Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(19):3593-3602. doi:10.7150/jca.26465 This issue Cite
Research Paper
LncRNA PVT1 promotes malignant progression in squamous cell carcinoma of the head and neck
1. Department of Otolaryngology Head and Neck Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450000, People's Republic of China
2. Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, People's Republic of China
3. Otolaryngology Major Disease Research Key Laboratory of Hunan Province, 87 Xiangya Road, Changsha, Hunan 410008, People's Republic of China
Received 2018-4-3; Accepted 2018-8-2; Published 2018-9-8
Abstract
Long non-coding RNAs (lncRNAs) are potentially critical regulators of cancer malignant behaviours. Aberrant expression and dysfunction of lncRNA PVT1 have been reported in multiple human cancers. However, its role in squamous cell carcinoma of the head and neck (SCCHN) remains largely unknown. Our current study demonstrated that PVT1 expression was increased in SCCHN. High PVT1 expression was positively correlated with SCCHN clinical parameters including T classification, clinical stages and cervical lymph node metastasis. More importantly, high PVT1 expression predicted a poor prognosis in SCCHN patients. Gain-of function and loss-of function studies further indicated that PVT1 promoted the proliferation and invasion of SCCHN both in vitro and in vivo, which was accompanied by epithelial-mesenchymal transition and enhanced cancer stem cell-like properties. Further mechanistic investigation revealed that PVT1 activated Wnt/β-catenin signalling pathway, and inhibition of Wnt/β-catenin signalling reversed the malignant progression caused by PVT1 overexpression. Together, our study reveals that PVT1 accelerates the malignant progression of SCCHN and represents a potential biomarker and therapeutic target in SCCHN.
Keywords: Squamous cell carcinoma of the head and neck, Plasmacytoma variant translocation 1, metastasis, epithelial-mesenchymal transition, stemness, Wnt/β-catenin
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) accounts for approximately 90% of all head and neck cancers, and ranks as the six leading cancer worldwide. Despite considerable improvements in diagnosis and therapies have been achieved for decades, life quality and survival rate of SCCHN patients are far from satisfactory. Unlimited proliferation and metastasis are major challenges in the treatment of SCCHN patients [1, 2]. Molecular mechanisms underlying the tumorigenesis and progression of SCCHN are complicated, which are intensively studied in diverse gene types including protein coding genes and non-coding genes [3, 4]. Hence, a comprehensive and profound understanding of the molecular mechanisms underlying cancer development and progression is critical for the discovery of novel and promising treatment strategies for the management of SCCHN patients.
Long non-coding RNAs (lnc RNAs) are a class of RNAs longer than 200 nucleotides with no or limited protein-coding potential. It is well established that lnc RNAs play critical roles in the regulation of various biological and pathological processes, such as proliferation, apoptosis, cell cycle progression, migration and invasion etc [5]. Emerging studies have shown that lncRNAs are frequently deregulated in a wide type of malignancies including SCCHN, and are involved in the development and progression of cancer [6]. For example, two well-studied lncRNAs including MALAT1 and HOTAIR are increased in numerous solid cancers including SCCHN, and is important in cancer malignant progression [7, 8].
Plasmacytoma variant translocation 1 (PVT1), as one lncRNA with a length of 1716 nucleotides, is located adjacent to oncogene c-Myc on chromosome 8q24.21 region [9]. Aberrant PVT1 expression has been detected in a variety of malignancies, including non-small-cell lung cancer [10-12], breast cancer [13], hepatocellular carcinoma [14, 15], clear cell renal cell carcinoma [16] and prostate cancer [17], etc. Available evidence indicates that PVT1 has broad functions in cancer cell proliferation, apoptosis, cell cycle arrest, invasion, radioresistance and chemoresistance [18, 19]. Recent studies suggest that PVT1 serves as a competing endogenous RNA (ceRNA) model to sponging microRNA, such as miR-200 family, and then collaborate together to regulate genes expression including oncogenes and/or suppressors in cancer [20, 21]. Taken together, PVT1 is a potential oncogene and a valuable biomarker in human cancers. However, the functions and molecular mechanisms of PVT1 in SCCHN are still waiting for investigation.
In our current study, PVT1 was found to be significantly increased in SCCHN and its overexpression was tightly associated with T classification, clinical stages and metastasis in SCCHN patients. Importantly, PVT1 overexpression predicted a poor prognosis. Further in vitro and in vivo experiments revealed that PVT1 promoted proliferation, migration and invasion of SCCHN via the activation of Wnt/β-catenin signaling pathway, which induced epithelial-mesenchymal transition (EMT) and enhanced cancer stem cell-like properties. These results suggest that PVT1 may serve as a potential prognostic biomarker and therapeutic target in SCCHN.
Materials and methods
Cell lines and cell culture
The SCCHN cell line Tu686, established from a primary tongue tumor, was kindly provided by Dr. Zhuo G. Chen (Emory University Winship Cancer Institute, Atlanta, Georgia, USA). The human hypopharyngeal carcinoma cell line FaDu was purchased from the American Type Culture Collection (ATCC®, Manassas, VA, USA). Tu686 cells and FaDu cells were maintained in Dulbecco's modified Eagle's medium (DMEM)/F12 medium (1:1) (Gibco) and DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco) at 37°C in a humidified incubator with 5% CO2. Cells in the logarithmic growth phase were used in all the subsequent experiments.
StarBase V2.0 data analysis
Public available RNA-seq data were analyzed from starBase V2.0 project (http://starbase.sysu.edu.cn/) [22], in which 425 SCCHN and 42 adjacent non-cancerous epithelial (ANCE) samples were included.
Preparation of human SCCHN cancer tissues
A total of 83 SCCHN and their corresponding ANCE tissues were obtained from patients who received surgery in Xiangya Hospital, Central South University (Changsha, China) from January 2008 to December 2011. SCCHN patients enrolled in the study had to follow these inclusion criteria: (i) primary squamous cell carcinoma without other malignancies; (ii) no history of previous radiotherapy or chemotherapy. The main clinicopathological features of patients were listed in Table 1. Pathological tumor-node-metastasis (TNM) stage was determined based on the criteria of the 7th American Joint Committee on Cancer staging system in 2010. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at -80°C until RNA extraction. The follow-up range was from 13 to 60 months. Clinical physical examination, imaging evaluation, and pathological examination were employed to confirm the recurrence or metastasis. Overall survival (OS) was calculated from the day of surgery to the date of death. The study was approved by Research Ethics Committee of Xiangya Hospital, Central South University, Changsha, China, and written informed consent was obtained from each patient before surgery.
Quantitative reverse transcription-polymerase chain reaction analysis (qRT-PCR)
Total RNAs from SCCHN tissues and cell lines were extracted using Trizol reagent (Invitrogen) according to the manufacture's instruction. Briefly, 1 μg RNA was used to generate complementary DNA (cDNA) with the High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific Inc.). 25 μl reaction system was established and PCR amplification was performed using SYBR® Green PCR Master Mix (Thermo Fisher Scientific Inc.) on the Applied Biosystems 7500 Real-Time PCR System. qPCR primers were listed as follows: PVT1 [23], forward 5'-TGA GAA CTG TCC TTA CGT GAC C-3', reverse 5'-AGA GCA CCA AGA CTG GCT CT-3'; E-cadherin, forward 5'-GCT GGA CCG AGA GAG TTT CC-3', reverse 5'-CAA AAT CCA AGC CCG TGG TG-3'; Vimentin, forward 5'-TGT CCA AAT CGA TGT GGA TGT TTC-3', reverse 5'-TTG TAC CAT TCT TCT GCC TCC TG-3'; GAPDH, forward 5'-TCC AAA ATC AAG TGG GGC GA-3', reverse 5'-AGT AGA GGC AGG GAT GAT GT-3'. Relative gene expression was calculated using the 2-ΔΔCq method [24] and normalized to GAPDH.
Overexpression or knockdown of PVT1
For PVT1 overexpression, full-length human PVT1 cDNA was amplified by PCR and then cloned into pLV lentiviral plasmid (Addgene). Empty vector was used as a control. For PVT1 knockdown, two shRNAs against PVT1 were used and the target sequences of PVT1 were showed as follows [23]: sh1: 5'-CCC AAC AGG AGG ACA GCT T-3'. sh2: 5'-GCU UGG AGG CUG AGG AGT T-3'. shRNAs were cloned into shRNA-expressing lentiviral vector pLKO.1 (Addgene). The above vectors together with lentivirus packaging vectors were cotransfected into 293T cells using FuGENE® 6 (Promega). Lentiviral particles were harvested 48 hours after transfection and freshly used for cell transfection. Stable cells were screened by puromycin for 2 weeks and efficiency of gene modulation was assayed by qPCR.
Cell proliferation
FaDu cells or Tu686 cells after PVT1 regulation were seeded into 6-well plate for cell counting at different time points, or into 96-well plate for proliferation assay via Cell Counting Kit-8 (Beyotime Institute of Biotechnology) [25].
Transwell migration and invasion assays
Transwell cell migration and invasion assays were performed as we previously described [25-27]. In brief, 4.0x104 Tu686 cells or FaDu cells in serum-free medium were seeded in the upper chamber of transwell plates (Corning). Medium containing 10% FBS was added into the lower chambers. Cells that migrated through 8 μm pores were fixed by methanol and stained with hematoxylin and eosin. Stained cells were visualized under a microscope and counted from 5 random fields. For cell invasion experiments, the upper chambers were precoated with 1mg/ml Matrigel (BD Biosciences) and then subjected to the protocol as cell migration assay.
Western blotting analysis
The western blotting analysis was performed as we previously described [25, 27]. In brief, 50-80 μg total protein was separated by 8-12% SDS-PAGE, and then transferred to polyvinylidene fluoride membranes (Millipore). After blocked with 5% skimmed milk at room temperature for 1 hour, the membranes were incubated with primary antibodies at 4 ˚C overnight. Primary antibodies were listed as follows: Mouse monoclonal antibody against E-cadherin (1:400; Santa Cruz), mouse monoclonal antibody against Vimentin (1:200; Santa Cruz), mouse monoclonal antibody against p-GSK3β (1:500; Santa Cruz) and mouse monoclonal antibody against β-catenin (1:500; Santa Cruz). Then the membranes were washed three times in TBST and incubated with horseradish peroxidase (HRP)-labeled goat Anti-mouse IgG (H+L) (1:1,000; Beyotime Institute of Biotechnology) for 1 hour at room temperature. For normalization of protein loading, mouse monoclonal antibody against β-actin (1:1,000; Beyotime Institute of Biotechnology) was used.
Tumorsphere formation assays
FaDu cells were cultured as a monolayer before being harvested as single cell suspension. Cells were suspended in tumorsphere medium (serum-free DMEM medium supplemented with 20 ng/mL human recombinant epidermal growth factor (EGF, Millipore), 20 ng/mL human recombinant basic fibroblast growth factor (bFGF, PeproTech) and 2% B27), and then subsequently plated in ultra-low attachment 24-well plates (Corning) at a density of no more than 2,000 cells/well. Culture medium was changed every 3 days. Spheres with a diameter >75 μm were counted after 14 days [28].
Xenograft tumor model
Athymic male nude mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. Changsha, Hunan. Mice were housed in specific pathogen-free laboratory. Animal procedures were reviewed and approved by the Animal Ethics Committee of Central South University, Changsha, Hunan. Mice at the age of 5-7 weeks were subcutaneously injected into the position of hind flank with 2.0×106 Tu686/PVT1 shRNA1 or Tu686/Vector cells that suspended in 200 µl cold PBS (4 mice for each group). Basic condition and individual mouse weight were monitored during the whole period. Tumor volume was determined every 3-5 days by calliper measurements and was calculated using the modified ellipse formula (volume = length × width2/2). Thirty days after cancer cell injection, all mice were euthanized and sacrificed. Xenograft tumors in all groups were removed and final weight of each tumor was measured. Each tumor sample was spliced into 2 parts and stored in nitrogen for the following qPCR and western blotting assays, respectively.
Statistical analysis
All statistical analyses were performed using IBM SPSS 19.0 software. Quantitative data in this study were expressed as the mean ± SD. Two-tailed Student's t-test was employed to determine the significant difference. Correlations between PVT1 expression and clinicopathological parameters in SCCHN patients were analyzed using Student's t-test. The Cox proportional hazards regression model was used to identify factors that were associated with overall survival. Survival curves were plotted with the Kaplan-Meier method and compared by the log-rank test. P-value < 0.05 was considered to be statistically significant.
Results
Increased PVT1 expression predicts a poor prognosis in SCCHN patients
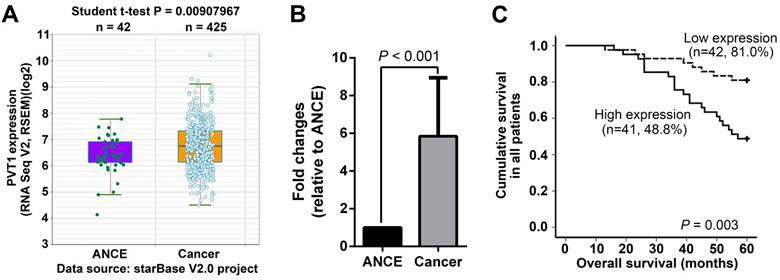
PVT1 expression was initially analyzed in the RNA-seq data from starBase V2.0 project (http://starbase.sysu.edu.cn/), in which 425 SCCHN and 42 ANCE samples were included [22]. SCCHN RNA-seq data clearly indicated that PVT1 transcripts were significantly increased in SCCHN compared to ANCE samples (Figure 1A). To validate the above online result and further explore the potential associations between PVT1 expression and clinical variables in SCCHN patients, PVT1 expression was assayed by qPCR in 83 SCCHN patients with intact clinical and prognostic information. As expected, PVT1 expression was dramatically elevated in SCCHN tissues (Figure 1B). Subsequently, these SCCHN patients were divided into two groups, high PVT1 expression group (n = 42) and low expression group (n = 41), in which PVT1 expression was greater or less than the median value in the 83 SCCHN patients cohort. As statistically summarized via Student t-test in Table 1, PVT1 expression was positively correlated with higher T classification (P = 0.041), advanced clinical stages (P < 0.001) and lymph node metastasis in SCCHN patients (P < 0.001). Importantly, survival analysis via Kaplan-Meier method and a log-rank test demonstrated that SCCHN patients with a higher PVT1 expression displayed a worse prognosis than patients with low PVT1 expression (Figure 1C, P = 0.003). Univariate Cox regression analyses demonstrated that T classification, clinical stages, lymph node metastasis, and PVT1 expression were significantly associated with the overall survival in SCCHN patients (Table 2; All P < 0.05). However, only metastasis was determined to be an independent prognostic factor for the overall survival in SCCHN patients (Table 2; P = 0.004). Collectively, these clinical data suggest that PVT1 overexpression is associated with a progressive behavior and indicates a poor prognosis in SCCHN patients.
Correlations between lncRNA PVT1 expression and clinicopathological parameters in SCCHN patients
| Parameters | No. of patients | PVT1 expression | t value | P-value* |
|---|---|---|---|---|
| Age | ||||
| <59 | 41 | 6.429±3.067 | 1.721 | 0.089 |
| ≥59 | 42 | 5.269±3.073 | ||
| Gender | ||||
| Female | 4 | 6.068±4.361 | 0.148 | 0.883 |
| Male | 79 | 5.831±3.068 | ||
| Smoking | ||||
| Yes | 47 | 6.002±2.871 | -0.533 | 0.595 |
| No | 36 | 5.633±3.421 | ||
| Histological grade | ||||
| G1+G2 | 29 | 5.927±3.064 | 0.181 | 0.857 |
| G3 | 54 | 5.797±3.157 | ||
| T classification | ||||
| T1+T2 | 46 | 5.219±3.020 | -2.080 | 0.041 |
| T3+T4 | 37 | 6.617±3.078 | ||
| Clinical stage | ||||
| I+ II | 36 | 4.434±2.139 | -4.139 | 0.000 |
| III+ IV | 47 | 6.921±3.314 | ||
| Lymph node metastasis | ||||
| N0 | 54 | 4.595±2.535 | -5.945 | 0.000 |
| N+ | 29 | 8.165±2.741 |
NOTE: *. P < 0.05 was considered to be statistical significance.
PVT1 overexpression indicates a poor prognosis in SCCHN patients. (A) PVT1 expression was analyzed based on RNA-seq data from starBase V2.0 project (http://starbase.sysu.edu.cn/), in which 425 SCCHN and 42 ANCE samples were included. (B) PVT1 expression was assayed by qPCR in 83 SCCHN and ANCE samples. (C) Kaplan-Meier analysis and log-rank test for overall survival in SCCHN patients with different PVT1 expression. ANCE, adjacent non-cancerous epithelial samples.
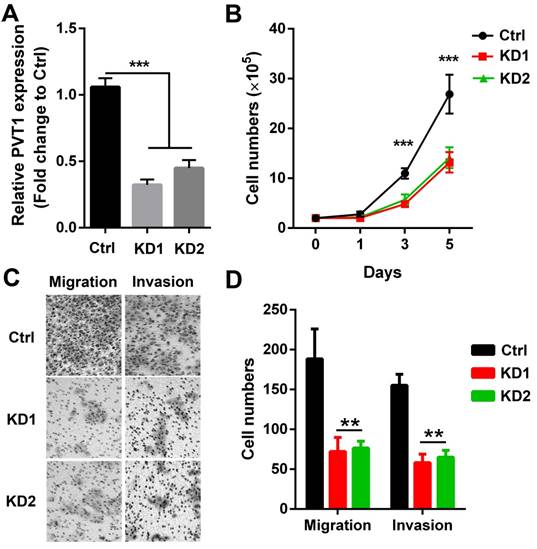
PVT1 knockdown inhibits SCCHN progression in vitro. Lentivirus harboring PVT1 shRNA1, shRNA2 and control shRNA were used to transfect Tu686 cells, and (A) qPCR was applied to confirm PVT1 knockdown efficiency. (B) Cells in each group were counted. (C) Representative images of transwell migration and invasion assays (×200). (D) Quantitative results of transwell migration and invasion assays. **, P < 0.01;***, P < 0.001. Ctrl, Tu686 cells transfected with control shRNA; KD1, Tu686 cells transfected with PVT1 shRNA1; KD2, Tu686 cells transfected with PVT1 shRNA2.
Cox model analysis of overall survival
| Parameters | Relative risk (95%CI) | P-value |
|---|---|---|
| Univariate | ||
| Age | 0.787(0.378-1.636) | 0.521 |
| Gender | 0.766(0.182-3.223) | 0.716 |
| Smoking | 0.457(0.202-1.033) | 0.060 |
| Histological grade | 0.661(0.293-1.493) | 0.319 |
| T classification | 0.210(0.089-0.494) | 0.000 |
| Clinical stage | 0.113(0.034-0.374) | 0.000 |
| Lymph node metastasis | 0.137(0.060-0.312) | 0.000 |
| PVT1 expression | 0.307(0.136-0.695) | 0.005 |
| Multivariate | ||
| Lymph node metastasis | 0.179(0.056-0.570) | 0.004 |
NOTE: All the clinicopathological variables listed in the table were included in the univariate and multivariate analyses.
Abbreviation: 95% CI, 95% confidence interval.
PVT1 promotes the progression of SCCHN in vitro
To directly confirm whether PVT1 participated in SCCHN malignant progression, 2 PVT1 shRNAs carried by lentivirus vector were used to knockdown the expression of PVT1 in SCCHN cells. As presented in Figure 2A, both 2 shRNAs efficiently inhibited PVT1 expression in Tu686 cells. Subsequently, alterations including growth capacity, migration and invasion were examined. We found that both PVT1 shRNAs inhibited the in vitro proliferation from day 3 (Figure 2B). In addition, PVT1 inhibition slowed down the migration and impeded the invasion of Tu686 cells (Figure 2C and 2D). These data indicates that PVT1 knockdown inhibits the malignant SCCHN progression including proliferation, migration and invasion.
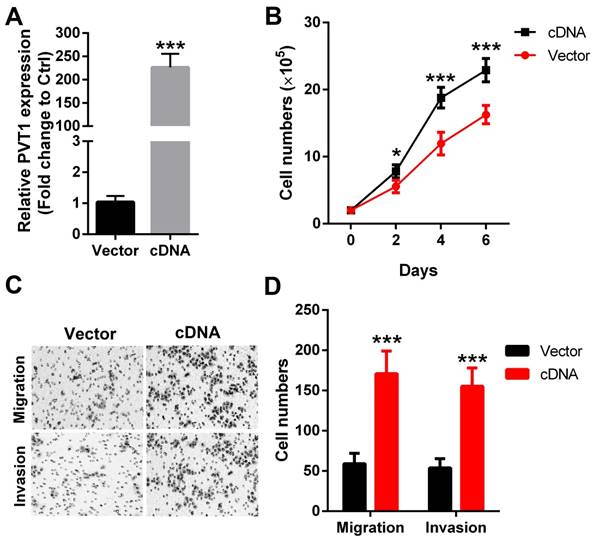
To further strengthen the above results, PVT1 was overexpressed in another SCCHN FaDu cells (Figure 3A). Our data revealed that forced expression of PVT1 correspondingly enhanced the proliferation (Figure 3B), migration (Figure 3C) and invasion (Figure 3D) in FaDu cells. Together, the above gene gain-of function and loss-of function analyses clearly indicate that PVT1 is an oncogene in SCCHN and functions critically in SCCHN malignant progression.
PVT1 induces EMT and enhances stem cell-like properties of SCCHN in vitro
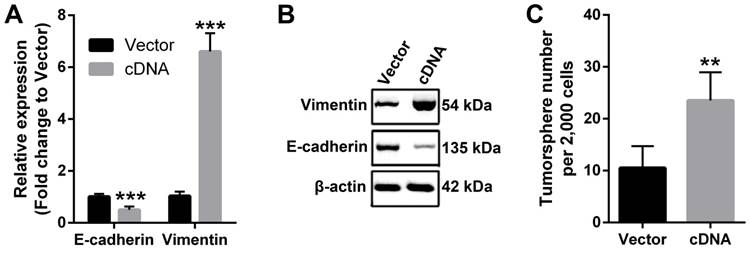
EMT is a critical molecular reprogram in cancer cells that enhances the malignancy in numerous cancer types [29]. Therefore, we used qPCR and western blotting to check the molecular changes that involved in EMT. Our data demonstrated that ectopic expression of PVT1 in FaDu cells downregulated the expression of epithelial marker E-cadherin, while increased the expression of mesenchymal marker Vimentin at both mRNA and protein levels (Figure 4A and 4B), indicating that PVT1 overexpression induced EMT in SCCHN cells. Moreover, recent evidence has shown that EMT reprogram enhanced cancer stemness [28], we also studied the effect of PVT1 on the stem cell-like phenotype by tumorsphere formation assay. The results indicated that forced expression of PVT1 enhanced the sphere-forming ability of SCCHN FaDu cells in vitro (Figure 4C). Taken together, our data suggest that PVT1 promotes SCCHN progression via inducing EMT and enhancing its stem cell-like properties.
Forced expression of PVT1 promotes SCCHN progression in vitro. (A) qPCR was applied to check the level of PVT1. (B) Cells in each group were counted. (C) Representative images of transwell migration and invasion assays (×200). (D) Quantitative results of transwell migration and invasion assays. *, P < 0.05; ***, P < 0.001. Vector, FaDu cells transfected with empty lentivirus; cDNA, FaDu cells transfected by lentivirus mediated PVT1 cDNA.
PVT1 induces EMT and enhances stem cell-like properties of SCCHN in vitro. (A) qPCR and (B) western blotting assays were performed to check the expression of EMT markers including E-cadherin and Vimentin. (C) Tumorsphere formation assays were used to determine cancer stem cell-like properties after PVT1 overexpression. **, P < 0.01; ***, P < 0.001. Vector, FaDu cells transfected with empty lentivirus; cDNA, FaDu cells transfected by lentivirus mediated PVT1 cDNA.
Wnt/β-catenin signaling pathway is involved in PVT1- mediated SCCHN progression. Specific inhibitor iCRT14 (20μmol/L) was used to impede Wnt/β-catenin signaling pathway in FaDu cells infected with PVT1 cDNA and empty vector, and then: (A) western blotting assays were used to check alterations of Wnt/β-catenin signaling molecules. (B) Cell proliferation was assayed by CCK8. (C) Transwell migration and (D) invasion assays were used to examine the changes of migration and invasion. (E) E-cadherin and (F) Vimentin mRNAs were examined by qPCR.
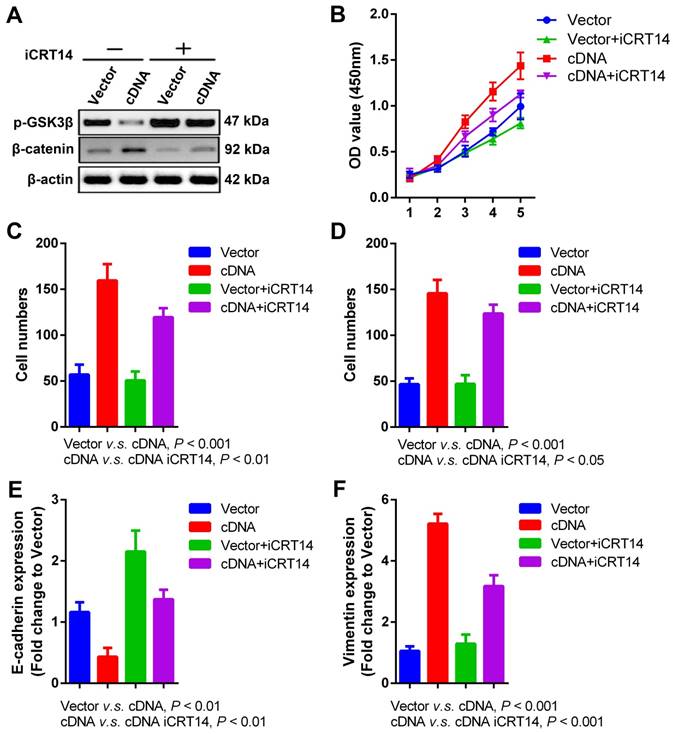
Wnt/β-catenin signaling pathway contributes to PVT1-mediated SCCHN progression
Wnt/β-catenin signaling pathway is a key driver for cancer behaviors in numerous malignancies [30], which is also intensively studied in our previous researches about radioresistance and metastasis of SCCHN [26, 31, 32]. Recent publication indicates that PVT1 promotes chemoresistance in bladder urothelial carcinoma via Wnt/β-catenin signaling pathway [33]. In our current study, we found that PVT1 overexpression in FaDu cells significantly repressed the expression of phosphorylated-GSK3β (p-GSK3β) and upregulated the level of β-catenin (Figure 5A), indicating the activation of Wnt/β-catenin signaling pathway caused by PVT1 overexpression. To validate whether Wnt/β-catenin is a downstreaming signaling pathway in PVT1-mediated SCCHN progression, iCRT14 as a specific β-catenin/TCF complex inhibitor was applied in PVT1 overexpressed FaDu cells. Our data demonstrated that the blocking of Wnt/β-catenin signaling pathway partially inhibited the proliferation (Figure 5B), migration (Figure 5C) and invasion (Figure 5D) of FaDu cells caused by PVT1 overexpression. At molecular level, iCRT14 also reversed the mRNA expression of E-cadherin and Vimentin induced by PVT1 (Figure 5E and 5F). These data reveal that PVT1 promotes SCCHN progression via the activation of Wnt/β-catenin signaling pathway.
PVT1 accelerates SCCHN tumorigenesis in vivo
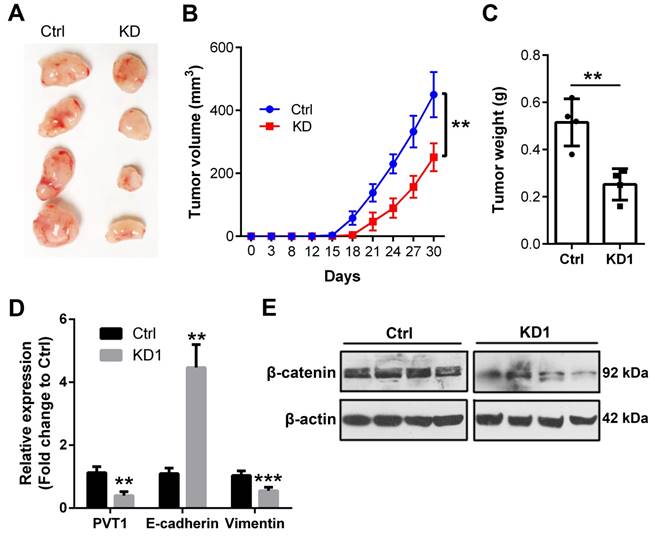
In the end, we extended to the in vivo environment to further confirm the above in vitro results. Therefore, shRNA1 was used to knockdown PVT1 expression in Tu686 cells that were subjected to puromycin screening for 2 weeks. Consequently, 2.5×106 Tu686 cells were subcutaneously injected into the flank of nude mice. Tumor size was monitored every 3-5 days and mice were euthanized at day 30 post cell injection. As shown in Figure 6A and 6B, PVT1 knockdown led to decreased tumor size and impeded the speed of tumor growth in nude mice (P < 0.01). Final weight of xenograft tumors were declined from (0.52 ± 0.10) g in control Tu686 cells to (0.25 ± 0.07) g in PVT1 knockdown Tu686 cells (Figure 6C, P < 0.01). Moreover, PVT1 knockdown correspondingly upregulated the expression of E-cadherin and inhibited the expression of Vimentin at mRNA level (Figure 6D), which was accompanied by the deactivation of Wnt/β-catenin signaling pathway (Figure 6E).Collectively, these in vivo data indicate that PVT1 knockdown inhibits the tumorigenesis of SCCHN via modulating Wnt/β-catenin signaling pathway.
Discussion
Available evidence indicates that PVT1 plays an important role in carcinogenesis and cancer progression. In the region of head and neck, PVT1 is remarkably upregulated in thyroid cancer, and PVT1 silencing significantly inhibited proliferation and cell cycle progression of cancer cells [34]. More recently, PVT1 knockdown induces apoptosis of nasopharyngeal carcinoma via preventing the DNA damage repair pathway after radiotherapy [19]. However, the role and clinical relevance of PVT1 in SCCHN is far from clear. In this study, PVT1 expression is obviously elevated in SCCHN tissues and high PVT1 expression is associated with T classification, clinical stages and cervical lymph node metastasis, which is tightly correlated with a poor prognosis in SCCHN patients. Further in vitro and in vivo experiments indicate that PVT1 promotes carcinogenesis and progression via activating Wnt/β-catenin signaling pathway, inducing EMT and enhancing the stem cell-like properties in SCCHN.
Knockdown of PVT1 inhibits SCCHN tumorigenesis in vivo. Lentivirus mediated PVT1 shRNA1 (KD1) and control shRNA (Ctrl) were used to transfect Tu686 cells, which were then used to establish xenograft tumors. Final tumor images (A), tumor size (B), and tumor weight (C) were shown. (D) qPCR was used to emaxine the expression of E-cadherin and Vimentin. (E) Western blotting was applied to check the expression of total β-catenin protein. **, P < 0.01; ***, P < 0.001.
One major finding of our study is that high PVT1 expression is tightly associated with several clinical parameters and a poor prognosis in SCCHN patients, which is consistent with reports in other solid tumors including non-small-cell lung cancer [10, 12], hepatocellular carcinoma [15], clear renal cell carcinoma [35], prostate cancer [17], glioma [36, 37], gastric [23] and cervical cancer [38], etc. We have to mention that our current clinical analysis is based on a relative small sample size, therefore, verification in a larger patient cohort will further strengthen the clinical significance of PVT1 in SCCHN patients. However, these data still suggest that PVT1 is a promising prognostic indicator for SCCHN patients. Together with the fact that increased expression of PVT1 exists in a wide spectrum of cancer types, it highlights that PVT1 is a valuable prognostic marker in human cancers. In line with our clinical results, both in vitro and in vivo data also reveal that PVT1 promotes the unlimited proliferation and invasion of SCCHN, indicating that it is also a potential therapeutic molecular target in SCCHN.
Metastasis provides critical guidance information for the determination of treatment strategy, and also contributes to the unfavorable prognosis in cancer. EMT and enhanced cancer stemness are widely accepted to participate in the process of metastasis [28]. In our current study, forced PVT1 expression induced EMT and maintained stem cell-like properties of SCCHN, which partially explains the mechanisms for PVT1-mediated metastasis in SCCHN. In accordance with our data, PVT1 also promotes stem cell-like properties of hepatocellular carcinoma cells by stabilizing NOP2 [15]. In another aspect, PVT1 has also gradually confirmed to be an important regulator in the process of EMT [39-42]. It is not surprising to find that PVT1 can modulate both the processes of EMT and cancer stem cell, because cancer cells that experienced EMT generates more cells with properties of stem cell [28]. However, how PVT1 remodels the EMT and cancer stem cell properties is still in mystery.
Canonical activation of Wnt/β-catenin signaling pathway leads to series of cellular responses through activation of β-catenin/TCF target genes [30]. Once activation, β-catenin accumulates in cytoplasm and translocate to nucleus, where it engages DNA bounding TCF transcription factors to activate the expression of Catenin Responsive Transcription (CRT) reporter genes and other endogenous genes [43]. In our study, PVT1 overexpression activated β-catenin signaling pathway, as reflected by decreased p-GSK3β and increased β-catenin expression, indicating that PVT1 overexpression leads to the activation of Wnt/β-catenin signaling pathway, which is also a major finding of our study. To confirm whether Wnt/β-catenin pathway is actually involved in PVT1-mediated effects, we used iCRT14, a potent inhibitor of β-catenin-responsive transcription, to block Wnt/β-catenin pathway. Upon the treatment of iCRT14, PVT1-mediated changes were coordinately reversed, which confirmed that PVT1 exerted its functions through Wnt/β-catenin signaling pathway.
However, we have to note that Wnt/β-catenin inhibition could not completely restore the phenotypes caused by PVT1 overexpression in our study, which indicates that other latent pathways may also contribute to this process. In addition, lncRNAs can function as competing endogenous RNAs (ceRNAs, also known as miRNA “decoy” or miRNA “sponges”), which are RNA transcripts that compete for the binding to specific miRNAs. Subsequently, lncRNAs depress the effect of miRNAs on their binding target genes [44]. PVT1, as one member of lncRNAs, has been reported to function as a sponge to inhibit the effects of miRNAs such as miR-152, miR-488-3p, miR-203 and miR-195 etc [45-49]. This potential ceRNA regulatory network composed by PVT1 and its binding miRNAs reveals the complexity of PVT1-mediated effects, which may be also involved in the progression of SCCHN caused by PVT1 overexpression.
In conclusion, we have demonstrated that PVT1 is increased in SCCHN and significantly associated with a poor prognosis and enhanced SCCHN progression. However, detailed investigation of molecular mechanisms is urgent and indispensable to further clarify the function of PVT1 in SCCHN.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (Nos. 81874133, 81402232, 81772903 and 81602684), the Huxiang Young Talent Project and the Natural Science Foundation of Hunan Province (Nos. 2018JJ2630 and 2017JJ3488).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Allen CT, Law JH, Dunn GP, Uppaluri R. Emerging insights into head and neck cancer metastasis. Head Neck. 2013;35:1669-78
2. Dragovic AF, Caudell JJ, Spencer SA, Carroll WR, Nabell LA, Bonner JA. Locoregional failure and the risk of distant metastasis after modern radiotherapy for head and neck cancer. Head Neck. 2013;35:381-7
3. Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:6348
4. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
5. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61
6. Sannigrahi MK, Sharma R, Panda NK, Khullar M. Role of non-coding RNAs in head and neck squamous cell carcinoma: A narrative review. Oral Dis. 2017 [Epub ahead of print]
7. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y. et al. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep. 2015;5:15972
8. Sun S, Wu Y, Guo W, Yu F, Kong L, Ren Y. et al. STAT3/HOTAIR signaling axis regulates HNSCC growth in an EZH2-dependent manner. Clin Cancer Res. 2018;24:2665-77
9. Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H. et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82-6
10. Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37:4127-34
11. Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929-35
12. Huang C, Liu S, Wang H, Zhang Z, Yang Q, Gao F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8:5025-34
13. Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J. et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745-55
14. Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng C. et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9:955-63
15. Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C. et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278-90
16. Yang T, Zhou H, Liu P, Yan L, Yao W, Chen K. et al. lncRNA PVT1 and its splicing variant function as competing endogenous RNA to regulate clear cell renal cell carcinoma progression. Oncotarget. 2017;8:85353-67
17. Yang J, Li C, Mudd A, Gu X. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci Biotechnol Biochem. 2017;81:2301-6
18. Cui M, You L, Ren X, Zhao W, Liao Q, Zhao Y. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun. 2016;471:10-4
19. He Y, Jing Y, Wei F, Tang Y, Yang L, Luo J. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235
20. Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One. 2017;12:e0171661
21. Wang C, Han C, Zhang Y, Liu F. LncRNA PVT1 regulate expression of HIF1alpha via functioning as ceRNA for miR199a5p in nonsmall cell lung cancer under hypoxia. Mol Med Rep. 2018;17:1105-10
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-7
23. Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM. et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82
24. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8
25. Liu Y, Yu C, Qiu Y, Huang D, Zhou X, Zhang X. et al. Downregulation of EphA2 expression suppresses the growth and metastasis in squamous-cell carcinoma of the head and neck in vitro and in vivo. J Cancer Res Clin Oncol. 2012;138:195-202
26. Liu C, Li G, Ren S, Su Z, Wang Y, Tian Y. et al. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett. 2017;13:2631-6
27. Yu C, Liu Y, Tan H, Li G, Su Z, Ren S. et al. Metadherin regulates metastasis of squamous cell carcinoma of the head and neck via AKT signalling pathway-mediated epithelial-mesenchymal transition. Cancer Lett. 2014;343:258-67
28. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
29. Jung HY, Fattet L, Yang J. Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res. 2015;21:962-8
30. Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985-99
31. Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y. et al. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. Eur J Cancer. 2013;49:2596-607
32. Liu C, Li G, Yang N, Su Z, Zhang S, Deng T. et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017;17:2
33. Liu Z, Zhang H. LncRNA plasmacytoma variant translocation 1 is an oncogene in bladder urothelial carcinoma. Oncotarget. 2017;8:64273-82
34. Zhou Q, Chen J, Feng J, Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR). Tumour Biol. 2016;37:3105-13
35. Bao X, Duan J, Yan Y, Ma X, Zhang Y, Wang H. et al. Upregulation of long noncoding RNA PVT1 predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Cancer Biomark. 2017;21:55-63
36. Zou H, Wu LX, Yang Y, Li S, Mei Y, Liu YB. et al. lncRNAs PVT1 and HAR1A are prognosis biomarkers and indicate therapy outcome for diffuse glioma patients. Oncotarget. 2017;8:78767-80
37. Yang A, Wang H, Yang X. Long non-coding RNA PVT1 indicates a poor prognosis of glioma and promotes cell proliferation and invasion via target EZH2. Biosci Rep. 2017:37
38. Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS One. 2016;11:e0156274
39. Zheng X, Hu H, Li S. High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2016;12:2357-62
40. Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long Noncoding RNA PVT1 Promotes EMT and Cell Proliferation and Migration Through Downregulating p21 in Pancreatic Cancer Cells. Technol Cancer Res Treat. 2017:1533034617700559
41. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25:637-44
42. Chang Z, Cui J, Song Y. Long noncoding RNA PVT1 promotes EMT via mediating microRNA-186 targeting of Twist1 in prostate cancer. Gene. 2018;654:36-42
43. Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW. et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100-7
44. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
45. Li T, Meng XL, Yang WQ. Long Noncoding RNA PVT1 Acts as a "Sponge" to Inhibit microRNA-152 in Gastric Cancer Cells. Dig Dis Sci. 2017;62:3021-8
46. Li Y, Li S, Luo Y, Liu Y, Yu N. LncRNA PVT1 Regulates Chondrocyte Apoptosis in Osteoarthritis by Acting as a Sponge for miR-488-3p. DNA Cell Biol. 2017;36:571-80
47. Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233:4044-55
48. Li PD, Hu JL, Ma C, Ma H, Yao J, Chen LL. et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget. 2017;8:34164-76
49. Zhou Q, Chen F, Zhao J, Li B, Liang Y, Pan W. et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7:82620-33
Author contact
![]() Corresponding authors: Hua Cao, Department of Otolaryngology Head and Neck Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450000, People's Republic of China. Email: chcarencom and Yong Liu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, People's Republic of China. Email: liuyongentedu.cn
Corresponding authors: Hua Cao, Department of Otolaryngology Head and Neck Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450000, People's Republic of China. Email: chcarencom and Yong Liu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, People's Republic of China. Email: liuyongentedu.cn

 Global reach, higher impact
Global reach, higher impact