3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(20):3651-3659. doi:10.7150/jca.27117 This issue Cite
Research Paper
miRNA-21 and miRNA-223 expression signature as a predictor for lymph node metastasis, distant metastasis and survival in kidney renal clear cell carcinoma
1. Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning 530007, P. R. China
2. Department of Medical Oncology of Gastroenterology, Affiliated Tumor Hospital of Guangxi Medical University, Nanning 530021, P. R. China
# These authors contributed equally to this work.
Received 2018-5-6; Accepted 2018-8-2; Published 2018-9-8
Abstract
Purpose: The aim of this study was to generate a novel miRNA expression signature to effectively assess nodal metastasis, distant metastasis and predict prognosis for patients with kidney renal clear cell carcinoma (KIRC) and explore its potential mechanism of affecting the prognosis.
Method: Using expression profiles downloaded from the Cancer Genome Atlas database, we identified multiple miRNAs with differential expression between KIRC and paired normal tissues. The diagnostic values of the differentially expressed miRNAs for nodal metastasis and distant metastasis were evaluated by Receiver Operating Characteristic (ROC) curve analysis. Then, we evaluated the impact of miRNAs on overall survival (OS) by univariate and multivariate COX regression analyzes. This analysis was ultimately used to construct a miRNA signature that effectively assessed nodal metastasis, distant metastasis and predicted prognosis. The functional enrichment analysis of the miRNAs included in the signatures was used to explore its potential molecular mechanism in KIRC.
Results: Based on our cutoff criteria (P < 0.05 and |log2FC| > 1.0), we identified 104 differentially expressed microRNAs (miRNAs), including 43 that were up-regulated in KIRC tissues and 61 that were down-regulated. We found 12 miRNAs were potentially diagnostic biomarkers of nodal metastasis and distant metastasis by ROC curve analysis. Two miRNAs (miRNA-21 and miRNA-223) were significant miRNAs independently associated with OS based on Cox univariate and multivariate analysis. We generated a signature index based on expression of these two miRNAs, and the two-miRNA signature is promising as a biomarker for diagnosing nodal metastasis, distant metastasis and predicting 5-year survival rate of KIRC with areas under the curve (AUC)=0.738, 0.659 and 0.731, respectively. Patients were stratified into high-risk and low-risk groups, according to median of the signature prognosis indexes. Patients in the high-risk group had significantly shorter survival times than those in the low-risk group (P = 0.000). The functional enrichment analysis suggested that the target genes of two miRNAs may be involved in various pathways related to cancer, p53 signaling pathway, apoptosis, and MAPK signaling pathway.
Conclusion: The two-miRNA signature could assess nodal metastasis, distant metastasis and predict survival of KIRC. As a promising prediction tool, the mechanism of the two miRNAs in KIRC deserves further study.
Keywords: Renal clear cell carcinoma, miRNA signature, prognosis
Introduction
Renal cell carcinoma (RCC) is the most common form of kidney cancer and is responsible for up to 85% of cases [1]. Kidney renal clear cell carcinoma (KIRC) is the most common subtype of RCC, occurring in 70-75% of cases [2]. KIRC is associated with radiotherapy and chemotherapy resistance [3], and the overall prognosis is still poor, particularly for patients who present with high-stage disease [4]. Tumor stage (TNM), defined by the anatomic involvement of disease, is recognized as one of the strongest prognostic factors in the clinical outcome of patients with KIRC, as described in the eighth edition of the American Joint Commission on Cancer (AJCC) Cancer Staging Manual [5]. Because surgical resection of KIRC remains curative for a proportion of patients with localized disease, several models based on TNM and nuclear grade have been developed to predict outcomes after surgery [6-8]. So, it is important to know the status of nodal metastasis and distant metastasis for KIRC patients. Even though imaging examinations such as computed tomography (CT) scan and magnetic resonance imaging (MRI) are available, the status of lymph node metastasis or distant metastasis in a number of patients with KIRC cannot be assessed and is marked as Nx and Mx. Therefore, the treatment and follow-up assessment in KIRC patient may be affected. There is an urgent need for additional biomarkers to help clinicians assess lymph node metastasis and distant metastasis in KIRC which is marked as Nx and Mx.
MicroRNAs (miRNAs) are endogenous, highly conserved, non-coding RNAs 20-24 bp in length that function in a variety of biological processes at the transcriptional or post-transcriptional level. Abnormal expression levels of miRNAs are recognized as an important issue in cancer development. miRNAs are promising candidates as markers for the diagnosis and prognosis and as targeted therapies for cancers [9].
The Cancer Genome Atlas Project (TCGA) is an effort by the National Cancer Institute to profile different tumor types with genomic platforms and to make raw and processed data available to all researchers [10]. In the present study, we analyzed high-throughput miRNA data from the TCGA database to identify differentially expressed miRNAs (DEmiRs) in KIRC tissues and paired normal kidney tissues and found a two-miRNA signature that effectively assesses nodal metastasis, distant metastasis and predicts prognosis. The functional enrichment analysis suggested that the target genes of two miRNAs were involved in various pathways related to cancer.
Materials and Methods
Data processing
The KIRC preprocessed miRNA stem loop expression profiles in TCGA database, displayed as log2 converted reads per million (log2(RPM+1)), and clinical information were downloaded from the UCSC Xena (https://xenabrowser.net/datapages/, version 09-15-2017). If the same patient has more than two samples of the same tissue, the average expression value was extracted. The inclusion criteria were as follows: (1) the sample included both miRNA expression profiles and clinical information, (2) the sample included prognosis information, and (3) miRNAs with expression levels of zero in less than 50% of the samples. The DEmiRs between KIRC and normal tissues were analyzed by “limma” package in R [11]. The fold changes (FCs) in the expression of individual miRNA were calculated and differentially expressed miRNAs with log2|FC| > 1.0 and P < 0.05 were considered to be significant.
Diagnostic value for nodal metastasis, distant metastasis and predictive value for survival of differentially expressed miRNAs
Patients with N0 and N1 were extracted then ROC curve analysis was performed to evaluated the diagnostic value of the DEmiRs for nodal metastasis. Similarly, patients with M0 and M1 were extracted to evaluated the diagnostic value of the DEmiRs for distant metastasis. ROC curves analysis was performed by “pROC” package in R [12]. P-values of <0.05 were considered significant. Then, we evaluated the association between the common miRNA markers and OS by univariate and multivariate COX regression analyzes in all patients. The patients were separated into high- and low-level groups based on median, followed by the Cox univariate and multivariate analysis. Finally, a miRNA-based prognosis index score was constructed on the basis of a linear combination of the expression level multiplied regression coefficient derived from the multivariate cox regression model (β) with the following formula.
Prognosis Index (PI) = expmiRNA*β1 + expmiRNA*β2 + expmiRNA*β3+…
The “β” value is the estimated regression coefficient of miRNA and is derived from the multivariate Cox regression analysis, and “exp” indicates the expression profiles of the miRNAs. The KIRC patients were divided into two groups of low-risk and high-risk, according to median PI. ROC curve analysis was used to assess the diagnostic value of the miRNA signature for nodal metastasis and distant metastasis. Time-dependent ROC was used to assess the miRNA signature's predictive value for 5-year survival of KIRC and performed with “survivalROC” package [13] in R.
lncRNA-miRNA-target network construction, target prediction and function analysis
The target genes of two miRNAs (miR-21 and miR-223) were predicted using miRNet [14] online analysis tools (http://www.mirnet.ca/faces/home.xhtml). The miRNet is an easy-to-use tool with comprehensive support for statistical analysis and functional interpretation of data generated in miRNAs studies. MiRNet offers networks on miRNA-gene and miRNA-long non-coding RNA (lncRNA) modifier. A lncRNA-miRNA-gene network was constructed. The network was further optimized using Cytoscape software [15] to improve visualization. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the target genes were performed using miRNet by Hypergeometric test algorithm.
Statistical analysis
We analyzed the expression levels of the miRNAs in KIRC and matched normal tissues by unpaired t tests. We performed Kaplan-Meier survival analysis with the log-rank method and univariate/multivariate Cox proportional hazard regression analysis to compare the expression (low vs. high expression levels) and prognostic significance (low-risk vs. high-risk) of each miRNA. We considered P-values < 0.05 to be statistically significant. Statistical analysis was performed using IBM SPSS statistics software program version 22.0 (IBM, North Castle, NY, USA).
Clinicopathological features of 516 KIRC.
| Characteristics | Case | Percentage |
|---|---|---|
| Sex | ||
| Male | 335 | 64.92% |
| Female | 181 | 35.08% |
| Age (years) | ||
| <60 | 238 | 46.12% |
| ≥60 | 278 | 53.88% |
| Pathologic stage | ||
| Stage I-II | 308 | 59.69% |
| Stage III-IV | 205 | 39.73% |
| Not known | 3 | 0.58% |
| T stage | ||
| T1+T2 | 326 | 63.18% |
| T3+T4 | 190 | 36.82% |
| Lymph node status | ||
| N0 | 228 | 44.19% |
| N1 | 17 | 3.29% |
| Nx | 271 | 52.52% |
| Metastatic | ||
| M0 | 406 | 78.68% |
| M1 | 78 | 15.12% |
| Mx | 30 | 5.81% |
| Not known | 2 | 0.39% |
| Grade | ||
| G 1-2 | 231 | 44.77% |
| G 3-4 | 277 | 53.68% |
| Not known | 8 | 1.55% |
Results
Differentially expressed miRNAs in KIRC and healthy tissues
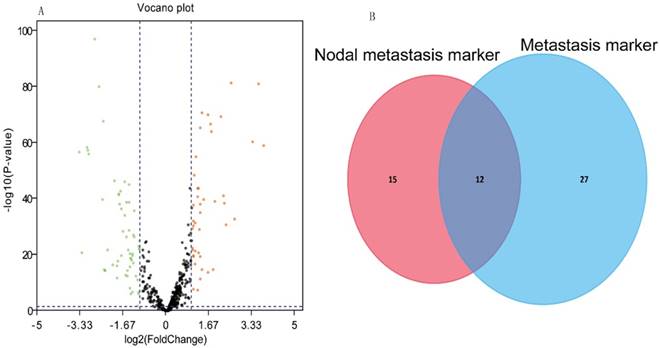
Our analysis included 516 KIRC tissues and 71 matched normal tissues. Detailed clinical characteristics, including gender, age at diagnosis, nodal metastasis, distant metastasis, pathological stage, nuclear grade, and tumor size (T stage), are listed in Table 1. As shown in Table 1, the nodal metastasis status could not be assessed (Nx) in more than 50% of 516 patients, and distant metastasis could not be assessed (Mx) in 6.59%. According to the cut-off criteria (P < 0.05 and |log2FC| > 1), 104 miRNAs were differentially expressed between KIRC and the matched normal tissues. 43 miRNAs were up-regulated in the KIRC tissue and 61 miRNAs that were down-regulated. The results of the expression analysis are presented as a volcano plot (Fig. 1A) to demonstrate that the distributions of P-values and |log2FC| were reasonable with respect to each other.
Diagnostic value for nodal metastasis, distant metastasis and predictive value for prognosis of differentially expressed miRNAs in KIRC
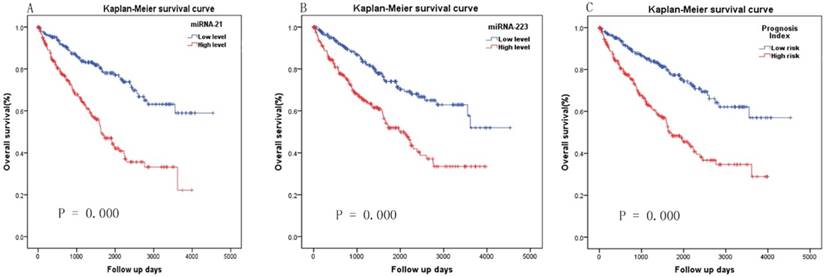
The ROC curve analysis showed 27 and 39 differential miRNAs that may diagnose nodal metastasis and distant metastasis of KIRC respectively (P<0.05). We created a Venn diagram (Fig. 1B) and found 12 overlapping miRNAs that could diagnose nodal metastasis and distant metastasis. For the 12 miRNAs, we used median as cutoff point to classify the 516 patients in two groups: high and low level. Patients were divided into two groups and subjected to univariate/multivariate Cox proportional hazard regression analysis. MiRNA-21 and miRNA-223 were shown to be independent prognostic indicators for KIRC (Table 2). MiR-21and miR-223 were up-regulated in KIRC. And we got the PI = 0.658*expmiRNA-21+0.601*expmiRNA-223. The Kaplan-Meier survival curve analysis of two miRNAs in overall survival were shown in Fig.2A-B.
Diagnostic value for nodal metastasis, distant metastasis and predictive value for 5-year-survival of two-miRNA signature Prognosis Index in KIRC
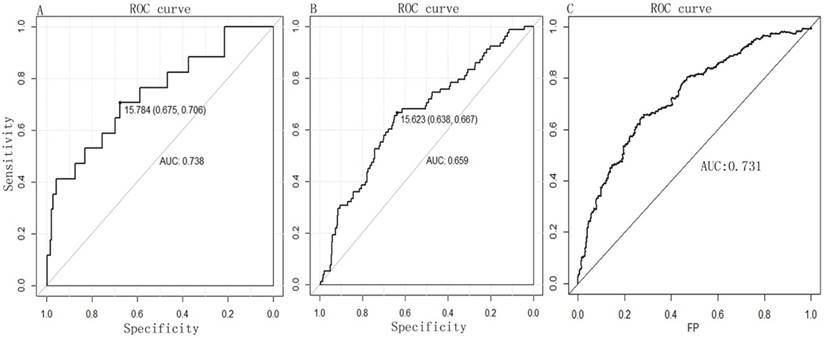
A PI score for each patient was calculated and ranked. Thus, the 516 patients were classified into a high- (n= 258) and low-risk (n= 258) groups, according to the median. Survival analysis using the Kaplan-Meier method with a Log-rank statistical test showed that patients in high-risk group have a significantly worse OS than patients in the low-risk group (P = 0.000, Fig. 2C). The diagnostic value of the two-miRNA signature PI for nodal metastasis, distant metastasis was evaluated with “pROC” package, and the results are shown in Fig. 3A-B. The predictive value of the two-miRNA signature PI for 5-year-survival rate was evaluated with “survivalROC” package, and the results are shown in Fig. 3C. The two-miRNA signature is a promising biomarker for diagnosing nodal metastasis, distant metastasis and predicting 5-year-survival rate of KIRC with AUCs=0.738, 0.659 and 0.731, respectively.
(A) Volcano plot of the differentially expressed miRNAs between KIRC and normal tissues. Red dots represent significantly up-regulated miRNAs, and green dots represent significantly down-regulated miRNAs (log2|FC|>1.0, P<0.05). (B) Venn analysis of miRNAs that could potentially diagnose nodal metastasis and distant metastasis.
Kaplan-Meier survival curve analysis of two miRNAs ang the two-miRNA signature in overall survival. (A) miRNA-21, (B) miRNA-223 and (C) two-miRNA signature prognosis index.
Univariate and multivariate analysis of 12 miRNAs in KIRC patients.
| miRNA | Univariate analysis | multivariate analysis | ||||
|---|---|---|---|---|---|---|
| P value | HR (95%CI) | β | P* value | HR (95%CI) | ||
| miR-155 | 0.059 | 0.989-1.808 | ||||
| miR-183 | 0.000 | 1.486 -2.777 | 0.344 | 0.059 | 0.987 -2.015 | |
| miR-193a | 0.011 | 1.095- 2.003 | -0.091 | 0.596 | 0.651 -2.280 | |
| miR-21 | 0.000 | 2.038 -3.876 | 0.658 | 0.003 | 1.256 -2.557 | |
| miR-223 | 0.000 | 1.632 -3.062 | 0.601 | 0.001 | 1.300- 1.606 | |
| miR-4677 | 0.018 | 1.064 -1.951 | 0.153 | 0.349 | 0.846 -1.266 | |
| miR-144 | 0.003 | 0.460-0.850 | -0.353 | 0.240 | 0.390-1.241 | |
| miR-451a | 0.003 | 0.459-0.849 | -0.383 | 0.210 | 0.375-1.241 | |
| miR-149 | 0.000 | 1.335-2.474 | 0.309 | 0.067 | 0.978-1.897 | |
| miR-204 | 0.000 | 0.363-0.677 | -0.105 | 0.607 | 0.604-1.342 | |
| miR-31 | 0.000 | 1.278-2.364 | 0.087 | 0.614 | 0.779-1.528 | |
| miR-200c | 0.405 | 0.841-1.533 | ||||
Abbreviations: KIRC, Kidney renal clear cell carcinoma; HR, hazard ratio;
CI, confidence interval. * p value of <0.05 was considered statistically significant difference.
The diagnostic value of two-miRNA for (A) nodal metastasis and (B) distant metastasis by ROC curve analysis. (C) The predictive value of the two-miRNA signature PI for 5-year-survival rate. FP, False Positive.
Univariate/multivariate analysis of clinicopathological features and two-miRNA signature.
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| P value | HR (95% CI) | β | P value | HR (95% CI) | ||
| Sex | ||||||
| Male | 0.608 | 0.676-1.258 | ||||
| Female | ||||||
| Age(years) | ||||||
| <60 | 0.000 | 1.367-2.581 | 0.353 | 0.110 | 0.983-2.337 | |
| ≥60 | ||||||
| Pathologic stage | ||||||
| Stage I-II | 0.000 | 2.715-5.146 | 0.276 | 0.560 | 0.555-3.379 | |
| Stage III-IV | ||||||
| T stage | ||||||
| T1+T2 | 0.000 | 2.251-4.145 | 0.191 | 0.650 | 0.569-2.976 | |
| T3+T4 | ||||||
| Nodal metastasis | ||||||
| N0 | 0.000 | 1.860-6.324 | 0.450 | 0.189 | 0.862-3.277 | |
| N1 | ||||||
| Distant metastatic | ||||||
| M0 | 0.000 | 3.081-5.762 | 0.960 | 0.000 | 1.355-3.633 | |
| M1 | ||||||
| Grade | ||||||
| G 1-2 | 0.000 | 1.877-3.748 | 0.363 | 0.125 | 0.896-2.453 | |
| G 3-4 | ||||||
| Two-miRNA signature PI | ||||||
| Low risk | 0.000 | 1.882-3.568 | 0.873 | 0.000 | 1.312-3.606 | |
| High risk | ||||||
Abbreviations: HR, hazard ratio; CI, confidence interval; PI, prognosis Index
* p value of <0.05 was considered statistically significant difference.
Comparison between the two-miRNA signatures that remained independent prognostic factors and routine clinicopathological factors
To test its predictive value for prognosis, we added the two-miRNA signature PI to other clinicopathological features for univariate and multivariate cox analyzes. The two-miRNA signature was found to be associated with OS and an independent prognostic factor in KIRC patients (Table 3). The results suggest that the two-miRNA signature could be used as a prognostic biomarker.
Apply the miRNA-based signature to diagnosis the patient with Nx and Mx respectively
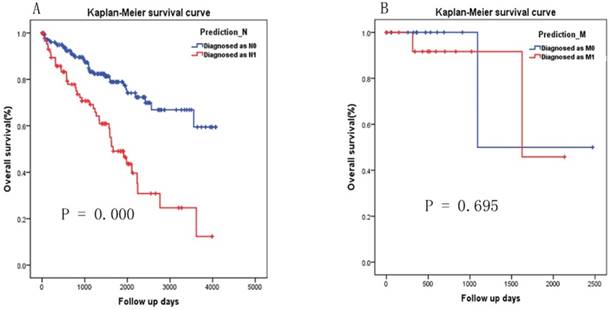
As there were 271 patients with Nx and 32 patients with Mx or not known, we applied the two-miRNA signature to diagnosis the patient with Nx and Mx respectively. According to the cutoff selected by ROC curve analysis (Fig. 3A), 182 and 89 patients were diagnosed as N0 and N1 respectively. Patients diagnosed as N1 have significantly poorer survival than patients diagnosed as N0 (Fig.4A, P=0.000). According to the cutoff selected by ROC curve analysis (Fig. 3B), 16 and 16 patients were diagnosed as M1 and M0 respectively. There was no significant difference in survival between patients diagnosed with M1 and those diagnosed with M0 (Fig. 4B, P=0.695).
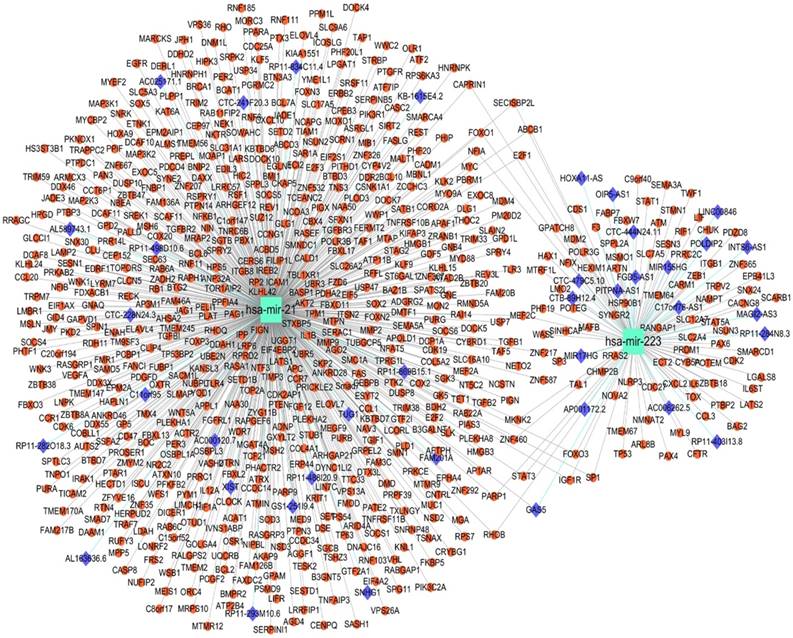
lncRNA-miRNA-target network constructed, target prediction and function analysis of the two-miRNA signature
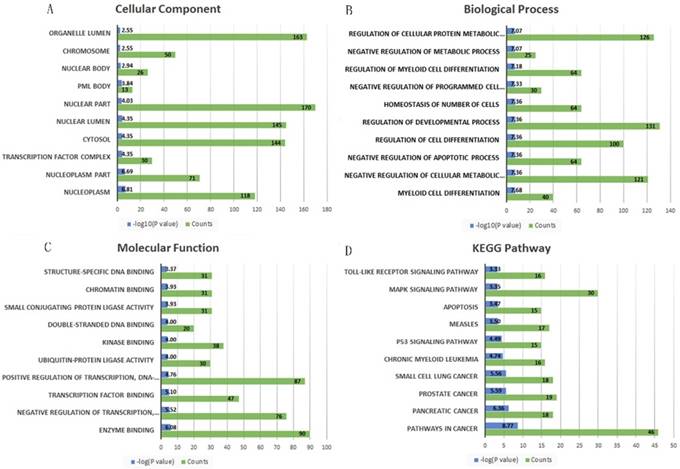
A total 695 genes and 38 lncRNAs were selected by using miRNet online analysis tools based on the two miRNAs (miRNA-21 and miRNA-223). The miRNA-gene network and lncRNA-miRNA network were exported and an lncRNA-miRNA-gene network was constructed by using cytoscape software. 748 edges and 735 nodes were included in the network (Fig. 5). GO and KEGG pathway enrichment analyses of the target genes were performed using miRNet with Hypergeometric test algorithm. GO enrichment analyses included biological process (BP), molecular function (MF), and cellular component (CC). The results of GO and KEGG pathway enrichment analysis were ranked by P value and the top 10 terms and pathways were showed in Fig. 5. The analysis suggested that the target genes of two miRNAs may be involved in various pathways related to cancer, like p53 signaling pathway, apoptosis, and MAPK signaling pathway. Table 4 shows the target genes of miRNA-21 and miRNA-223 in the relevant pathway.
Kaplan-Meier survival curve analysis of the two-miRNA signature's diagnosis results in patients with Nx and Mx. (A) Diagnosed as N0 versus N1, (B) Diagnosed as M0 versus M1.
lncRNA-microRNA-gene networks constructed on miRNet based on miRNA-21 and miRNA-223. Small red dots indicate genes, azure squares indicate miRNAs and blue diamonds indicate lncRNAs.
The target genes of miRNA-21 and miRNA-223 in the relevant pathways
| KEGG pathway | miRNA | targets |
|---|---|---|
| p53 signaling pathway | miRNA-21 | APAF1, FAS, CASP8, CCNG1, CDK6, MDM4, SERPINB5, PTEN, TNFRSF10B, SESN1 |
| miRNA-223 | ATM, CDK2, MDM2, TP53, SESN3 | |
| Apoptosis | miRNA-21 | AKT2, APAF1, FAS, FASLG, BCL2, CASP8, IL1B, IRAK1, MYD88, NFKB1, PIK3R1, TNFRSF10B |
| miRNA-223 | ATM,TP53,CHUK | |
| MAPK signaling pathway | miRNA-21 | FASLG, ATF2, DAXX, DUSP8, EGFR, FGF12, MKNK2, IL1B, MEF2C, MAP3K1, MYC, NFKB1, NTF3, MAP2K3, RASA1, RPS6KA3, TGFB1, TGFB2, TGFBR2, RASGRP1, MAP3K2, DUSP10, RASGRP3 |
| miRNA-223 | STMN1,CACNG8, RRAS2, CHUK, TP53, MEF2C, MKNK2 | |
| Cell cycle | miRNA-21 | E2F1, CDC25A, CDK6, E2F2, E2F3, MYC, ORC4, RB1, SKP2, TGFB1, TGFB2, SMC1A, STAG2 |
| miRNA-223 | CDC27, ATM, MDM2, TP53, CDK2, E2F1 |
The top 10 significantly enriched Gene Ontology annotation and Kyoto encyclopedia of genes and genomes pathway analysis of the target genes of the two miRNAs. (A) cellular component, (B) biological process, (C) molecular function and (D) KEGG pathway.
Discussion
KIRC patients with stage I disease have a 5-year recurrence free-survival of > 92%, whereas the risk of recurrence for those with stage II and III disease is up to 40% [16, 17]. Several models have been developed to predict outcomes after surgery. The Leibovich score classified patients with KIRC into low-, intermediate-, or high-risk categories, according to stage, tumor size, nuclear grade, and the presence of tumor necrosis [6, 7]. The University of California, Los Angeles, Integrated Staging System (UISS) defines low-, intermediate-, and high-risk prognostic groups based on stage, Fuhrman nuclear grade, and Eastern Cooperative Oncology Group performance status [8]. Both models require explicit status of nodal metastasis and metastasis, but patients with Nx or Mx account for a considerable portion and that do limit the application of the two models. In addition, the status of nodal metastasis and distant metastasis are two factors of prognosis. In just a few short years miRNAs have become firmly established as key molecular components of the cell in both normal and pathologic states [18]. Tumor miRNA profiles can define relevant subtypes, patient survival, and treatment response [19-21]. In the present study, we comprehensively analyzed the miRNA sequencing data downloaded from TCGA datasets. We identified 104 differentially expressed miRNAs, of which 43 were upregulated and 61 were down-regulated. We evaluated the diagnostic value of each differentially expressed miRNA for nodal metastasis and distant metastasis then developed a two-miRNA signature with diagnostic value for these two clinicopathological features. Further, the two-miRNA signature, as an independent factor of prognosis, successfully separated patients into low- and high-risk groups. The results suggest that the two-miRNA signature could be used as a prognostic biomarker of KIRC prognosis. In addition, the diagnosis for patients with Nx based on our two-miRNA signature, the overall survival rate of patients diagnosed as N1 was significantly poorer than that of patients diagnosed as N1. It did not obtained similar results in patients with Mx or not known because of the small sample size (n=32) or the lower diagnostic value of the two-miRNA signature for distant metastasis (AUC=0.659).
Several studies had performed to investigate the relationship between miRNA expression patterns and the prognosis of patients with KIRC. Cairns et al. found a consistent signature that included significant upregulation of miRNA-21-5p, miRNA-142-3p, let-7g-5p, let-7g-5p and miRNA-424-5p, as well as downregulation of miRNA-204-5p, to be associated with KIRC of high stage, or high grade, or progression [22]. Yann Christinat's study unveiled a KIRC-specific five-miRNA (miRNA-10b, miRNA-21, miRNA-143, miRNA-183, and miRNA-192) signature that predicted KIRC outcome more accurately than TNM staging alone using a computational approach [23]. Wang et al. reported a three-microRNA (miRNA-21, miRNA-584, and miRNA-155) signature as a diagnostic and prognostic marker in KIRC [24]. One of their miRNAs, miRNA-21 overlap with our two-miRNA signature. The different results of these studies may result from differences in the different main purpose and methods of these studies. One advantage of our present study, compared with previous studies, is that our signature is a leaner one containing only two miRNAs, another advantage of our present study is the two-miRNA signature could not only assess important pathological features but also predict survival of KIRC. This two-miRNA signature could act as a new effective tool for clinicians dealing with patients with KIRC, especially, the patients in whom the status of nodal metastasis and distant metastasis cannot be assessed by routine examination.
Increasing evidence shows that miRNA-21 acts as an oncogene by targeting many tumor suppressor genes related to proliferation, apoptosis, and invasion [25, 26]. miRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in KIRC[27], and up-regulation of miRNA-21 correlated with lower kidney cancer survival[28], which is similar with our results. As for miRNA-223, Wang et al. reported that miRNA-223 promotes the biological behavior of prostate cancer by targeting SEPT6 [29], while its molecular mechanism in KIRC has few studies. Zhang et al. reported miR-223 could enhance radiation sensitivity of U87MG cells in vitro and in vivo by targeting ataxia telangiectasia [30]. MiRNA-223 is highly expressed in KIRC and may serve as a potential therapeutic target, but its role needs further study. In our current study, the lncRNA-miRNA-gene network was constructed, which would help us and other researchers to explore KIRC further and deeper. The results of GO and KEGG pathway enrichment analysis suggested that the target genes of two miRNAs may be involved in various pathways related to cancer, included p53 signaling pathway [31], apoptosis [32], and MAPK signaling pathway [33]. Our study suggests that two miRNAs are potential predictors of nodal metastasis and distant metastasis, suggesting that these two miRNAs may be related to tumor metastasis. In particular, there are few studies about miRNA-223 in KIRC. Our results suggested that miRNA-223 may play a role in pathways related to cancer by targeting ATM, CDK2, MDM2, TP53, SESN3, CHUK, MKNK2, E2F1, CDC27, STMN1, CACNG8, MEF2C and MEF2C. Further experimental verification is needed.
Although cross-validation was lack in our present study, a two-miRNA signature successfully was constructed in a relatively large sample size. Since we did not find data sets that meet the inclusion criteria in other databases, we regret that we could not add more samples or use an independent external data set to validate our results. Future studies with independent cohorts of large samples from different sample types are needed to validate our findings for clinical practice. The main weakness for our study is lack of direct experimental validation, further functional investigation is required to explore the molecular functions of the two miRNAs in KIRC. Based on our present study, our research team is considering collecting samples in our hospital for validation and further research.
In summary, our study identified a two-miRNA signature from differentially expressed miRNAs in KIRC and matched normal tissues. The two-miRNA signature could not only assess nodal metastasis and distant metastasis. It also predicted survival in KIRC. Ultimately, we hope that this miRNA signature can be used as a new prognostic tool for clinicians dealing with patients with KIRC, especially those patients for whom the status of nodal metastasis and metastasis was not clear.
Acknowledgements
This study was supported by the Key Research and Development project of Guangxi (Grant No. Guike AB17195002), Guangxi Natural Science Foundation (Grant No. 2017GXNSFAA198249), the Scientific Research Funding from Population and Family Planning Commission of Guangxi Zhuang Autonomous Region (Grant No. S2017009), the funding from Department of Education of Guangxi Zhuang Autonomous Region (2017JGA159), the Talents Highland of Emergency and Medical Rescue of Guangxi Zhuang Autonomous Region.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A. et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67:85-97
3. Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089-100
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
5. Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK. et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-9
6. Pichler M, Hutterer GC, Chromecki TF, Jesche J, Kampel-Kettner K, Rehak P. et al. External validation of the Leibovich prognosis score for nonmetastatic clear cell renal cell carcinoma at a single European center applying routine pathology. J Urol. 2011;186:1773-7
7. Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED. et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97:1663-71
8. Patard JJ, Kim HL, Lam JS, Dorey FJ, Pantuck AJ, Zisman A. et al. Use of the University of California Los Angeles integrated staging system to predict survival in renal cell carcinoma: an international multicenter study. J Clin Oncol. 2004;22:3316-22
9. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-9
10. Chandran UR, Medvedeva OP, Barmada MM, Blood PD, Chakka A, Luthra S. et al. TCGA Expedition: A Data Acquisition and Management System for TCGA Data. PLoS One. 2016;11:e0165395
11. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47
12. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77
13. Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337-44
14. Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135-41
15. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-504
16. Janowitz T, Welsh SJ, Zaki K, Mulders P, Eisen T. Adjuvant therapy in renal cell carcinoma-past, present, and future. Semin Oncol. 2013;40:482-91
17. Stephenson AJ, Chetner MP, Rourke K, Gleave ME, Signaevsky M, Palmer B. et al. Guidelines for the surveillance of localized renal cell carcinoma based on the patterns of relapse after nephrectomy. J Urol. 2004;172:58-62
18. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515-24
19. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
20. Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A. et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378-82
21. Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387-99
22. Gowrishankar B, Ibragimova I, Zhou Y, Slifker MJ, Devarajan K, Al-Saleem T. et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15:329-41
23. Christinat Y, Krek W. Integrated genomic analysis identifies subclasses and prognosis signatures of kidney cancer. Oncotarget. 2015;6:10521-31
24. Liang B, Zhao J, Wang X. A three-microRNA signature as a diagnostic and prognostic marker in clear cell renal cancer: An In Silico analysis. PLoS One. 2017;12:e0180660
25. Mlcochova H, Machackova T, Rabien A, Radova L, Fabian P, Iliev R. et al. Epithelial-mesenchymal transition-associated microRNA/mRNA signature is linked to metastasis and prognosis in clear-cell renal cell carcinoma. Sci Rep. 2016;6:31852
26. Liang T, Hu XY, Li YH, Tian BQ, Li ZW, Fu Q. MicroRNA-21 Regulates the Proliferation, Differentiation, and Apoptosis of Human Renal Cell Carcinoma Cells by the mTOR-STAT3 Signaling Pathway. Oncol Res. 2016;24:371-80
27. Cao J, Liu J, Xu R, Zhu X, Liu L, Zhao X. MicroRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in clear cell renal cells. Mol Med Rep. 2016;13:75-82
28. Zaman MS, Shahryari V, Deng G, Thamminana S, Saini S, Majid S. et al. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7:e31060
29. Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu Z. et al. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. 2014;4:7546
30. Liang L, Zhu J, Zaorsky NG, Deng Y, Wu X, Liu Y. et al. MicroRNA-223 enhances radiation sensitivity of U87MG cells in vitro and in vivo by targeting ataxia telangiectasia mutated. Int J Radiat Oncol Biol Phys. 2014;88:955-60
31. Fukawa T, Ono M, Matsuo T, Uehara H, Miki T, Nakamura Y. et al. DDX31 regulates the p53-HDM2 pathway and rRNA gene transcription through its interaction with NPM1 in renal cell carcinomas. Cancer Res. 2012;72:5867-77
32. Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87
33. Huang D, Ding Y, Luo WM, Bender S, Qian CN, Kort E. et al. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res. 2008;68:81-8
Author contact
![]() Corresponding authors: Jianfeng Zhang, The Second Affiliated Hospital of Guangxi Medical University, No 166 Daxuedong Road, Nanning, Guangxi 530007, People's Republic China. Office number: 867713277199; Fax: 867713277285; Email: drzhangjfcom and Dr. Zhuoqing Lei, The Second Affiliated Hospital of Guangxi Medical University, No 166 Daxuedong Road, Nanning, Guangxi 530007, People's Republic China. Office number: 867713277199; Fax: 867713277285; Email: l_zhuoqingcom
Corresponding authors: Jianfeng Zhang, The Second Affiliated Hospital of Guangxi Medical University, No 166 Daxuedong Road, Nanning, Guangxi 530007, People's Republic China. Office number: 867713277199; Fax: 867713277285; Email: drzhangjfcom and Dr. Zhuoqing Lei, The Second Affiliated Hospital of Guangxi Medical University, No 166 Daxuedong Road, Nanning, Guangxi 530007, People's Republic China. Office number: 867713277199; Fax: 867713277285; Email: l_zhuoqingcom

 Global reach, higher impact
Global reach, higher impact