Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(17):3138-3148. doi:10.7150/jca.25377 This issue Cite
Research Paper
Role of CXCR7 as a Common Predictor for Prognosis in Solid Tumors: a Meta-Analysis
1. Biotherapy Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China
2. Cancer Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China
3. Imaging Department, the Third Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China
4. School of Life Sciences, Zhengzhou University, Zhengzhou 450052, Henan, China
5. Henan Key Laboratory for Tumor Immunology and Biotherapy, Zhengzhou, Henan 450052, P.R. China
* These authors have contributed equally to this work
Received 2018-2-5; Accepted 2018-7-7; Published 2018-8-6
Abstract
Background: Accumulating evidence indicated that the CXC chemokine receptor (CXCR) 7 (CXCR7) was overexpressed in a variety of tumors. However, the value of the CXCR7 expression in predicting prognosis in solid tumors remains controversial. Therefore, we performed this meta-analysis to evaluate the correlation between CXCR7 expression and lymph node metastasis (LNM), tumor pathological grade and survival, including overall survival (OS), disease-free survival (DFS) and recurrence-free survival (RFS).
Methods: Eligible studies were searched in PubMed, Web of Science, and PMC up to April 2018. A total of 27 studies were included in this meta-analysis. Odds ratio (OR), hazard ratio (HR) and 95 % confidence intervals (CI) were used as effect measures.
Results: The meta-analysis showed that high expression of CXCR7 predicted a high risk of LNM (pooled OR = 2.22, 95%CI: 1.41-3.50), high tumor grade (pooled OR = 1.94, 95%CI: 1.20-3.13), poor OS (pooled HR = 1.66, 95%CI: 1.30-2.03), and poor DFS/RFS (pooled HR = 1.82, 95%CI: 1.21-2.43). Subgroup analysis showed that CXCR7 expression had a positive correlation with LNM in pan-adenocarcinoma subgroup (pooled OR = 3.73, 95%CI: 2.21-6.30), while no correlation was found in pan-squamous cancer subgroup (pooled OR = 1.29, 95%CI: 0.56-2.96). Subgroup analysis of tumor grade revealed that high expression of CXCR7 predicted high tumor grade both in pan-squamous cancer and pan-adenocarcinoma (pooled OR = 3.58, 95%CI: 1.39-9.22, pooled OR = 2.25, 95%CI: 1.20-4.20). As in OS group, we divided the data based on analysis method and it turned out that overexpressed CXCR7 predicted worse OS both in multivariate analysis (pooled HR =1.57, 95%CI: 1.12-2.01) and univariate analysis subgroup (pooled HR =1.86, 95%CI: 1.23-2.49).
Conclusions: Our meta-analysis revealed that high expression of CXCR7 predicted unfavorable prognosis and may serve as potential targets of cancer therapy.
Keywords: CXCR7, lymph node metastasis, tumor grade, survival, solid tumors, meta-analysis
Introduction
Cancer is becoming a severe health public problem for its increasing incidence and mortality whether in developing or developed countries. In 2012, about 14.1 million new cancer cases and 8.2 million deaths occurred worldwide [1]. Most cancers have a poor survival rate because of late diagnosis, metastasis and drug resistance [2, 3]. Therefore, it is essential to find a common and valuable molecular marker to predict prognosis in tumor patients and provides a therapeutic target for the treatment of cancer patients.
It is well known that CXCR4 and its ligand CXCL12 play a significant role in tumorigenesis and metastasis of various tumors [4]. Similarly, high expression of CXCR7 has been confirmed in various types of tumors as well and is involved in tumorigenesis, metastasis, and drug resistance [5-7]. CXCR7 has been identified as a novel receptor for CXCL12 with higher affinity of binding CXCL12 than CXCR4 in recent years [8]. Both CXCR4 and CXCR7 are seven-span transmembrane G-protein coupled receptors, while CXCR7 does not only play typical G-protein signaling pathways but also activate MAP kinases through beta-arrestins in transiently transfected cells [9-11]. Through bioinformatics analysis, our previous studies revealed that CXCR7 had a significantly increased expression in esophageal cancer than normal tissue [12]. Moreover, we previously confirmed that IL-6 derived from cancer associated fibroblast promoted chemoresistance via CXCR7 in esophageal squamous cell carcinoma [13]. Although most studies suggested that CXCR7 served as a positive regulator in tumor development, there were still some opposite points. Our study revealed that there was no relationship between CXCR7 expression and lymph node metastasis in esophageal squamous cell carcinoma [13]. Guo et al found there was no correlation between CXCR7 expression and survival outcomes [14]. Furthermore, Uto-Konomi et al revealed that overexpression of CXCR7 attenuated CXCR4-CXCL12 mediated response and made anti-tumor effect [15].
Based on these controversial findings, we performed this meta-analysis to evaluate the role of CXCR7 in predicting lymph node metastasis, tumor grade and survival outcomes in patients with solid tumors. Our results revealed that high expression of CXCR7 predicted a high risk of lymph node metastasis, high tumor grade and unfavorable outcomes of OS and DFS/RFS, indicating that CXCR7 can serve as a common prognostic indicator in solid tumors.
Methods
Search strategy
Two independent authors systematically searched PubMed, Web of Science, PMC up to April 2018. The following keywords were used in the search strategy: (“CXCR7” OR “chemokine (C-X-C motif) receptor 7” OR “CXC chemokine receptor 7” OR “chemokine receptor CXCR7” OR “RDC1”) AND (“neoplasms” OR “tumors” OR “neoplasias” OR “cancers” OR “malignancy” OR “carcinoma”) AND (“prognosis” OR “prognostic” OR “outcome” OR “survival” OR “metastasis” OR “metastatic” OR “grade”). A manual search for the references of articles was also performed to find out other potential studies. Discrepancies were discussed with consensus.
Inclusion and exclusion criteria
Two reviewers (ZQT and QGH) independently selected and carried out eligible study based on following inclusion criteria: (1) full-text original published in English; (2) CXCR7 expression was detected by immunohistochemistry in patients with solid tumors without restrictions of the types of the tumor; (3) clinicopathologic parameters of lymph node metastasis and tumor pathological grade were described in the study; (4) the correlation between CXCR7expression and prognosis in patients were evaluated, such as overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS); (5) reported the hazard ratio (HR) value and their 95%CI or sufficient data to calculate the HR and 95%CI based on survival curves and figures given in the articles. The exclusion criteria were included: (1) articles about cell lines or animals; (2) no full text, case reports, letters, reviews, duplicate articles; (3) didn't report sufficient data to extract or calculate the HR and their 95%CI; (4) survival analysis performed by using public dataset.
Data extraction and quality assessment
Two reviewers (ZQT and ZPH) independently extracted data from eligible studies. All disagreement was discussed with consensus. The following data were extracted: first author, the year of publication, country, cancer type, sample size, number of patients with high and low CXCR7 expression, number of patients with lymph node metastasis, number of patients with tumor grade, detection method of CXCR7 expression levels, cut off values, fellow up time, survival outcome, analysis method, HR estimation. If univariate and multivariate analysis of HR and 95%CI were both reported, we extracted multivariate results. In the absence of multivariate results, HR was extracted from the univariate analysis or calculated using the Kaplan-Meier survival curves [16]. The quality of all included studies was assessed by the Newcastle-Ottawa Scale (NOS), and all included studies were high quality [17].
Statistical analysis
The software STATA 12.0 (STATA Corporation, College Station, TX, USA) was used to carry out all the statistical analysis. Survival outcomes were divided into two groups, OS and DFS/RFS according to data acquired from included studies. CXCR7 expression levels were extracted from studies at high and low level. We defined CXCR7 positive expression as high level and CXCR7 negative expression as low level. The extracted HRs and 95%CIs were performed to obtain pooled HR and 95%CI. If HR>1 and its 95%CI did not overlap with 1 indicated a high risk of lymph node metastasis, high tumor grade and poor survival prognosis in tumor patients. The heterogeneity among studies was checked using Cochran's Q test and Higgins's-squared statistic and we defined heterogeneity as I2> 50% or p < 0.05. When heterogeneity was absent between studies, we used a fixed-effects model to analyze the results, otherwise, a random-effects model was applied. Sensitivity analysis was performed to assess the stability of the results. Publication bias was analyzed by Begg's and Egger's tests. Two-tailed p<0.05 was considered statistically significant in our results.
Flow diagram of study selection process.
Meta-analysis results
Study characteristics
Initially, a total of 2230 articles were selected by using search strategy (Figure 1). After excluding duplicates, 1894 articles were selected. By further reviewing, 10 articles were excluded for non-English articles, 1140 articles were excluded for no-full text, 124 articles were excluded for non-human research and 585 articles were excluded due to no data of lymph node metastasis or tumor grade or survival analysis. 35 articles were assessed for eligibility. 6 articles were excluded because the detection method was not immunohistochemistry (IHC), 1 article was excluded for insufficient data for estimating of HR and 95%CI of survival rate and 1 article was excluded for survival analyzing by public database. Finally, 27 articles were included in this meta-analysis.
These studies were performed among 6 countries (China, Italy, Germany, Japan, The Netherlands, Austria) published from 2010 to 2017, the number of sample sizes ranged from 37 to 232. In these studies, Polimeno M et al stratified the data into validated cohort and training cohort, while the outcome date of two studies was presented independently, and these data were analyzed separately. Xue T et al stratified the data based on the combination of CXCR7 expression with low HNF4α expression and relative degree of differentiation of cancer and these data were also analyzed separately. Among 27 studies, three studies each evaluated colorectal carcinoma (or rectal cancer) [18-20], hepatocellular carcinoma [21-23], esophageal carcinoma (or esophageal squamous cell carcinoma) [13, 24, 25], and thyroid carcinoma [26-28]. Two studies evaluated renal cell carcinoma [29, 30], pancreatic cancer [31, 32], cervical cancer (or cervical squamous cell carcinoma) [33, 34], nasopharyngeal carcinoma [35, 36], glioma or glioblastoma [37, 38] and gastric cancer [39, 40]. One study evaluated gallbladder cancer [14], cutaneous squamous cell carcinoma [41] and chondrosarcoma [42]. In survival analysis, nine studies had available HR and 95%CI data, four studies presented K-M curves, which can estimate HR and 95%CI, one study with survival analysis data was provided by our lab. The main characteristics of 27 studies were shown in Table 1, Table 2 and Table 3.
Association of CXCR7 expression with LNM
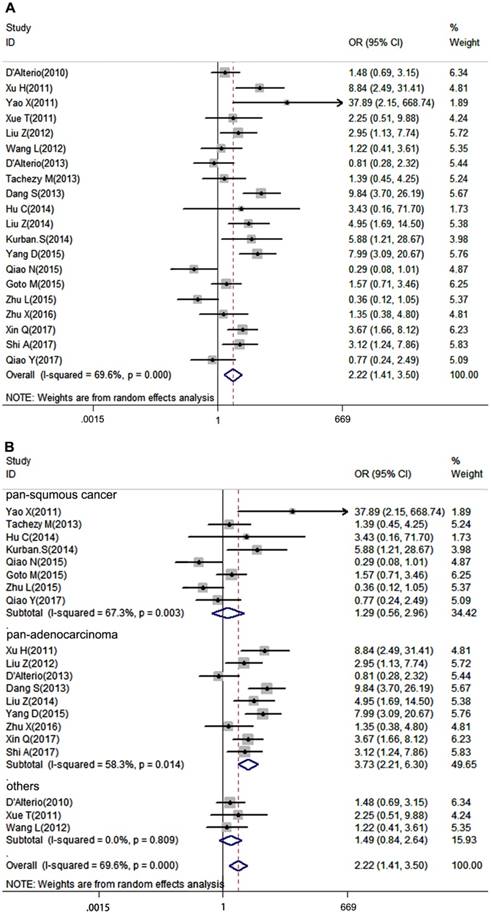
Lymph node metastasis is a very important clinicopathological parameter involved in tumor progression. A total of 20 studies provided LNM of patients based on different CXCR7 expression, a random-effects model was used to calculate the pooled OR due to obvious inter-study heterogeneity (I2=69.6%, P=0.000). The results revealed that high CXCR7 expression predicted a high risk of LNM in solid tumors (pooled OR = 2.22, 95%CI: 1.41-3.50, Figure 2A). Next, we performed a subgroup analysis based on the source of tumor origin. We divided the group into pan-squamous cancer and pan-adenocarcinoma subgroup. Pan-squamous cancer included esophageal squamous cell carcinoma, cutaneous squamous cell carcinoma, cervical squamous cell carcinoma, nasopharyngeal carcinoma, which mostly originate from squamous cells. Pan-adenocarcinoma included colorectal carcinoma, thyroid carcinoma, pancreatic adenocarcinoma, gastric cancer, which mostly originate from glandular epithelial cells. Notably, high expression of CXCR7 only predicted a high risk of LNM in pan-adenocarcinoma subgroup (pooled OR = 3.73, 95%CI: 2.21-6.30, Figure 2B). While, there was no correlation between CXCR7 expression and LNM in pan-squamous cancer (pooled OR = 1.29, 95%CI: 0.56-2.96, Figure 2B), suggesting that CXCR7 played a distinct role in predicting LNM in two types of tumors and it is more reliable that CXCR7 can predict LNM in adenocarcinoma. Meanwhile, more studies should investigate the molecular mechanisms contributing to these differences.
Characteristics of studies in the LNM group in this meta-analysis
| First author | Year | Country | Cancer type | Samples | CXCR7 Expression | ||||
|---|---|---|---|---|---|---|---|---|---|
| High expression | High with LNM | Low expression | Low with LNM | Method | |||||
| D'Alterio C | 2010 | Italy | RCC | 221 | 178 | 141 | 43 | 31 | IHC |
| Xu H | 2011 | China | CRC | 66 | 19 | 15 | 47 | 14 | IHC |
| Yao X | 2011 | China | GBC | 64 | 49 | 27 | 15 | 0 | IHC |
| Xue T | 2011 | China | HCC | 116 | 51 | 5 | 65 | 3 | IHC |
| Liu Z | 2012 | China | PTC | 79 | 52 | 33 | 27 | 10 | IHC |
| Wang L | 2012 | China | RCC | 97 | 71 | 57 | 26 | 20 | IHC |
| D'Alterio C | 2013 | Italy | CRC | 66 | 45 | 17 | 21 | 9 | IHC |
| Tachezy M | 2013 | Germany | EC | 52 | 26 | 11 | 26 | 9 | IHC |
| Dang S | 2013 | China | PTC | 135 | 33 | 27 | 102 | 32 | IHC |
| Hu C | 2014 | China | SCC | 40 | 28 | 3 | 12 | 0 | IHC |
| Liu Z | 2014 | China | PAC | 64 | 33 | 25 | 31 | 12 | IHC |
| Kurban S | 2014 | China | CSCC | 63 | 44 | 18 | 19 | 2 | IHC |
| Yang D | 2015 | China | CRC | 96 | 62 | 16 | 34 | 9 | IHC |
| Qiao N | 2015 | China | NPC | 72 | 38 | 26 | 34 | 30 | IHC |
| Goto M | 2015 | Japan | ESCC | 113 | 40 | 18 | 73 | 25 | IHC |
| Zhu L | 2015 | China | NPC | 62 | 38 | 10 | 24 | 12 | IHC |
| Zhu X | 2016 | China | PTC | 39 | 22 | 12 | 17 | 8 | IHC |
| Xin Q | 2017 | China | GC | 160 | 126 | 84 | 34 | 12 | IHC |
| Shi A | 2017 | China | GC | 83 | 52 | 36 | 31 | 13 | IHC |
| Qiao Y | 2017 | China | ESCC | 45 | 23 | 11 | 22 | 11 | IHC |
Abbreviations: RCC, renal cell carcinoma; CRC, colorectal carcinoma; GBC, gallbladder cancer; HCC, hepatocellular carcinoma; PTC, papillary thyroid carcinoma; EC, Esophageal carcinoma; SCC, cutaneous squamous cell carcinoma; PAC, pancreatic adenocarcinoma; CSCC, cervical squamous cell carcinoma; NPC, nasopharyngeal carcinoma; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer.
Characteristics of studies in tumor grade group in this meta-analysis
| First author | Year | Country | Cancer type | Samples | CXCR7 Expression | ||||
|---|---|---|---|---|---|---|---|---|---|
| High expression | High with high grade | Low expression | Low with high grade | Method | |||||
| D'Alterio C | 2010 | Italy | RCC | 221 | 178 | 42 | 43 | 11 | IHC |
| Xu H | 2011 | China | CRC | 66 | 19 | 7 | 47 | 3 | IHC |
| Yao X | 2011 | China | GBC | 72 | 49 | 21 | 23 | 5 | IHC |
| Xue T | 2011 | China | HCC | 116 | 51 | 17 | 65 | 26 | IHC |
| Wang L | 2012 | China | RCC | 97 | 72 | 25 | 25 | 5 | IHC |
| D'Alterio C | 2013 | Italy | CRC | 66 | 45 | 4 | 21 | 1 | IHC |
| Dang S | 2013 | China | PTC | 135 | 33 | 10 | 102 | 8 | IHC |
| Hu C | 2014 | China | SCC | 40 | 28 | 16 | 12 | 2 | IHC |
| Liu Z | 2014 | China | PAC | 64 | 33 | 8 | 31 | 6 | IHC |
| Yang D | 2015 | China | CRC | 96 | 62 | 10 | 34 | 2 | IHC |
| Polimeno M | 2015 | Italy | HCC | 77 | 69 | 19 | 8 | 3 | IHC |
| Li G | 2015 | China | CS | 60 | 49 | 32 | 11 | 0 | IHC |
| Xin Q | 2017 | China | GC | 120 | 99 | 34 | 21 | 8 | IHC |
| Shi A | 2017 | China | GC | 83 | 52 | 36 | 31 | 18 | IHC |
Abbreviations: CS, chondrosarcoma
Characteristics of studies in OS, DFS/RFS group in this meta-analysis.
| First author | Year | Country | Cancer type | Clinical stage | Samples | Cut off | Follow-up (month) | Method | Outcome | Analysis | HR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D'Alterio C | 2010 | Italy | RCC | I-IV | 170 | >20% | 3-180 | IHC | DFS | U/M | Reported |
| Yao X | 2011 | China | GBC | I-IV | 72 | Score≥2 | 3-120 | IHC | OS | M | Reported |
| Schrevel M | 2012 | The Netherlands | CC | NA | 101 | positive | 137 (5-266) | IHC | DFS | U | Reported |
| Wang L | 2012 | China | RCC | I-IV | 97 | ≥25% | 120 | IHC | OS/RFS | U/M | Reported |
| D'Alterio C | 2013 | Italy | CRC | I-III | 64 | >50% | 60 | IHC | RFS | U | Reported |
| Polimeno M (tc) | 2014 | Italy | HCC | I-IV | 80 | ≥60% | 126 | IHC | OS | U | Reported |
| Polimeno M (vc) | 2014 | Italy | HCC | I-IV | 39 | ≥60% | 87 | IHC | OS | U | Reported |
| Xue T (H) | 2014 | China | HCC | NA | 74 | High | 110 | IHC | OS/RFS | U | SC |
| Xue T (EH) | 2014 | China | HCC | NA | 48 | High | 110 | IHC | OS/RFS | U | SC |
| Birner P | 2015 | Austria | Glioma | NA | 37 | positive | 52 | IHC | OS | U | Reported |
| Li G | 2015 | China | CS | I-III | 60 | ≥10% | 60 | IHC | OS | U | SC |
| Yang D | 2015 | China | CRC | I-IV | 96 | Score≥2 | 60 | IHC | OS | M | Reported |
| Goto M | 2015 | Japan | ESCC | I-IV | 113 | ≥1+ | 94 | IHC | RFS | U | SC |
| Guo J | 2016 | China | PC | NA | 232 | Score4-9 | 2-87 | IHC | OS | M | SC |
| Deng L | 2017 | China | GBM | NA | 146 | High | 120 | IHC | OS | M | Reported |
| Qiao Y | 2017 | China | ESCC | I-IV | 45 | High | 45 | IHC | OS | U | SC |
Abbreviations: NSCLC, nonsmall cell lung cancer; CC cervical cancer; PC, pancreatic cancer; tc, training cohort; vc, validation cohort; H, low HNF4α expression; EH, Edmondson grade 1/2 and low HNF4α expression, U, univariate analysis; M, multivariate analysis; SC: survival
Forest plot diagrams of OR for the association between CXCR7 expression and LNM. (A) The correlation of CXCR7 expression with LNM in solid tumors. (B) Subgroup analysis of CXCR7 expression with LNM in pan-squamous cancer and pan-adenocarcinoma subgroup.
Association of CXCR7 expression with tumor pathological grade
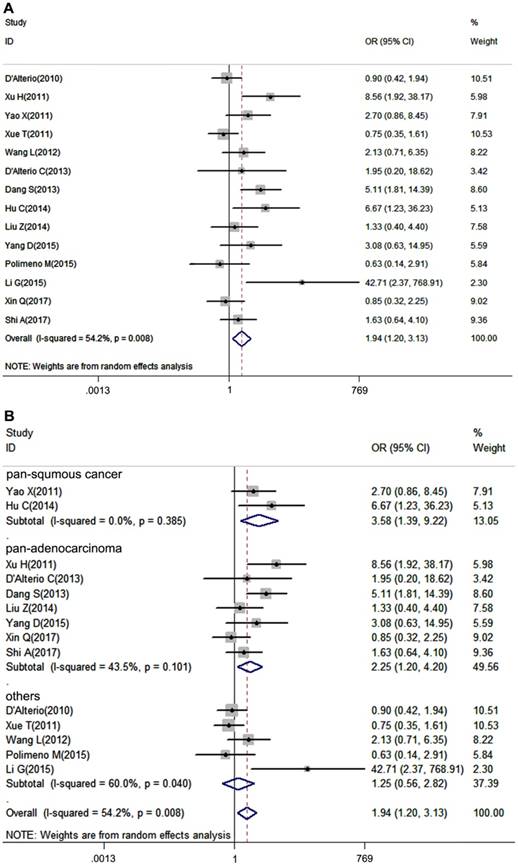
Tumor pathological grade is an essential index influencing prognosis of patients, thus we put the tumor grade in our analysis. 14 studies, which provided tumor grade information, were included in our meta-analysis. Due to the heterogeneity among studies was significant (I2=54.2%, P=0.008), a random-effects model was performed. It turned out that CXCR7 expression was positively correlated with high tumor grade (pooled OR = 1.94, 95%CI: 1.20-3.13). Additionally, subgroup analysis showed that overexpressed CXCR7 was correlated with high tumor grade both in pan-squamous cancer and pan-adenocarcinoma subgroup ((pooled OR = 3.58, 95%CI: 1.39-9.22, pooled OR = 2.25, 95%CI: 1.20-4.20, Figure 3B).
Association of CXCR7 expression with OS
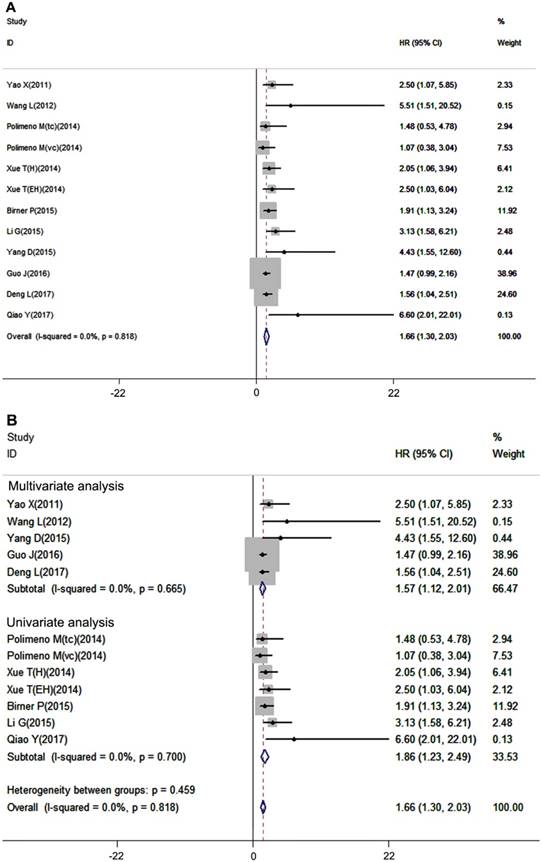
Overall survival is considered the most important prognostic outcome index. In our meta-analysis, overall survival analysis included 12 datasets with 1026 patients, and a fixed-effects model was used due to no inter-study heterogeneity (I2=0.0%, P=0.818). The results showed that high expression of CXCR7 was significantly associated with poor OS in patients with solid tumors (pooled HR =1.66, 95%CI: 1.30-2.03, Figure 4A). In the subgroup analysis, 5 studies used multivariate analysis and 7studies used univariate analysis. The results revealed that high expression of CXCR7 predicted worse survival both in multivariate analysis subgroup (pooled HR =1.57, 95%CI: 1.12-2.01) and univariate analysis subgroup (pooled HR =1.86, 95%CI: 1.23-2.49) (Figure 4B).
Forest plot diagrams of OR for the association between CXCR7 expression and tumor grade. (A) The correlation of CXCR7 expression with tumor grade in solid tumors. (B) Subgroup analysis of CXCR7 expression with tumor grade in pan-squamous cancer and pan-adenocarcinoma subgroup.
Forest plot diagrams of HR for the association between CXCR7 expression and OS (A) The correlation of CXCR7 expression with OS. (B) Subgroup analysis of the correlation between CXCR7 expression and OS in multivariate analysis and univariate analysis subgroup.
Association of CXCR7 expression with DFS/RFS
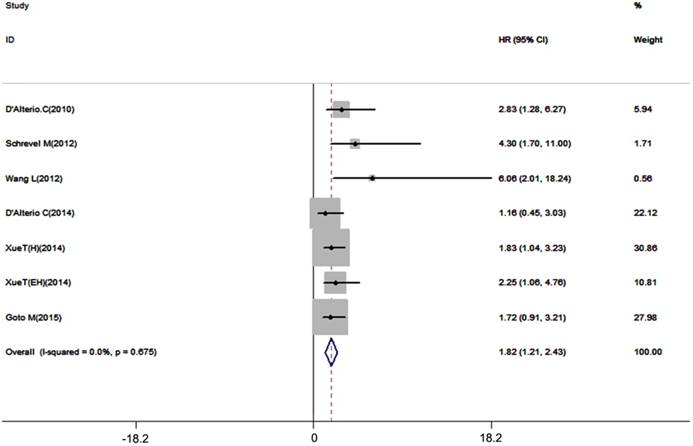
Disease-free survival and recurrence-free survival are also important prognostic outcome index. 7 datasets with 667 patients included in our meta-analysis, the results showed that high expression of CXCR7 had a potent correlation with unfavorable DFS/RFS (pooled HR = 1.82, 95%CI: 1.21-2.43, Figure 5). A fixed-effects model was used for no inter-study heterogeneity (I2=0.0%, p=0.675).
Sensitivity analysis
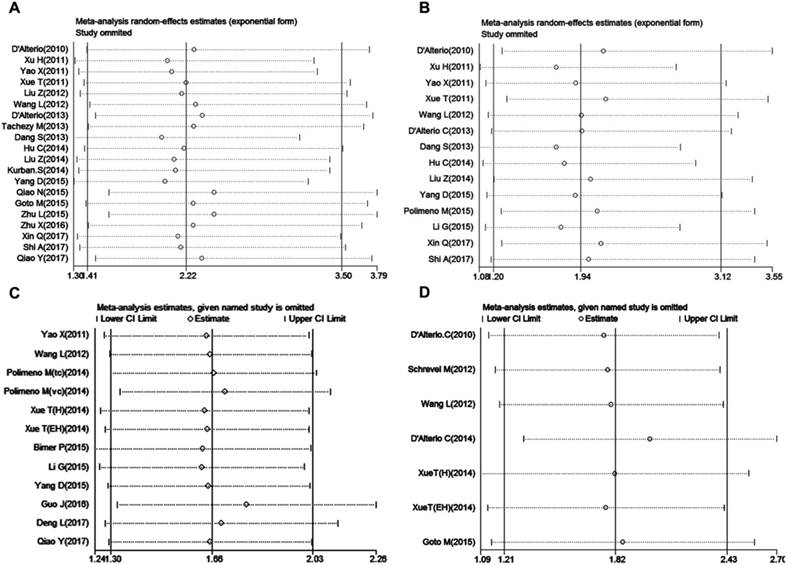
Sensitivity analysis was used to assess the stability and credibility of results by omitting one study sequentially at a time. The results showed that no individual study significantly influence the meta-analysis results in four groups (Figure 6 A-D), indicating that the results of this meta-analysis were stable and reliable.
Forest plot diagrams of HR for the association between CXCR7 expression and DFS/RFS.
Sensitivity analysis in this meta-analysis. (A) LNM group; (B) tumor grade group; (C) OS group and (D) DFS/RFS group.
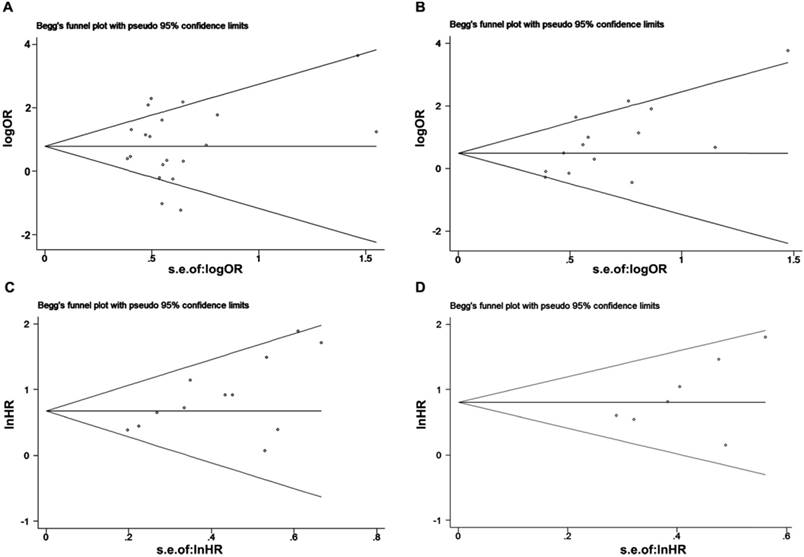
Begg's funnel plot for the evaluation of potential publication bias in this meta-analysis. (A) LNM group; (B) tumor grade group; (C) OS group and (D) DFS/RFS group.
Publication bias
We used Begg's and Egger's tests to evaluate publication bias of included studies. Both Begg's test and Egger's test revealed that no obvious publication bias in LNM analysis (Begg's test, P=0.57; Egger's test P=0.542, Figure 7A) and DFS/RFS (Begg's test, P=0.176; Egger's test, P=0.177 Figure 7D). While, there was statistically publication bias in the studies of tumor grade group (Begg's test, P=0.037; Egger's test, P=0.014, Figure 7B) and OS group (Begg's test, P=0.02; Egger's test, P=0.02, Figure 7C).
Discussion
CXCR4 has been considered as the only receptor of CXCL12 for a long time, but the theory was challenged by the fact that CXCR7 was identified as a novel receptor binding to CXCL12. CXCR7, formally known as RDC1, is a G-protein-coupled seven-transmembrane receptor. In fact, CXCR7 does not only mediate typical chemokine response, as chemotaxis, receptor-mediated calcium mobilization, activation of intracellular signaling cascades[11] but also mediates basic β-arrest signaling pathway. As a membrane-associated receptor, CXCR7 was present in all sorts of cell types, including dendritic cells, B cells, fibroblasts, activated endothelial cells, fetal liver cells, placenta, progenitor cells and tumor cells [43-47]. Similar to CXCR4, high expression of CXCR7 was also observed in various types of tumors, for insistence, renal cell carcinoma, pancreatic adenocarcinoma, cervical squamous cell carcinoma, papillary thyroid carcinoma, hepatocellular carcinoma and esophageal squamous cell carcinoma, etc. In most studies, CXCR7 was reported to promote tumor growth, metastasis, and drug resistance, leading to a poor prognosis in tumor patients. There were several signaling mechanisms involved in these processes, including aberrated expression of PI3K/AKT, MAPK/ERK, Rho/ROCK and mTOR signaling pathway [23, 32, 48]. However, the role of CXCR7 in tumor progression was challenged by some reports. Our studies revealed that CXCR7 expression had no correlation with LNM in patients with esophageal squamous cell carcinoma [13, 49]. In addition, CXCR7 may act as a negative regulator of CXCR4 and attenuates CXCR4-CXCL12- mediating tumor growth.
In our meta-analysis, 27 studies were included to evaluate the relationship between CXCR7 expression and LNM, tumor grade, OS and DFS/RFS. The approach of CXCR7 investigation was limited to IHC to improve the accuracy of the analysis. Furthermore, we performed sensitivity analyzes to evaluate the stability of the results. The results demonstrated that high expression of CXCR7 was positively correlated with LNM (pooled OR: 2.22, 95%CI: 1.41-3.50), high tumor grade (pooled OR = 1.94, 95%CI: 1.20-3.13) and poor survival outcomes, OS (pooled HR =1.69, 95%CI: 1.27-2.11) and DFS/RFS (pooled HR=1.82, 95%CI: 1.21-2.43). Pan-cancers have been reported to have similar molecular signatures and tumor biology, thus, we divided our studies based on tumor origin referred to as pan-adenocarcinoma subgroup and pan-squamous cancer subgroup, subgroup analyses were performed subsequently. Our findings revealed that high expression of CXCR7 predicted a high risk of LNM only in pan-adenocarcinoma subgroup, suggesting a different role of CXCR7 on mediating LNM in squamous tumor and adenocarcinoma. In addition, our results also revealed that it is more reliable to use CXCR7 to predict LNM in adenocarcinoma. Furthermore, overexpressed CXCR7 was positively correlated with tumor grade both in pan-adenocarcinoma and pan-squamous cancer subgroup. Notably, inconsistent results were found that the role of CXCR7 in predicting LNM and tumor grade in the pan-squamous subgroup. One reason may be due to a small number of studies (only 3 studies) in the pan-squamous subgroup in tumor grade group that decreased the accuracy of the results. Otherwise, CXCR7 may mediate intracellular signaling that promotes tumor cell differentiation, resisting to drug therapy. As described above, our lab proved that CXCR7 medicated drug resistance through IL-6/CXCR7/NF-κB axis, while, no correlation was found between CXCR7 and LNM in esophageal squamous cell carcinoma. Additionally, we also performed a subgroup analysis based on the analysis method in the OS group. It turned out that CXCR7 overexpression predicted a favorable outcome both in multivariate analysis subgroup and univariate analysis subgroup.
This is the first complete meta-analysis concerning on the role of CXCR7 in LNM, tumor grade and survival in various types of solid tumors. Although Qian Chen et al performed a meta-analysis of correlation between CXCR7 expression and clinicopathological features, the included studies were not enough as well as lacking survival data [50]. However, there were also some limitations should be acknowledged in our study. Firstly, the language of our included studies only limit to English, thus language bias may exist. Secondly, there were only 3 datasets in pan-squamous cancer subgroup in tumor grade group, 12 datasets in OS group, 7 datasets in DFS/RFS group and in some studies, the sample size was fairly small. Thirdly, some studies in this meta-analysis did not report HR and 95%CI, thus, these dates only can be extracted from survival curves, but these may be less reliable than directly report.
In conclusion, our meta-analysis suggested that high expression of CXCR7 could act as a common maker to predict a high risk of LNM, especially in adenocarcinoma, high tumor grade and poor OS or DFS/RFS in patients with the solid tumor. However, based on the limitations of this meta-analysis, large size and better design studies are needed to validate the clinical role of CXCR7.
Abbreviations
CXCR7: CXC chemokine receptor (CXCR) 7; LNM: lymph node metastasis; OS: overall survival; DFS: disease-free survival; RFS: recurrence-free survival; OR: odds ratio, HR: hazard ratios; RCC: renal cell carcinoma; CRC: colorectal carcinoma; GBC: gallbladder cancer; PTC: papillary thyroid carcinoma; HCC: hepatocellular carcinoma; EC: esophageal carcinoma; SCC: cutaneous squamous cell carcinoma; PAC: pancreatic adenocarcinoma; CSCC: cervical squamous cell carcinoma; BC: breast cancer; NPC: nasopharyngeal carcinoma; ESCC: esophageal squamous cell carcinoma; TC: thyroid cancer; CS: chondrosarcoma; NSCLC: non- small cell lung cancer; CC: cervical cancer; PC: pancreatic cancer; tc: training cohort; vc: validation cohort; H: low HNF4α expression; EH: Edmondson grade 1/2 and low HNF4α expression, U: univariate analysis; M: multivariate analysis; SC: survival curve.
Acknowledgements
This work was supported by National Natural Sciences Foundation of China (No. 81771781).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
4. Du Y, Long Q, Guan B, Mu L. Prognostic Value of High CXCR4 Expression in Renal Cell Carcinoma: A System Review and Meta-Analysis. Dis Markers. 2015;2015:568980
5. Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z. et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201-13
6. Zheng K, Li HY, Su XL, Wang XY, Tian T, Li F. et al. Chemokine receptor CXCR7 regulates the invasion, angiogenesis and tumor growth of human hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2010;29:31
7. Wang HX, Tao LY, Qi KE, Zhang HY, Feng D, Wei WJ. et al. Role of CXC chemokine receptor type 7 in carcinogenesis and lymph node metastasis of colon cancer. Mol Clin Oncol. 2015;3:1229-32
8. Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B. et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760-6
9. Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226-44
10. Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP. et al. Beta-arrestin- but not G protein-mediated signaling by the "decoy" receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628-32
11. Thelen M, Thelen S. CXCR7, CXCR4 and CXCL12: an eccentric trio? J Neuroimmunol. 2008;198:9-13
12. Yue Y, Song M, Qiao Y, Li P, Yuan Y, Lian J. et al. Gene function analysis and underlying mechanism of esophagus cancer based on microarray gene expression profiling. Oncotarget. 2017;8:105222-37
13. Qiao Y, Zhang C, Li A, Wang D, Luo Z, Ping Y. et al. IL6 derived from cancer-associated fibroblasts promotes chemoresistance via CXCR7 in esophageal squamous cell carcinoma. Oncogene. 2017
14. Yao X, Zhou L, Han S, Chen Y. High expression of CXCR4 and CXCR7 predicts poor survival in gallbladder cancer. J Int Med Res. 2011;39:1253-64
15. Uto-Konomi A, McKibben B, Wirtz J, Sato Y, Takano A, Nanki T. et al. CXCR7 agonists inhibit the function of CXCL12 by down-regulation of CXCR4. Biochem Biophys Res Commun. 2013;431:772-6
16. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815-34
17. Maxwell L, Santesso N, Tugwell PS, Wells GA, Judd M, Buchbinder R. Method guidelines for Cochrane Musculoskeletal Group systematic reviews. J Rheumatol. 2006;33:2304-11
18. Xu H, Wu Q, Dang S, Jin M, Xu J, Cheng Y. et al. Alteration of CXCR7 expression mediated by TLR4 promotes tumor cell proliferation and migration in human colorectal carcinoma. PLoS One. 2011;6:e27399
19. D'Alterio C, Avallone A, Tatangelo F, Delrio P, Pecori B, Cella L. et al. A prognostic model comprising pT stage, N status, and the chemokine receptors CXCR4 and CXCR7 powerfully predicts outcome in neoadjuvant resistant rectal cancer patients. Int J Cancer. 2014;135:379-90
20. Yang D, Dai T, Xue L, Liu X, Wu B, Geng J. et al. Expression of chemokine receptor CXCR7 in colorectal carcinoma and its prognostic significance. Int J Clin Exp Pathol. 2015;8:13051-8
21. Xue TC, Han D, Chen RX, Zou JH, Wang Y, Tang ZY. et al. High expression of CXCR7 combined with Alpha fetoprotein in hepatocellular carcinoma correlates with extra-hepatic metastasis to lung after hepatectomy. Asian Pac J Cancer Prev. 2011;12:657-63
22. Neve Polimeno M, Ierano C, D'Alterio C, Simona Losito N, Napolitano M, Portella L. et al. CXCR4 expression affects overall survival of HCC patients whereas CXCR7 expression does not. Cell Mol Immunol. 2015;12:474-82
23. Xue TC, Jia QA, Bu Y, Chen RX, Cui JF, Tang ZY. et al. CXCR7 correlates with the differentiation of hepatocellular carcinoma and suppresses HNF4alpha expression through the ERK pathway. Oncol Rep. 2014;32:2387-96
24. Tachezy M, Zander H, Gebauer F, von Loga K, Pantel K, Izbicki JR. et al. CXCR7 expression in esophageal cancer. J Transl Med. 2013;11:238
25. Goto M, Yoshida T, Yamamoto Y, Furukita Y, Inoue S, Fujiwara S. et al. CXCR4 Expression is Associated with Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Ann Surg Oncol. 2017;24:832-40
26. Liu Z, Sun DX, Teng XY, Xu WX, Meng XP, Wang BS. Expression of stromal cell-derived factor 1 and CXCR7 in papillary thyroid carcinoma. Endocr Pathol. 2012;23:247-53
27. Dang S, Peng Y, Ye L, Wang Y, Qian Z, Chen Y. et al. Stimulation of TLR4 by LMW-HA induces metastasis in human papillary thyroid carcinoma through CXCR7. Clin Dev Immunol. 2013;2013:712561
28. Zhu X, Bai Q, Lu Y, Lu Y, Zhu L, Zhou X. et al. Expression and function of CXCL12/CXCR4/CXCR7 in thyroid cancer. International journal of oncology. 2016;48:2321-9
29. D'Alterio C, Consales C, Polimeno M, Franco R, Cindolo L, Portella L. et al. Concomitant CXCR4 and CXCR7 expression predicts poor prognosis in renal cancer. Curr Cancer Drug Targets. 2010;10:772-81
30. Wang L, Chen W, Gao L, Yang Q, Liu B, Wu Z. et al. High expression of CXCR4, CXCR7 and SDF-1 predicts poor survival in renal cell carcinoma. World J Surg Oncol. 2012;10:212
31. Liu Z, Teng XY, Meng XP, Wang BS. Expression of stromal cell-derived factor 1 and CXCR7 ligand receptor system in pancreatic adenocarcinoma. World J Surg Oncol. 2014;12:348
32. Guo JC, Li J, Zhou L, Yang JY, Zhang ZG, Liang ZY. et al. CXCL12-CXCR7 axis contributes to the invasive phenotype of pancreatic cancer. Oncotarget. 2016;7:62006-18
33. Kurban S, Tursun M, Kurban G, Hasim A. Role of CXCR7 and effects on CXCL12 in SiHa cells and upregulation in cervical squamous cell carcinomas in Uighur women. Asian Pac J Cancer Prev. 2014;15:9211-6
34. Schrevel M, Karim R, ter Haar NT, van der Burg SH, Trimbos JB, Fleuren GJ. et al. CXCR7 expression is associated with disease-free and disease-specific survival in cervical cancer patients. Br J Cancer. 2012;106:1520-5
35. Qiao N, Wang L, Wang T, Li H. Inflammatory CXCL12-CXCR4/CXCR7 axis mediates G-protein signaling pathway to influence the invasion and migration of nasopharyngeal carcinoma cells. Tumour Biol. 2016;37:8169-79
36. Zhu L, Luo K, Gu XH, Hou N, Huang CP, Lou Q. et al. CXCR7 expression in nasopharyngeal carcinoma tissues correlates with disease severity. Int J Clin Exp Med. 2015;8:21257-61
37. Birner P, Tchorbanov A, Natchev S, Tuettenberg J, Guentchev M. The chemokine receptor CXCR7 influences prognosis in human glioma in an IDH1-dependent manner. J Clin Pathol. 2015;68:830-4
38. Deng L, Zheng W, Dong X, Liu J, Zhu C, Lu D. et al. Chemokine receptor CXCR7 is an independent prognostic biomarker in glioblastoma. Cancer Biomark. 2017;20:1-6
39. Xin Q, Zhang N, Yu HB, Zhang Q, Cui YF, Zhang CS. et al. CXCR7/CXCL12 axis is involved in lymph node and liver metastasis of gastric carcinoma. World J Gastroenterol. 2017;23:3053-65
40. Shi A, Shi H, Dong L, Xu S, Jia M, Guo X. et al. CXCR7 as a chemokine receptor for SDF-1 promotes gastric cancer progression via MAPK pathways. Scand J Gastroenterol. 2017;52:745-53
41. Hu SC, Yu HS, Yen FL, Chen GS, Lan CC. CXCR7 expression correlates with tumor depth in cutaneous squamous cell carcinoma skin lesions and promotes tumor cell survival through ERK activation. Exp Dermatol. 2014;23:902-8
42. Li G, Yang Y, Xu S, Ma L, He M, Zhang Z. Slug signaling is up-regulated by CCL21/CCR7 [corrected] to induce EMT in human chondrosarcoma. Med Oncol. 2015;32:478
43. Infantino S, Moepps B, Thelen M. Expression and regulation of the orphan receptor RDC1 and its putative ligand in human dendritic and B cells. J Immunol. 2006;176:2197-207
44. Raggo C, Ruhl R, McAllister S, Koon H, Dezube BJ, Fruh K. et al. Novel cellular genes essential for transformation of endothelial cells by Kaposi's sarcoma-associated herpesvirus. Cancer Res. 2005;65:5084-95
45. Mazzinghi B, Ronconi E, Lazzeri E, Sagrinati C, Ballerini L, Angelotti ML. et al. Essential but differential role for CXCR4 and CXCR7 in the therapeutic homing of human renal progenitor cells. J Exp Med. 2008;205:479-90
46. Circelli L, Sciammarella C, Guadagno E, Tafuto S, del Basso de Caro M, Botti G. et al. CXCR4/CXCL12/CXCR7 axis is functional in neuroendocrine tumors and signals on mTOR. Oncotarget. 2016;7:18865-75
47. Kremer D, Cui QL, Gottle P, Kuhlmann T, Hartung HP, Antel J. et al. CXCR7 Is Involved in Human Oligodendroglial Precursor Cell Maturation. PLoS One. 2016;11:e0146503
48. Zhang Y, Yang CQ, Gao Y, Wang C, Zhang CL, Zhou XH. Knockdown of CXCR7 inhibits proliferation and invasion of osteosarcoma cells through inhibition of the PI3K/Akt and beta-arrestin pathways. Oncol Rep. 2014;32:965-72
49. Bianco AM, Uno M, Oba-Shinjo SM, Clara CA, de Almeida Galatro TF, Rosemberg S. et al. CXCR7 and CXCR4 Expressions in Infiltrative Astrocytomas and Their Interactions with HIF1alpha Expression and IDH1 Mutation. Pathol Oncol Res. 2015;21:229-40
50. Cheng Q, Wen Y. Meta-analysis of CXCR7 Expression Related to Clinical Prognosis in Cancers. Journal of Integrative Oncology. 2016:05
Author contact
![]() Corresponding author: Yi Zhang, Biotherapy Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China. E-mail: yizhangedu.cn
Corresponding author: Yi Zhang, Biotherapy Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China. E-mail: yizhangedu.cn

 Global reach, higher impact
Global reach, higher impact