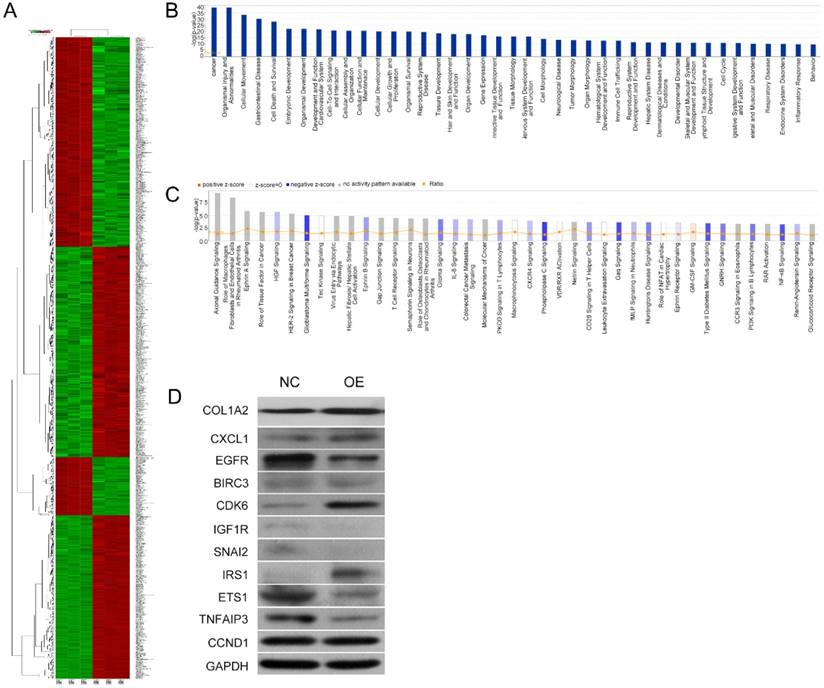
COL1A2 overexpression regulated critical signaling pathways and downstream target genes
To further investigate the underlying mechanisms for COL1A2-mediated CRC development, a cDNA microarray assay was performed to analyze the whole transcriptome in COL1A2-overexpressing and control SW480 cells. The results revealed that 1592 genes were differentially expressed (P < 0.05 and absolute fold-change [FC Absolute] > 3). Among these genes, 927 were downregulated, whereas 665 were upregulated (Fig. 5A). In addition, IPA biological function analysis showed that COL1A2 overexpression mediated various cellular behaviors and processes such as malignancy, gastrointestinal disease, cell movement, cellular organization and assembly, cell-cell interaction and signaling, maintenance of cellular homeostasis, cellular growth, proliferation and death, and organismal survival (Fig. 5B). IPA analysis also demonstrated that COL1A2 regulated a large panel of signaling pathways involved in cancer development and progression, including hepatocyte growth factor (HGF) signaling, glioblastoma multiforme signaling, ephrin B signaling, interleukin 8 (IL-8) signaling, phosphoinositide 3-kinase (PI3K) signaling, and nuclear factor-kappa B (NF-κB) signaling (Fig. 5C). Moreover, we identified many downstream target genes of COL1A2. Among them, expression of CXCL1, EGFR, BIRC3, CDK6, IGF1R, SNAI2, ETS1, TNFAIP3, and CCND1 was significantly downregulated by COL1A2, while IRS1 was significantly upregulated in COL1A2 expressing SW480 cells (Fig. 5D). Consistent results were observed in western blot analyses regarding protein expression of EGFR, BIRC3, IGF1R, SNAI2, ETS1, TNFAIP3 and IRS1, but not of CXCL1, CDK6, and CCND1. The comparison between western blot and qPCR results has been summarized in Table 4. These data suggest that COL1A2 may be a master regulator in CRC development, and regulation of these target genes by COL1A2 may provide valuable clues for a better understanding of the roles of COL1A2 in CRC and the development of novel therapeutic approaches against CRC.
Comparison of qPCR and western blot result of downstream target molecules of COL1A2
| Molecules | Change (Genechip) | Change (WB) | Consistency |
|---|---|---|---|
| CXCL1 | down | up | N |
| EGFR | down | down | Y |
| BIRC3 | down | down | Y |
| CDK6 | down | up | N |
| IGF1R | down | down | Y |
| SNAI2 | down | down | Y |
| IRS1 | up | up | Y |
| ETS1 | down | down | Y |
| TNFAIP3 | down | down | Y |
| CCND1 | down | up | N |
Y, Yes; N, No
Abbreviations: COL1A2: collagen type 1 alpha 2 chain; CXCL1:chemokine (C-X-C motif) ligand 1;EGFR: epidermal growth factor receptor;BIRC3: baculoviral IAP repeat containing 3;CDK6: cyclin dependent kinase 6;IGF1R: insulin like growth factor 1 receptor;SNAI2: snail family transcriptional repressor 2;IRS1: insulin receptor substrate 1;ETS1: ETS proto-oncogene 1;TNFAIP3: TNF alpha induced protein 3;CCND1: cyclin D1
Discussion
Despite the advances in CRC research, it is still a major cause of morbidity and mortality worldwide. A previous study revealed a role for high-density collagen in promoting fast and persistent cell migration, enabling cells to switch to a highly invasive motility behavior in solid human cancers. 3D collagen characterized by short fibers and small pores has been identified as an inducer of cancer transdifferentiation associated with a vasculogenic mimicry-like phenotype [11]. COL1A2 has been found to play a prognostic role in various cancers [12, 13]. However, it is not clear whether COL1A2 has a role in CRC progression. In this study, we found that COL1A2 expression was significantly weaker in primary CRC tissues compared to adjacent normal tissues, which is consistent with previous findings [8, 13]. Furthermore, COL1A2 mRNA expression in CRC tissues was positively correlated with tumor differentiation, invasion depth, and lymph node metastasis. These results suggest that COL1A2 may be a potential biomarker for distinguishing CRC tissues from non-malignant tissues.
We next investigated the possible role of COL1A2 in colorectal carcinogenesis and progression in vitro, and found that COL1A2 overexpression inhibited CRC cell proliferation, migration, and invasion. These findings suggest an anticarcinogenic role of COL1A2 in CRC development, which is consistent with studies highlighting the tumor-suppressive role of COL1A2 in certain malignancies [14-16]. However, some studies have demonstrated that COL1A2 can promote the development of other malignancies [17-20]. The distinct roles of COL1A2 in various cancers may be partially attributable to specific genetic features of the different malignancies.
To reveal the underlying mechanisms of COL1A2-regulated CRC development and progression, we analyzed the microarray data and identified numerous COL1A2-regulated genes. These genes are closely associated with transcriptional regulation and a series of cellular processes in cancer-like cell movement and communication. A panel of key signaling pathways involved in cancer progression in response to COL1A2 overexpression were also identified, including protein kinase A signaling, HGF signaling, glioblastoma multiforme signaling, ephrin B signaling, IL-8 signaling, and NF-κB signaling. These pathways actively participate in CRC development.
In addition, the microarray data revealed that 10 genes (CXCL1, EGFR, BIRC3, CDK6, IGF1R, SNAI2, ETS1, TNFAIP3, CCND1 and IRS1) were significantly upregulated or downregulated by COL1A2. The expression pattern of most of these genes was further validated by Western blot analysis. Among them, CXCL1 encodes a member of the CXC subfamily of chemokines that serves as a chemoattractant for neutrophils and plays a critical role in inflammation. CXCL1 has been found to be upregulated in CRC, bladder cancer, and gastric cancer, and is associated with oncogenesis and malignancy [21-23]. EGFR is a transmembrane receptor. The interaction of EGFR with epidermal growth factor induces EGFR dimerization and autophosphorylation, which promotes cell proliferation. EGFR mutations are correlated with the development of breast and lung cancers [24, 25]. Of note, EGFR is increasingly recognized as a biomarker of drug resistance in tumors. BIRC3 encodes a protein that inhibits serum deprivation-induced apoptosis, but does not have an effect on apoptosis resulting from exposure to menadione, an inducer of free radicals. BIRC3 has been identified as a mediator of therapeutic resistance and a prognostic marker in cancer cells [26, 27]. CDK6 is a catalytic subunit of CMGC serine/threonine protein kinase, and regulates G1 phase progression and G1/S transition. Up-regulated CDK6 activity is associated with the development of several types of malignancies [28-30]. CDK6 mutation may result in reduced cell proliferation, as well as impaired cell motility and polarity. IGF1R is overexpressed in most cancer tissues and acts as an antiapoptotic factor by promoting cell survival. It is also an indicator of poor prognosis in breast cancer [31, 32]. SNAI2 is involved in the epithelial-mesenchymal transition pathway important during tumorigenesis, and functions by downregulating E-cadherin expression [33]. SNAI2 is regarded as a cancer promoting factor in breast cancer [34]. ETS1 contributes to stem cell development, cell senescence, death, and tumorigenesis, and also promotes the proliferation, migration, and invasion of tumor cells [35]. High ETS1 expression is correlated with poor response to chemotherapy, and targeting ETS1 appears to be a promising approach to reverse drug resistance of cancer cells during chemotherapy [36]. TNFAIP3 is responsible for cytokine-mediated immune and inflammatory responses by inhibiting both NF-κB signaling and tumor necrosis factor-mediated apoptosis. The downregulation of TNFAIP3 is correlated with distant metastasis and poor patient prognosis in many types of cancer [37, 38]. CCND1, a member of the cyclin family, is a regulatory component of CDK4 and CDK6. CCND1 contributes to tumorigenesis through crosstalk with the retinoblastoma tumor suppressor protein and alteration of cell cycle phases. Mutation and overexpression of CCND1 are frequently observed in various cancer types [39-41]. IRS-1 is a substrate of the insulin receptor tyrosine kinase, and its high expression is correlated with low tumor cell invasiveness in some cancer types. IRS-1 triggers cell aggregation and inhibits cancer cell invasiveness [42]. IRS-1-negative cancer cells have been found to exhibit high metastatic potential in prostate cancer [43]. Consistently, breast cancers with low levels of IRS-1 are related to well-differentiated and reduced metastatic phenotypes [44].
In this study, we demonstrated that COL1A2 overexpression was able to downregulate the expression of many genes, such as EGFR, BIRC3, IGF1R, SNAI2, ETS1, and TNFAIP3 in SW480 cells. On the other hand, IRS-1, CCND, CXCL1, and CDK6 were all upregulated by COL1A2 overexpression. These data demonstrate that overexpression of COL1A2 can inhibit cellular growth and metastasis in SW480 cells via a molecular network of these proteins. The molecules regulated by COL1A2 are also associated with cellular behaviors and signaling pathways in CRC, which may be a key reason of high recurrence and poor prognosis in CRC patients with low expression of COL1A2. Our study also indicated that the protein expression patterns of CCND1, CXCL1, and CDK6 were not consistent with their mRNA levels, which may be due to post-translational modifications of these proteins.
Collectively, our study demonstrated for the first time that overexpression of COL1A2 suppressed the proliferation, migration, and invasion abilities of CRC cells by regulating multiple cancer-associated pathways, which indicates that COL1A2 could be used as a potential biomarker for the prognosis of CRC. Additionally, by microarray analysis we identified the differentially expressed genes downstream of COL1A2. Interactions between COL1A2 and these molecules require further investigation.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of China Medical University affliated Shengjing Hospital. This article does not contain any studies with animals performed by any of the authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stewart BW WC. International Agency for Research on Cancer. In: (ed.) edition. World Cancer Report 2014. Lyon, France: World Health Organization. 2014
2. Misawa K, Kanazawa T, Misawa Y, Imai A, Endo S, Hakamada K. et al. Hypermethylation of collagen alpha2 (I) gene (COL1A2) is an independent predictor of survival in head and neck cancer. Cancer biomarkers: section A of Disease markers. 2011;10:135-44
3. Enokida. CpG hypermethylation of collagen type I α 2 contributes to proliferation and migration activity of human bladder cancer. International Journal of Oncology. 2009:34
4. Bonazzi VF, Nancarrow DJ, Stark MS, Moser RJ, Boyle GM, Aoude LG. et al. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PloS one. 2011;6:e26121
5. Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX. et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. Journal of hepatology. 2010;52:690-7
6. Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432-40
7. Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH(2)-terminal kinase 1 and up-regulating N-cadherin expression. Cancer research. 2006;66:11745-53
8. Kalmar A, Peterfia B, Hollosi P, Galamb O, Spisak S, Wichmann B. et al. DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1, PTGDR and SFRP1 in colorectal adenoma and cancer. BMC cancer. 2015;15:736
9. Zou X, Feng B, Dong T, Yan G, Tan B, Shen H. et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. Journal of proteomics. 2013;94:473-85
10. Rodia MT, Ugolini G, Mattei G, Montroni I, Zattoni D, Ghignone F. et al. Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget. 2016;7:30295-306
11. Velez DO, Tsui B. et al. 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat Commun. 2017;8:1651
12. Misawa K, Mochizuki D, Imai A. et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget. 2016;7:26087-98
13. Sengupta PK, Smith EM, Kim K, Murnane MJ, Smith BD. DNA hypermethylation near the transcription start site of collagen alpha2(I) gene occurs in both cancer cell lines and primary colorectal cancers. Cancer research. 2003;63:1789-97
14. Lauvrak SU, Munthe E, Kresse SH, Stratford EW, Namlos HM, Meza-Zepeda LA. et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. British journal of cancer. 2013;109:2228-36
15. Misawa K, Mochizuki D, Imai A, Endo S, Mima M, Misawa Y. et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget. 2016;7:26087-98
16. Mori K, Enokida H, Kagara I, Kawakami K, Chiyomaru T, Tatarano S. et al. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int J Oncol. 2009;34:1593-602
17. Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World journal of surgical oncology. 2016:14
18. Li S, Li H, Xu Y, Lv X. Identification of candidate biomarkers for epithelial ovarian cancer metastasis using microarray data. Oncology letters. 2017;14:3967-74
19. Shi W, Zhang Z, Yang B, Guo H, Jing L, Liu T. et al. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Medicine. 2017;96:e7764
20. Zhuo C, Li X, Zhuang H, Tian S, Cui H, Jiang R. et al. Elevated THBS2, COL1A2, and SPP1 Expression Levels as Predictors of Gastric Cancer Prognosis. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;40:1316-24
21. Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer research. 2017;77:3655-65
22. Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S. et al. CXCL1-Mediated Interaction of Cancer Cells with Tumor-Associated Macrophages and Cancer-Associated Fibroblasts Promotes Tumor Progression in Human Bladder Cancer. Neoplasia (New York, NY). 2016;18:636-46
23. Wang L, Zhang C, Xu J, Wu H, Peng J, Cai S. et al. CXCL1 gene silencing inhibits HGC803 cell migration and invasion and acts as an independent prognostic factor for poor survival in gastric cancer. Molecular medicine reports. 2016;14:4673-9
24. Tufman A, Kahnert K, Duell T, Kauffmann-Guerrero D, Milger K, Schneider C. et al. Frequency and clinical relevance of EGFR mutations and EML4-ALK translocations in octogenarians with non-small cell lung cancer. OncoTargets and therapy. 2017;10:5179-86
25. Gonzalez-Conchas GA, Rodriguez-Romo L, Hernandez-Barajas D, Gonzalez-Guerrero JF, Rodriguez-Fernandez IA, Verdines-Perez A. et al. Epidermal growth factor receptor overexpression and outcomes in early breast cancer: A systematic review and a meta-analysis. Cancer treatment reviews. 2017;62:1-8
26. Mendoza-Rodriguez M, Arevalo Romero H, Fuentes-Panana EM, Ayala-Sumuano JT, Meza I. IL-1beta induces up-regulation of BIRC3, a gene involved in chemoresistance to doxorubicin in breast cancer cells. Cancer letters. 2017;390:39-44
27. Wang D, Berglund AE, Kenchappa RS, MacAulay RJ, Mule JJ, Etame AB. BIRC3 is a biomarker of mesenchymal habitat of glioblastoma, and a mediator of survival adaptation in hypoxia-driven glioblastoma habitats. Scientific reports. 2017;7:9350
28. Wang G, Zheng L, Yu Z, Liao G, Lu L, Xu R. et al. Increased cyclin-dependent kinase 6 expression in bladder cancer. Oncology letters. 2012;4:43-6
29. Whiteway SL, Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM. et al. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. Journal of neuro-oncology. 2013;111:113-21
30. Xiao F, Zhang W, Zhou L, Xie H, Xing C, Ding S. et al. microRNA-200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6. Oncology reports. 2013;30:2203-10
31. Gohr K, Hamacher A, Engelke LH, Kassack MU. Inhibition of PI3K/Akt/mTOR overcomes cisplatin resistance in the triple negative breast cancer cell line HCC38. BMC cancer. 2017;17:711
32. Walsh LA, Damjanovski S. IGF-1 increases invasive potential of MCF 7 breast cancer cells and induces activation of latent TGF-beta1 resulting in epithelial to mesenchymal transition. Cell communication and signaling: CCS. 2011;9:10
33. Atmaca A, Wirtz RW, Werner D, Steinmetz K, Claas S, Brueckl WM. et al. SNAI2/SLUG and estrogen receptor mRNA expression are inversely correlated and prognostic of patient outcome in metastatic non-small cell lung cancer. BMC cancer. 2015;15:300
34. de Herreros AG, Peiro S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. Journal of mammary gland biology and neoplasia. 2010;15:135-47
35. Sun Q, Jiang CW, Tan ZH, Hou LY, Dong H, Liu K. et al. MiR-222 promotes proliferation, migration and invasion of lung adenocarcinoma cells by targeting ETS1. European review for medical and pharmacological sciences. 2017;21:2385-91
36. Wu M, Liu X, Jin W, Li Y, Li Y, Hu Q. et al. Targeting ETS1 with RNAi-based supramolecular nanoassemblies for multidrug-resistant breast cancer therapy. Journal of controlled release: official journal of the Controlled Release Society. 2017;253:110-21
37. Hadisaputri YE, Miyazaki T, Yokobori T, Sohda M, Sakai M, Ozawa D. et al. TNFAIP3 overexpression is an independent factor for poor survival in esophageal squamous cell carcinoma. Int J Oncol. 2017;50:1002-10
38. Ghadban T, Schmidt-Yang M, Uzunoglu FG, Perez DR, El Gammal AT, Miro JT. et al. Evaluation of the germline single nucleotide polymorphism rs583522 in the TNFAIP3 gene as a prognostic marker in esophageal cancer. Cancer genetics. 2015;208:595-601
39. Shi R, Sun J, Sun Q, Zhang Q, Xia W, Dong G. et al. Upregulation of FAM83D promotes malignant phenotypes of lung adenocarcinoma by regulating cell cycle. American journal of cancer research. 2016;6:2587-98
40. Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M. et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. American journal of cancer research. 2017;7:1820-34
41. Zagouri F, Kotoula V, Kouvatseas G, Sotiropoulou M, Koletsa T, Gavressea T. et al. Protein expression patterns of cell cycle regulators in operable breast cancer. PloS one. 2017;12:e0180489
42. Reiss K, Wang JY, Romano G, Tu X, Peruzzi F, Baserga R. Mechanisms of regulation of cell adhesion and motility by insulin receptor substrate-1 in prostate cancer cells. Oncogene. 2001;20:490-500
43. Nagle JA, Ma Z, Byrne MA, White MF, Shaw LM. Involvement of insulin receptor substrate 2 in mammary tumor metastasis. Molecular and cellular biology. 2004;24:9726-35
44. Sisci D, Morelli C, Garofalo C, Romeo F, Morabito L, Casaburi F. et al. Expression of nuclear insulin receptor substrate 1 in breast cancer. Journal of clinical pathology. 2007;60:633-41
Author contact
![]() Corresponding author: Baolin Liu, Department of General Surgery, Shengjing Hospital of China Medical University, Shenyang, Liaoning Province 110004, P. R. China, Phone /Fax: 0086-18940259560, E-mail: liublorg
Corresponding author: Baolin Liu, Department of General Surgery, Shengjing Hospital of China Medical University, Shenyang, Liaoning Province 110004, P. R. China, Phone /Fax: 0086-18940259560, E-mail: liublorg

 Global reach, higher impact
Global reach, higher impact