Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(19):3456-3466. doi:10.7150/jca.26112 This issue Cite
Research Paper
PI3K/Akt/HIF-1α signaling pathway mediates HPV-16 oncoprotein-induced expression of EMT-related transcription factors in non-small cell lung cancer cells
1. Institute of Biochemistry and Molecular Biology, Guangdong Medical University, Zhanjiang 524023, P.R. China
2. Collaborative innovation center for antitumor active substance research and development, Guangdong Medical University, Zhanjiang 524023, P.R. China
3. Dongguan Key Laboratory of Medical Bioactive Molecular Developmental and Translational Research, Guangdong Medical University, Dongguan 523808, P.R. China
4. Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics, Guangdong Medical University, Dongguan 523808, P.R. China.
* These authors contribute equally to this work.
Received 2018-3-16; Accepted 2018-8-3; Published 2018-9-8
Abstract
Background: Our previous studies have demonstrated that human papillomaviruse (HPV)-16 oncoproteins promoted epithelial-mesenchymal transition (EMT), leading to non-small cell lung cancer (NSCLC) progression, but the underlying molecular mechanisms still remain unclear. PI3K/Akt/HIF-1α signaling pathway has been reported to mediate hypoxia-induced EMT. In this study, we further explored the role of PI3K/Akt/HIF-1α signaling pathway in HPV-16 oncoprotein-induced EMT in NSCLC cells.
Methods: A549 and NCI-H460 NSCLC cells were transiently transfected with pEGFP-HPV-16 E6 or E7 constructs. Western blotting and RT-qPCR were respectively performed to determine the protein and mRNA expression of EMT-related transcription factors. HPV-16 E6 or E7-transfected NSCLC cells were co-transfected with specific HIF-1α-siRNA or pretreated with different concentrations of LY294002, a specific PI3K inhibitor, followed by the analysis of expression of EMT-related transcription factors. The correlation between HIF-1α and EMT-related transcription factors in NSCLC tissues was analyzed by immunohistochemical staining and Spearman rank correlation coefficient.
Results: HPV-16 E6 and E7 oncoproteins upregulated the expression of Slug and Twist1, the EMT-related transcription factors, at both protein and mRNA levels in A549 and NCI-H460 cells. The co-transfection with specific HIF-1α-siRNA, but not the non-specific (NS)-siRNA, significantly abrogated HPV-16 oncoprotein-induced upregulation of ZEB1, Snail1, Slug, and Twist1 at both protein and mRNA levels. Additionally, pretreatment with LY294002 obviously blocked HPV-16 E6- and E7-induced Snail1, Slug, and Twist1 protein expression in A549 and NCI-H460 cells. Further analysis of clinical specimens showed that HIF-1α protein was strongly expressed in NSCLC tissues, which was positively correlated with ZEB1, Snail1, Slug, and Twist1 protein expression.
Conclusions: PI3K/Akt/HIF-1α may contribute to the progression of HPV-associated NSCLC via mediating the expression of EMT-related transcription factors in NSCLC cells.
Keywords: PI3K/Akt/HIF-1α, human papillomaviruse (HPV)-16, non-small cell lung cancer (NSCLC), epithelial-mesenchymal transition (EMT), transcription factor
Introduction
According to World Health Organization (WHO), lung cancer is the most common cancer-related death, and has no effective treatment worldwide. Lung cancer is divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), while the later accounts for about 85% of all lung cancer cases. Long-term exposure to tobacco smoke is the most important cause of lung cancer. However, smoking has been banned in many public places, but lung cancer incidence and mortality rates have been increasing in the past 10 years. Moreover, 15% of men and 53% of women with lung cancer were not caused by smoking, which accounted for 25% of all lung cancer cases [1], and approximately 300,000 never-smokers died of lung cancer each year [2]. Other factors such as exposure to asbestos, radon elements, fumes, and environmental pollution have also been considered as risk factors of lung cancer [3-5]. However, the causes for many cases of lung cancer still remain largely unknown.
Human papillomaviruses (HPVs), the double-stranded DNA viruses, are divided into high-risk and low-risk types. In 1979, Syrjänen first hypothesized the role of HPV in the progression of bronchial squamous cell carcinoma [6]. Recently, accumulating evidence has demonstrated that high-risk HPV infection may be associated with non-smoking-associated lung cancer [7-12]. Moreover, HPV-16 E6 and E7 oncoproteins were found to enhance lung cancer progression by upregulating the expression of angiogenic factors including MMP-2, MMP-9, interleukin (IL)-6, IL-8, and IL-17 [13-15]. Furthermore, our previous study has demonstrated that HPV-16 E6 and E7 oncoproteins promote lung cancer angiogenesis by inducing hypoxia-inducible factor-1α (HIF-1α) protein accumulation and vascular endothelial growth factor (VEGF) expression [16]. These findings suggest that HPV-16 E6 and E7 oncoproteins may play a role in the progression of lung cancer. However, HPV infection was also reported to have no association of HPV status with lung cancer [17], and the underlying molecular mechanisms by which HPV-16 oncoproteins mediate the progression of non-smoking-associated lung cancer are not completely clear. Therefore, the effects of HPV-16 oncoproteins on the progression of non-smoking-associated lung cancer and the underlying molecular mechanisms need to be further studied.
Epithelial-mesenchymal transition (EMT), the crucial step of invasion and metastasis, mediates the progression of various cancers including lung cancer [18]. EMT is a process that epithelial cell markers (e.g. E-cadherin, cytokeratin, and ZO-1) decrease, mesenchymal cell markers (e.g. N-cadherin, vimentin, and fibroblast-specific protein-1) increase, actin cytoskeletal rebuilds, and cell to cell adhesion and polarity are lost. Moreover, EMT is regulated by several key transcription factors with the structures of zinc finger protein and basic helix-loop-helix (bHLH), including ZEB family members such as ZEB1 and ZEB2, Snail family members such as Snail1 and Snail2 (Slug), and bHLH factors such as Twist. Slug activates ZEB1 transcription, and Twist1 promotes Slug transcription by combining with the E-box near the promoter of Slug, leading to the indirect inhibition of E-cadherin expression and the induction of EMT [19,20]. ZEB1 was found to induce LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis [21], and the ERK-ZEB1 pathway mediated EMT in pemetrexed resistant lung cancer cells [22]. Snail1 and Slug were regarded as the important regulators of EMT in lung cancer [23,24]. Twist1 was reported to determine the occurrence of EMT in EGFR mutated lung adenocarcinoma [25].These reports indicate that EMT-related transcription factors play a crucial role in the progression of lung cancer. Interestingly, our previous study has demonstrated that HPV-16 E6 and E7 oncoproteins promote EMT in A549 and NCI-H460 NSCLC cells through upregulating the expression of ZEB1 and Snail1 [26], suggesting that HPV-16 E6 and E7 oncoproteins may promote NSCLC progression by regulating EMT-related transcription factors, but the underlying mechanisms still remain unclear.
HIF-1α, a bHLH-PAS transcription factor, is regulated by cellular oxygen concentration. Under normoxic conditions, HIF-1α protein is hydroxylated at Pro402 and Pro564 residues, triggering HIF-1α protein ubiquitination and degradation. However, the hydroxylation is blocked under hypoxic conditions, thus leading to stabilization and accumulation of HIF-1α protein. HIF-1α plays a key role in tumor angiogenesis, invasion, and metastasis. Recently, HIF-1α has also been demonstrated to mediate EMT via regulating EMT-related transcription factors including ZEB1, Snail, and Twist [27-30], thus promoting cancer progression. Interestingly, our previous study has found that HPV-16 E6 and E7 oncoproteins enhance HIF-1α protein accumulation and HIF-1α-dependent VEGF and IL-8 expression in NSCLC cells [16]. Recently, Fan et al. further demonstrated that overexpression of HPV-16 E6 and E7 oncoproteins upregulated the expression of GLUT1 possibly by increasing HIF-1α protein level in lung cancer cells [31]. These findings indicate that HIF-1α is a key transcription factor in mediating the progression of HPV-associated lung cancer. However, it remains to be elucidated whether HIF-1α contributes to HPV-associated lung cancer progression via mediating the expression of EMT-related transcription factors.
PI3K/Akt signaling pathway has been demonstrated to mediate EMT process [32,33]. Furthermore, the activation of the PI3K/Akt/HIF-1α signaling pathway has been found to play a pivotal role in mediating hypoxia-induced EMT transformation and invasion in rheumatoid arthritis-fibroblast-like synoviocytes (RA-FLSs) [34] and hypoxia-induced EMT and chemoresistance in hepatocellular carcinoma [35]. Our previous study has demonstrated that PI3K/Akt signaling pathway is involved in HPV-16 E6 and E7 oncoprotein-induced HIF-1α protein accumulation and HIF-1α- dependent VEGF and IL-8 expression in NSCLC cells [36]. However, the role of PI3K/Akt/HIF-1α signaling pathway in HPV-16 oncoprotein-induced EMT process in NSCLC cells is not clear.
In this study, we investigated the role of PI3K/Akt/HIF-1α signaling pathway in HPV-16 oncoprotein-induced EMT process in A549 and NCI-H460 NSCLC cells. We demonstrated for the first time, to the best of our knowledge, that PI3K/Akt/HIF-1α signaling pathway mediated HPV-16 E6 and E7 oncoprotein-induced expression of EMT transcription factors in A549 and NCI-H460 cells.
Materials and Methods
Reagents
Transfection reagent (LipofectamineTM 2000) was obtained from Invitrogen Corporation (Carlsbad, CA). Complete protease inhibitor cocktail was from Roche (Mannheim, Germany). Mouse anti-human HIF-1α monoclonal antibody was from BD Transduction Laboratories (San Diego, CA, USA). Rabbit anti-human ZEB1, Snail1, Slug, and Twist1 primary antibodies and goat anti-rabbit HRP-conjugated secondary antibody were purchased from Cell Signaling Technology (Beverly, MA, USA). Mouse anti-human β-actin antibody, goat anti-mouse HRP-conjugated secondary antibody, LY294002, and lysis buffer were purchased from Beyotime Biotechnology Corporation, Shanghai (Shanghai, China). PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) and SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) qPCR reagent Kit were purchased from TaKaRa Biotechnology (Dalian) Co., LTD (Dalian, China).
Cell lines and Cell Culture
A549 NSCLC cell line was obtained from American Type Culture Collection (ATCC, Rockville, MD), and NCI-H460 NSCLC cell line was purchased from Chinese Academy of Sciences Cell Bank of Type Culture Collection (Shanghai, China). A549 and NCI-H460 cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2.
NSCLC tissue specimens and Ethic approval
NSCLC tissue specimens were collected from 45 patients with NSCLC in Affiliated Hospital of Guangdong Medical University (Zhanjiang, Guangdong, China) from 2007 to 2010. Meanwhile, the matched non-tumor adjacent lung tissues (1 cm from NSCLC tissues) were obtained from the same patients [37]. The criteria that patients should be met were according to our previous study [37]. This study was received the ethic approval from the local committee of Affiliated Hospital of Guangdong Medical University, and all clinical studies were performed according to the principles defined by the Declaration of Helsinki [37].
Transient transfection
A series of plasmids including pEGFP-HPV-16 E6, E7, E6-mutant, and E7-mutant were constructed by our lab. A549 and NCI-H460 NSCLC cells at about 80 % confluence were respectively transfected with the above plasmids for 4 h using LipofectamineTM 2000 transfection reagent according to the manufacturer's instructions. The cells treated with transfection reagent alone served as mock transfection controls, and the cells transfected with pEGFP empty vector or mutant plasmids were regarded as negative controls. The transfection efficiency was analyzed by flow cytometry and the expression of HPV-16 E6 and E7 oncoproteins was determined by Western blot analysis. 24 h post-transfection, transfected cells were harvested to observe the effects of HPV-16 E6 and E7 oncoproteins on Slug (Snail2) and Twist1 expression. Slug and Twist1 protein and mRNA levels were analyzed by Western blot analysis and RT-qPCR, respectively.
RNA interference
The method was as described previously by us [36]. The target sequences of RNA interference (RNAi) against HIF-1α were synthesized by Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China). The sequences of sense strand-directed siRNA against HIF-1α are as follows: HIF-1α-siRNA 5′-GCCGCUCAAUUUAUG- AAUATT-3′; HIF-1α non-specific control siRNA (NS-siRNA) 5′-UUCUCCGAACGUGUCACGUTT-3′. HPV-16 E6 or E7-transfected NSCLC cells were co-transfected for 4 h with HIF-1α-siRNA or NS-siRNA via lipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. 48 h post-transfection, the cells were harvested and subjected to Western blot analysis.
Western blot analysis
The method was as described previously by us [16,26,36]. Briefly, total proteins were extracted from transfected or non-transfected cells with lysis buffer (Beyotime Biotechnology Corporation) supplemented complete protease inhibitor cocktail (Sigma). The proteins were separated on 10% PAGE-SDS gel and electro-blotted onto polyvinylidene difluoride (PVDF) membranes. After blocking with TBS or 5% skim milk, the membranes were respectively incubated with their specific primary antibodies overnight at 4°C, followed by the incubation with HRP-conjugated secondary antibodies. As loading controls, the blots were stripped and re-probed with anti-β-actin antibody.
RT-qPCR
The method was as described previously by us [16,26,36]. Total RNA was extracted by homogenization in 1 ml TRIZOL Reagent (Invitrogen), followed by chloroform extraction and isopropanol precipitation. The ZEB-1, Snail1, Slug, and Twist1 relative mRNA levels were analyzed using PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) and SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) qPCR reagent Kit according to the manufacturer's instructions (Dalian, China). The primers used were as follows. ZEB1: forward 5'- TCCCCATCACCTCTAAACCTT-3' and reverse 5'-CCCTGTTGCTTTGGTAGTGAA-3' (Genbank No: NM_001128128.2); Snail1: forward 5'-TTTCTGGTTCTGTGTCCTCTG-3' and reverse 5'-TGTCAGCCTTTGTCCTGTAGC-3' (Genbank No: HC-290415); Slug: forward 5'-CTACAGCGAACTGGACACACA-3' and reverse 5'-GCCCCAAAGATGAGGAGTATC-3' (Genbank No: HC-199495); Twist1: forward 5'-AGTCCGCAGTCATACGAGGAG-3' and reverse 5'-GACCTAGTAGAGGAAGTCGATG-3' (Genbank No: HC_201503); β-actin: forward 5'-TGGCACCCAGCACAATGAA-3' and reverse 5'- CTAAGTCATAGTCCGCCTAG-3' (Genbank No: NM_001101.3). All primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China). The qPCR conditions: 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, and 60°C for 30 s. The relative ZEB-1, Snail1, Slug, and Twist1 mRNA levels were normalized to β-actin.
Immunohistochemical staining
The method was as described previously by us [37]. The results were observed and analyzed in a double-blind manner by two senior pathologists. Ten randomly selected fields were observed and 100 cancer cells were counted in each field (total 10,000 cells) to determine the proportion of positive cells [37]. The semi-quantitative analysis of HIF-1α and ZEB-1, Snail1, Slug, and Twist1 protein expression and the correlation analysis were according to our previous study [37].
Statistical analysis
All experiments were repeated in triplicate, and all data were shown as mean ± SD for three independent experiments. One way ANOVA, LSD, chi-square (χ2) test, and Spearman rank correlation coefficient were performed for statistical analysis using SPSS 19.0. P<0.05 was considered statistically significant.
Results
Evaluation of the transfection efficiency
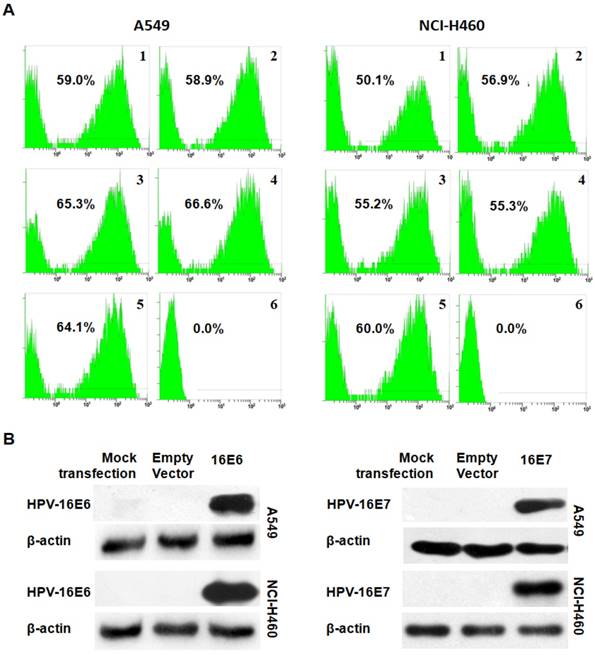
The transfection efficiency was first analyzed by flow cytometry. The results showed a transfection efficiency ranged from 50.1% to 66.6% in A549 (Figure 1A) and NCI-H460 (Figure 1B) NSCLC cells. Furthermore, the expression of HPV-16 E6 or E7 protein in transfected A549 and NCI-H460 cells was confirmed by Western blot analysis (Figure 1C and D). These results indicated that pEGFP-HPV-16 E6 and E7 plasmids were effectively transfected into A549 and NCI-H460 cells.
HPV-16 oncoproteins upregulated Slug and Twist1 expression in NSCLC cells
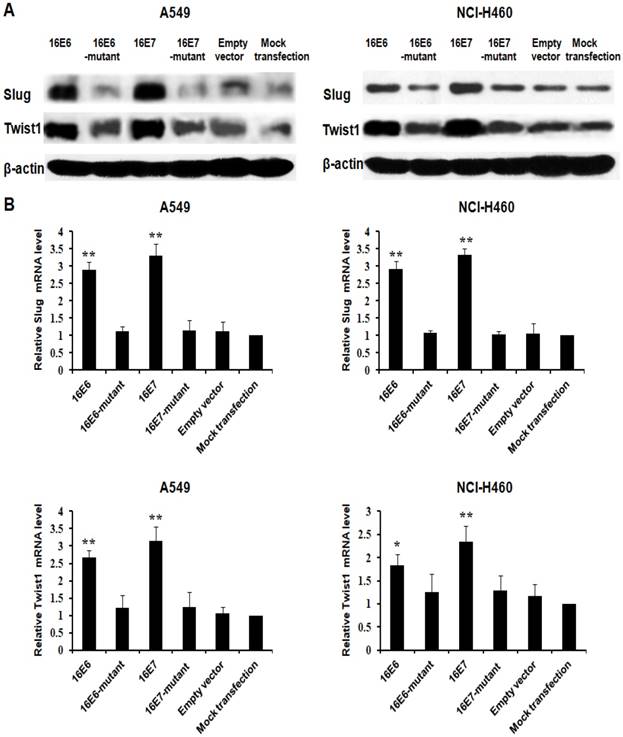
EMT has been demonstrated to mediate lung cancer progression [18], and EMT transcription factors including ZEB1, Snail1, Slug, and Twist1 promoted lung cancer progression by inducing EMT [21-25]. Our previous study has demonstrated that HPV-16 E6 and E7 oncoproteins enhance ZEB1 and Snail1 expression [26]. In this study, we further analyzed the effect of HPV-16 oncoproteins on the expression of Slug and Twist1. We further found that HPV-16 E6 and E7 oncoproteins promoted Slug and Twist1 protein expression in both A549 and NCI-H460 NSCLC cells as compared with empty vector or mutant controls (Figure 2A). Moreover, Slug and Twist1 mRNA levels were also upregulated by HPV-16 E6 and E7 oncoproteins in A549 and NCI-H460 cells (P<0.01, Figure 2B)
HPV-16 oncoprotein-induced upregulation of EMT-related transcription factors in NSCLC cells was HIF-1α-dependent
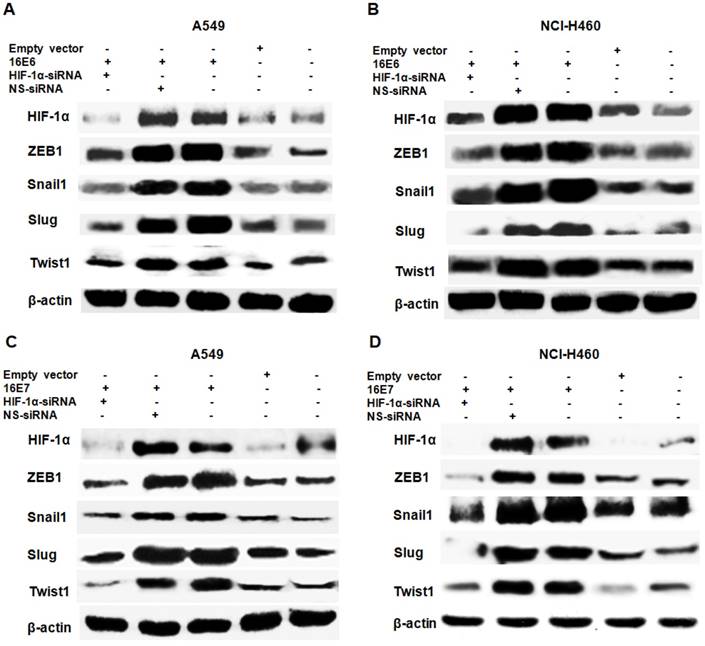
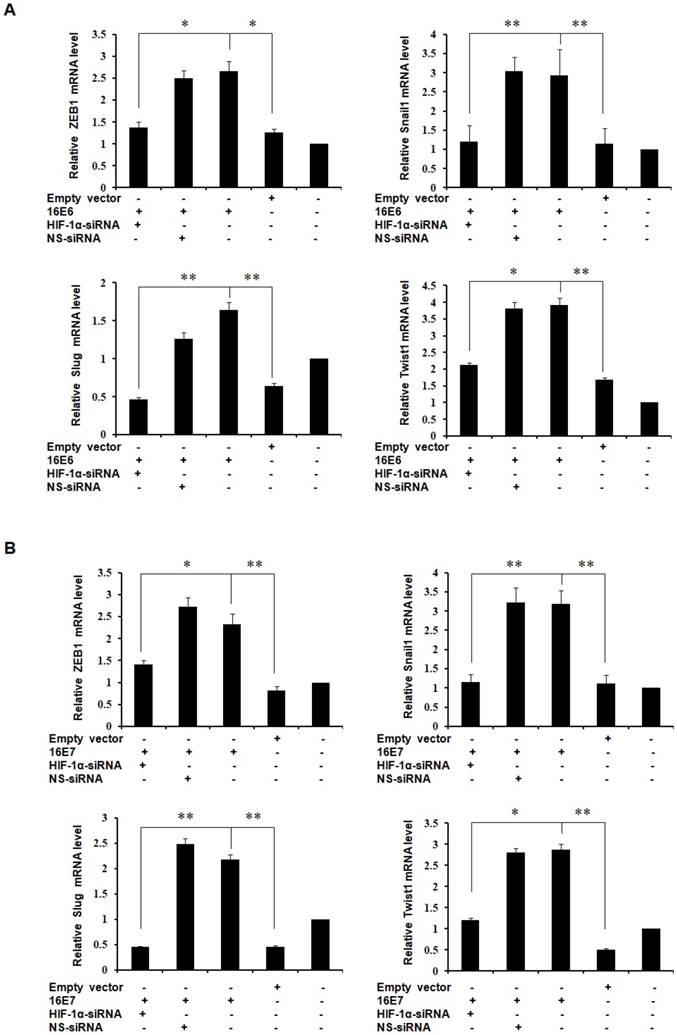
HIF-1α upregulates the expression of EMT-related transcription factors [27-30] and our previous studies have demonstrated that HPV-16 oncoproteins enhance HIF-1α protein accumulation in NSCLC cells [16, 38]. To further determine the role of HIF-1α in HPV-16 oncoprotein-induced expression of EMT-related transcription factors, HPV-16 E6- or E7-transfected NSCLC cells were co-transfected with HIF-1α siRNA or NS-siRNA. Our results showed that the increased protein expression of ZEB1, Snail1, Slug, and Twist1 induced by HPV-16 E6 oncoprotein was abrogated by HIF-1α siRNA co-transfection, but not by NS-siRNA co-transfection in both A549 (Figure 3A) and NCI-H460 (Figure 3B) cells. In addition, the knockdown of HIF-1α had a similar effect on HPV-16 E7 oncoprotein-induced Snail1, Slug, Twist1, and ZEB1 protein expression in both A549 (Figure 3C) and NCI-H460 (Figure 3D) cells. To further explore the role of HIF-1α in regulating the transcription levels of EMT-related transcription factors, RT-qPCR was performed to determine their mRNA levels after co-transfection. Our results showed that mRNA levels of ZEB1, Snail1, Slug, and Twist1 upregulated by HPV-16 E6 and E7 oncoproteins were also significantly blocked by HIF-1α-siRNA co-transfection, but not by NS-siRNA co-transfection in A549 cells (Figure 4). Taken together, these results indicated that HPV-16 oncoprotein-mediated up-regulation of ZEB1, Snail1, Slug, and Twist1 expression in NSCLC cells was HIF-1α-dependent.
Analysis of transfection efficiency. (A and B) The transfection efficiency was analyzed by flow cytometry in transfected A549 (A) and NCI-H460 (B) cells. 1. HPV-16E6-transfected cells; 2. HPV-16E6 mutant-transfected cells; 3. HPV-16E7- transfected cells; 4. HPV-16E7 mutant-transfected cells; 5. Empty vector-transfected control cells; 6. Mock-transfection control cells. (C and D) The expression of HPV-16E6 (C) and HPV-16E7 (D) oncoproteins was determined by Western blot analysis.
PI3K/Akt signaling pathway was involved in HPV-16 oncoprotein-mediated upregulation of EMT-related transcription factors in NSCLC cells
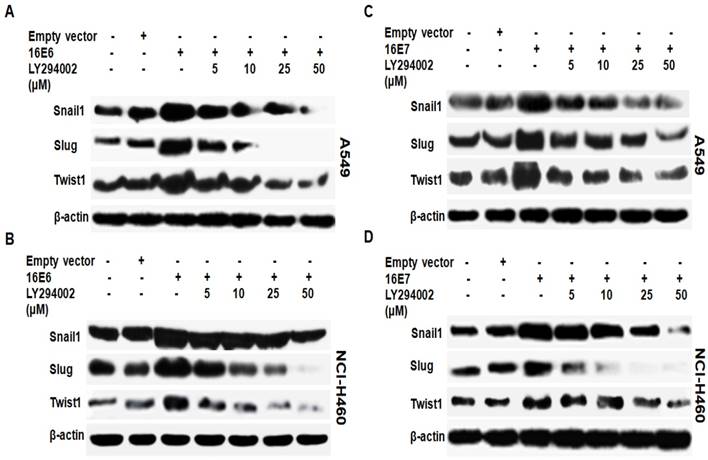
We have previously demonstrated that HPV-16 oncoproteins promote the activation of PI3K/Akt signaling pathway in NSCLC cells [36]. In this study, we further determined whether PI3K/Akt signaling pathway is involved in the expression of Snail1, Slug and Twist1 induced by HPV-16 E6 and E7 oncoproteins in NSCLC cells. HPV-16 E6- or E7-tranfected A549 and NCI-H460 cells were pretreated for 16 h with different concentrations of LY294002 (5, 10, 25, and 50 μM), a specific PI3K inhibitor, followed by Western blot analysis for Snail1, Slug, and Twist1 protein expression. As shown in Figure 5, the pretreatment with different concentrations of LY294002 inhibited HPV16 E6 (Figure 5A and B)- and E7 (Figure 5C and D)-induced Snail1, Slug, and Twist1 protein expression in both A549 and NCI-H460 cells, whereby the maximal inhibitory effect was noticed when the cells were pretreated with 50 μM of LY294002. These results suggest that PI3K/Akt signaling pathway is indispensable for HPV-16 E6 or E7 oncoprotein-induced upregulation of EMT-related transcription factors including Snail1, Slug, and Twist1.
HPV-16 oncoproteins enhanced Slug and Twist1 expression in NSCLC cells. A549 and NCI-H460 cells were transfected with plasmids harboring pEGFP-N1-HPV-16 E6 or E7, and the transfection with empty vector or mutant plasmids served as controls. (A) Western blot analysis was performed to detect Slug and Twist1 protein expression. (B) RT-qPCR was performed to determine Slug and Twist1 mRNA expression. All data are expressed as mean±SD of three independent experiments. Compared with empty vector and mutant controls, *P<0.05, **P<0.01.
HIF-1α protein expression was positively correlated with ZEB1, Snail1, Slug, and Twist1 protein expression in NSCLC tissues
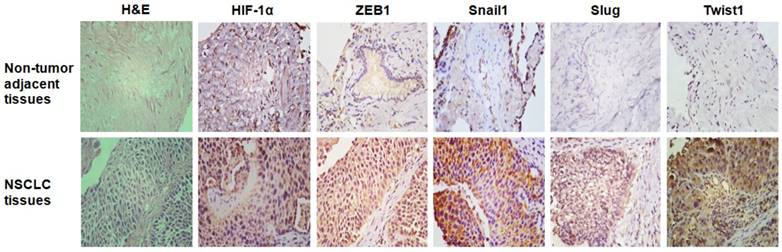
Our previous study demonstrated that EMT-related transcription factors including ZEB1, Snail1, Slug, and Twist were highly expressed in NSCLC tissues [37]. In this study, we further found that HIF-1α protein was robustly expressed in NSCLC tissues (Figure 6), and HIF-1α positive rate in NSCLC tissues (30/45, 66.7%) was much higher than that in the matched non-tumor adjacent lung tissues (10/45, 22.2%) (χ2=18.0, P < 0.01). Furthermore, Spearman rank correlation analysis showed that the abundant expression of HIF-1α protein in NSCLC tissues was positively correlated with ZEB1, Snail1, Slug, and Twist1 protein expression (P < 0.01, Table 1).
Discussion
HPV-16 E6 and E7 oncoproteins can inactivate tumor suppressors Rb and P53, respectively, leading to cervical cancer progression. Recently, HPV-16 E6 and E7 oncoproteins have been reported to promote lung cancer progression [13-15]. Our previous study has further demonstrated that HPV-16 E6 and E7 oncoproteins enhance the expression of HIF-1α, VEGF, IL-8, triggering lung cancer angiogenesis [16]. Recently, HIF-1α has been reported to regulate EMT by modulating the expression of EMT-related transcription factors including ZEB1, Snail, and Twist [27-30], and VEGF-A stimulated Snail expression in breast tumor cells [39] and VEGFR-1 activation led to an increase in the expression of Snail, Twist, and Slug in pancreatic carcinoma cells [40]. Furthermore, the inhibition of IL-8 expression and activity was found to prevent Snail-induced EMT in colorectal cancer cells [41]. These data suggest that HPV may affect the expression of EMT-related transcription factors by mediating HIF-1α, VEGF, and IL-8 in lung cancer cells. Interestingly, our previous study has demonstrated that HPV-16 E6 and E7 oncoproteins significantly upregulate ZEB1 and Snail1 expression in A549 and NCI-H460 NSCLC cells [26]. In this study, we further confirmed that HPV-16 E6 and E7 oncoproteins dramatically promoted Slug and Twist1 expression in A549 and NCI-H460 cells (Figure 2). However, the underlying molecular mechanisms by which HPV-16 oncoproteins enhanced the expression of EMT-related transcription factors are unclear.
Effect of HIF-1α knockdown on the protein expression of EMT-related transcription factors induced by HPV-16 oncoproteins in NSCLC cells. HPV-16 E6 (A and B)- or E7 (C and D)-transfected A549 and NCI-H460 cells were co-transfected with HIF-1α-siRNA or non-specific siRNA (NS-siRNA), followed by Western blot analysis for ZEB1, Snail1, Slug, and Twist1 protein expression. All results are representative of three independent experiments.
Effect of HIF-1α knockdown on the mRNA expression of EMT-related transcription factors induced by HPV-16 oncoproteins in A549 NSCLC cells. HPV-16 E6 (A)- and E7(B)- transfected A549 cells were co-transfected with HIF-1α-siRNA or non-specific siRNA (NS-siRNA), followed by RT-qRCR analysis for ZEB1, Snail1, Slug, and Twist1 mRNA expression. All data are expressed as mean±SD of three independent experiments. *P<0.05, **P<0.01.
Effect of LY294002 on HPV-16 oncoprotein-induced Snail1, Slug, and Twist1 protein expression in NSCLC cells. HPV-16 E6 (A and B)- and E7- (C and D) transfected A549 and NCI-H460 cells were pretreated for 24 h with different concentrations of LY294002, a specific PI3K inhibitor, followed by Western blot analysis for Snail1, Slug, and Twist1 protein expression. All results are representative of three independent experiments.
The representative results of immunohistochemical staining. Original magnification, ×400.
The correlation between HIF-1α expression and EMT Transcription factors in NSCLC
| HIF-1α | Total | rs | P | |||
|---|---|---|---|---|---|---|
| + | - | |||||
| ZEB1 | + | 23 | 1 | 24 | 0.778 | <0.01 |
| - | 7 | 14 | 21 | |||
| Snail1 | + | 25 | 3 | 28 | 0.837 | <0.01 |
| - | 5 | 12 | 17 | |||
| Slug | + | 24 | 1 | 25 | 0.818 | <0.01 |
| - | 6 | 14 | 20 | |||
| Twist | + | 26 | 3 | 29 | 0.821 | <0.01 |
| - | 4 | 12 | 16 | |||
| Total | 30 | 15 | 45 | |||
Hypoxia microenvironment can induce the occurrence of EMT regulated by HIF-1α [30,42,43]. Hypoxia is one of the inherent characteristics of tumor, and HIF-1α plays a key role in mediating tumor angiogenesis, invasion, and metastasis by regulating the transcription of multiple downstream genes. Recently, HIF-1α has been found to induce EMT by regulating the expression of EMT-related transcription factors including ZEB1, Snail, and Twist, and immortalizing mesenchymal phenotype, thereby accelerating the deterioration of tumors [27-30]. Our previous studies have demonstrated that HPV-16 E6- and E7-induced VEGF and IL-8 expression and angiogenesis in NSCLC cells is HIF-1α-dependent [16,36], indicating that HIF-1α plays an important role in lung cancer angiogenesis promoted by HPV-16 E6 and E7 oncoproteins. In this study, we further found that the knockdown of HIF-1α significantly inhibited HPV-16 E6- and E7-induced the expression of ZEB1, Snail1, Slug, and Twist1 proteins in A549 cells (Figure 3A and C). Consistent results were obtained in NCI-H460 cells (Figure 3B and D). To further understand the role of HIF-1α in ZEB1, Snail1, Slug, and Twist1 transcription, RT-qPCR was performed to analyze the effect of HIF-1α knockdown on ZEB1, Snail1, Slug, and Twist1 mRNA levels. Our results further demonstrated that the mRNA levels of ZEB1, Snail1, Slug, and Twist1 upregulated by HPV-16 oncoproteins were HIF-1α-dependent in A549 lung cancer cells (Figure 4). Moreover, we demonstrated that HIF-1α protein expression was positively correlated with ZEB1, Snail1, Slug, and Twist1 protein expression in NSCLC tissues (Figure 6 and Table 1). Taken together, these results support the notion that HIF-1α plays a crucial role in HPV-16 oncoprotein-induced EMT via regulating the expression of EMT-related transcription factors, triggering the progression of NSCLC.
Our previous study has demonstrated that HPV-16 E6 and E7 oncoproteins promote the activation of PI3K/Akt signaling pathway in A549 and NCI-H460 NSCLC cells [36]. Recently, PI3K/Akt signaling pathway has been reported to mediate EMT and metastasis by regulating the transcription of ZEB1 [46] and the expression of Snail [45-47] and Twist [48]. In the present study, we further demonstrated that the pretreatment with LY294002, a specific PI3K inhibitor, robustly downregulated HPV-16 E6 or E7 oncoprotein-induced Snail1, Slug, and Twist1 protein expression in A549 and NCI-H460 cells (Figure 5), suggesting that PI3K/Akt signaling pathway was involved in the expression of EMT-related transcription factors induced by HPV-16 oncoprotiens in NSCLC cells. Our previous study has demonstrated that PI3K/Akt signaling pathway mediates HPV-16 oncoprotein-induced HIF-1α protein accumulation [36], and PI3K/Akt/HIF-1α signaling pathway was found to be involved in EMT process [34,35]. In the present study, we confirmed that HPV-16 E6 and E7 oncoproteins in NSCLC cells promoted the expression of EMT-related transcription factors in a HIF-1α-dependent manner. Taken together, PI3K/Akt/HIF-1α signaling pathway may play a key role in HPV-16 E6 and E7 oncoprotein-induced expression of EMT-related transcription factors in NSCLC cells.
Conclusion
In summary, our present study demonstrated for the first time to the best of our knowledge that PI3K/Akt/HIF-1α signaling pathway mediated EMT process by regulating EMT-related transcription factors including ZEB1, Snial1, Slug, and Twist1, thus leading to the progression of NSCLC. These findings suggest that PI3K/Akt/ HIF-1α signaling pathway might be a potential target for the prevention and treatment of HPV-associated NSCLC.
Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China, 81372511 (to X.T.); Special Fund for Scientific and Technological Development (Basic and Applied Basic Research) of Guangdong Province in 2017 (Natural Science Foundation of Guangdong Province), 2017A030313539 (to X.T.); Guangdong Provincial Department of Science and Technology (Research and Development of Industrial Technology in Guangdong Province), 2013B031100002 (to X.T.); and “Sail plan” in Guangdong Province to cultivate high-level talents, 201635011 (to X.T.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Parkin DM, Bray F, Ferlay J. et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74-108
2. Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers-a different disease. Nat Rev Cancer. 2007;7(10):778-90
3. Grosche B, Kreuzer M, Kreisheimer M. et al. Lung cancer risk among German male uranium miners: A cohort study, 1946-1998. Br J Cancer. 2006;95(9):1280-7
4. Wang XR, Chiu YL, Qiu H. et al. The roles of smoking and cooking emissions in lung cancer risk among Chinese women in Hong Kong. Ann Oncol. 2009;20(4):746-51
5. Pallis AG, Syrigos KN. Lung cancer in never smokers: disease characteristics and risk factors. Crit Rev Oncol Hematol. 2013;88(3):494-503
6. Syrjänen KJ. Condylomatous Changes in Neoplastic Bronchial Epithelium. Report of a case. Respiration. 1979;38(5):299-304
7. Xiong WM, Xu QP, Li X. et al. The association between human papillomavirus infection and lung cancer: a system review and meta-analysis. Oncotarget. 2017;8(56):96419-32
8. Zhang EY, Tang XD. Human Papillomavirus Type 16/18 Oncoproteins: Potential Therapeutic Targets in Non-smoking Associated Lung Cancer. Asian Pac J Cancer Prev. 2012;13:5363-9
9. Bae JM, Kim EH. Human papillomavirus infection and risk of lung cancer in never-smokers and women: an 'adaptive' meta-analysis. Epidemiol Health. 2015;37:e2015052
10. Lin FC, Huang JY, Tsai SC. et al. The association between human papillomavirus infection and female lung cancer: A population-based cohort study. Medicine (Baltimore). 2016;95:e3856
11. de Freitas AC, Gurgel AP, de Lima EG. et al. Human papillomavirus and lung cancinogenesis: an overview. J Cancer Res Clin Oncol. 2016;142(12):2415-27
12. Robinson LA, Jaing CJ, Campbell CP. et al. Molecular evidence of viral DNA in non-small cell lung cancer and non-neoplastic lung. Br J Cancer. 2016;115(4):497-504
13. Shiau MY, Fan LC, Yang SC. et al. Human papillomavirus up-regulates MMP-2 and MMP-9 expression and activity by inducing interleukin-8 in lung adenocarcinomas. PLoS One. 2013;8(1):e54423
14. Chang YH, Yu CW, Lai LC. et al. Up-regulation of interleukin-17 expression by human papillomavirus type 16 E6 in non-small cell lung cancer. Cancer. 2010;116(20):4800-9
15. Cheng YW, Lee H, Shiau MY. et al. Human papillomavirus type 16/18 up-regulates the expression of interleukin-6 and antiapoptotic Mcl-1 in non-small cell lung cancer. Clin Cancer Res. 2008;14(15):4705-12
16. Li G, He L, Zhang E. et al. Overexpression of human papillomavirus (HPV) type 16 oncoproteins promotes angiogenesis via enhancing HIF-1α and VEGF expression in non-small cell lung cancer cells. Cancer Lett. 2011;311(2):160-70
17. Argyri E, Tsimplaki E, Marketos C. et al. Investigating the role of human papillomavirus in lung cancer. Papillomavirus Res. 2017;3:7-10
18. DauPhin M, Barbe C, Lemaire S. et al. Vimentin expression predicts the occurrence of metastases in non-small lung cell carcinomas. Lung Cancer. 2013;81(1):117-22
19. Wels C, Joshi S, Koefinger P. et al. Transcriptional Activation of ZEB1 by Slug Leads to Cooperative Regulation of the Epithelial-Mesenchymal Transition-Like phenotype in Melanoma. J Invest Dermatol. 2011;131(9):1877-1885
20. Casas E, Kim J, Bendesky A. et al. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71(1):245-54
21. Peng DH, Ungewiss C, Tong P. et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2017;36(14):1925-38
22. Chiu LY, Hsin IL, Yang TY. et al. The ERK-ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene. 2017;36(2):242-253
23. Yang X, Han M, Han H. et al. Silencing Snail suppresses tumor cell proliferation and invasion by reversing epithelial-to-mesenchymal transition and arresting G2/M phase in non-small cell lung cancer. Int J Oncol. 2017;50(4):1251-60
24. Shih JY, Yang PC. The EMT regulator Slug and lung carcinogenesis. Carcinogenesis. 2011;32(9):1299-1304
25. Pallier K, Cessot A, Côté JF. et al. TWIST1 a new determinant of epithelial to mesenchymal transition in EGFR mutated lung adenocarcinoma. PLoS One. 2012;7(1):e29954
26. Zhang W, Wu X, Hu L. et al. Overexpression of Human Papillomavirus Type 16 Oncoproteins Enhances Epithelial-Mesenchymal Transition via STAT3 Signaling Pathway in Non-Small Cell Lung Cancer Cells. Oncol Res. 2017;25(5):843-52
27. Chen Z, Zhu Y, Dong Y. et al. Overexpression of TrpC5 promotes tumor metastasis via the HIF-1α-Twist signaling pathway in colon cancer. Clin Sci (Lond). 2017;131(19):2439-50
28. Zhang W, Shi X, Peng Y. et al. HIF-1α Promotes Epithelial-Mesenchymal Transition and Metastasis through Direct Regulation of ZEB1 in Colorectal Cancer. PLoS One. 2015;10(6):e0129603
29. Luo D, Wang J, Li J. et al. Mouse snail is a target gene for HIF. Mol Cancer Res. 2011;9(2):234-45
30. Yang YJ, Na HJ, Suh MJ. et al. Hypoxia Induces Epithelial-Mesenchymal Transition in Follicular Thyroid Cancer: Involvement of Regulation of Twist by Hypoxia Inducible Factor-1α. Yonsei Med J. 2015;56(6):1503-14
31. Fan R, Hou WJ, Zhao YJ. et al. Overexpression of HPV16 E6/E7 mediated HIF-1α upregulation of GLUT1 expression in lung cancer cells. Tumour Biol. 2016;37:4655-63
32. Tian X, Zhou D, Chen L. et al. Polo-like kinase 4 mediates epithelial- mesenchymal transition in neuroblastoma via PI3K/Akt signaling pathway. Cell Death Dis. 2018;9(2):54
33. Wei S, Wang L, Zhang L. et al. ZNF143 enhances metastasis of gastric cancer by promoting the process of EMT through PI3K/AKT signaling pathway. Tumour Biol. 2016;37(9):12813-21
34. Li GQ, Zhang Y, Liu D. et al. PI3 kinase/Akt/HIF-1α pathway is associated with hypoxia-induced epithelial-mesenchymal transition in fibroblast-like synoviocytes of rheumatoid arthritis. Mol Cell Biochem. 2013;372(1-2):221-31
35. Jiao M, Nan KJ. Activation of PI3 kinase/Akt/HIF-1α pathway contributes to hypoxia-induced epithelial-mesenchymal transition and chemoresistance in hepatocellular carcinoma. Int J Oncol. 2012;40(2):461-8
36. Zhang E, Feng X, Liu F. et al. Roles of PI3K/Akt and c-Jun Signaling Pathways in Human Papillomavirus Type 16 Oncoprotein-Induced HIF-1α, VEGF, and IL-8 Expression and In Vitro Angiogenesis in Non-Small Cell Lung Cancer Cells. PLoS One. 2014;9(7):e103440
37. Liu F, Hu L, Ma Y. et al. Increased expression of monoamine oxidase A is associated with epithelial to mesenchymal transition and clinicopathological features in non-small cell lung cancer. Oncol Lett. 2018;15(3):3245-51
38. He L, Zhang E, Shi J. et al. (-)-Epigallocatechin-3-gallate inhibits human papillomavirus (HPV)-16 oncoprotein-induced angiogenesis in non-small cell lung cancer cells by targeting HIF-1α. Cancer Chemother Pharmacol. 2013;71(3):713-25
39. Wanami LS, Chen HY, Peiró S. et al. Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: implications for tumor progression. Exp Cell Res. 2008;314(13):2448-53
40. Yang AD, Camp ER, Fan F. et al. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66(1):46-51
41. Hwang WL, Yang MH, Tsai ML. et al. SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology. 2011;141(1):279-91
42. Li M, Wang YX, Luo Y. et al. Hypoxia inducible factor-1α-dependent epithelial to mesenchymal transition under hypoxic conditions in prostate cancer cells. Oncol Rep. 2016;36(1):521-7
43. Sun LL, Song Z, Li WZ. et al. Hypoxia facilitates epithelial-mesenchymal transition-mediated rectal cancer progress. Genet Mol Res. 2016:15 (4). doi: 10.4238/gmr15048936
44. Wu K, Fan J, Zhang L. et al. PI3K/Akt to GSK3β/β-catenin signaling cascade coordinates cell colonization for bladder cancer bone metastasis through regulating ZEB1 transcription. Cell Signal. 2012;24(12):2273-82
45. Dong J, Zhai B, Sun W. et al. Activation of phosphatidylinositol 3-kinase/AKT/snail signaling pathway contributes to epithelial-mesenchymal transition-induced multi-drug resistance to sorafenib in hepatocellular carcinoma cells. PLoS One. 2017;12(9):e0185088
46. Hu B, Xie S, Hu Y. et al. Hepatitis C virus NS4B protein induces epithelial-mesenchymal transition by upregulation of Snail. Virol J. 2017;14(1):83
47. Yang S, Ji Q, Chang B. et al. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8(4):5976-91
48. Rafael D, Doktorovová S, Florindo HF. et al. EMT blockage strategies: Targeting Akt dependent mechanisms for breast cancer metastatic behaviour modulation. Curr Gene Ther. 2015;15(3):300-12
Author contact
![]() Corresponding author: Xudong Tang, Institute of Biochemistry and Molecular Biology, Guangdong Medical University, 2 Wenming Donglu, Xiashan, Zhanjiang 524023, P.R. China. Tel: +86-759-2388581. Fax: +86-759-2284104. E-mail: tangxudong2599com, txdedu.cn.
Corresponding author: Xudong Tang, Institute of Biochemistry and Molecular Biology, Guangdong Medical University, 2 Wenming Donglu, Xiashan, Zhanjiang 524023, P.R. China. Tel: +86-759-2388581. Fax: +86-759-2284104. E-mail: tangxudong2599com, txdedu.cn.

 Global reach, higher impact
Global reach, higher impact