3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(20):3690-3698. doi:10.7150/jca.27263 This issue Cite
Research Paper
The prognostic value of long noncoding RNA ZEB1-AS1 on clinical outcomes in human cancer
1. Department of Respiratory Medicine, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, Jiangsu 210009, China
2. Department of Respiratory Medicine, Jinling Hospital, Nanjing University School of Medicine, Nanjing, Jiangsu 210002 , China
Received 2018-5-15; Accepted 2018-7-25; Published 2018-9-8
Abstract
Background: Although growing evidence have demonstrated that long non-coding RNA ZEB1-AS1 was aberrantly expressed in various types of cancers and can be used as a prognostic marker in cancer, the results remain inconclusive. Therefore, we performed this meta-analysis to evaluate the prognostic value of ZEB1-AS1 in human cancer.
Methods: A literature survey was conducted for all eligible studies by searching the following online databases: PubMed and Embase. The pooled odds ratios (ORs) or hazard ratios (HRs) with a 95 % confidence interval (95 % Cl) were computed to demonstrate its prognostic value.
Results: A total of 14 studies with 1096 individuals were included to evaluate the association of ZEB1-AS1 with clinicopathological features and overall survival (OS). In the pooled analyses stratified by clinicopathological features, ZEB1-AS1 expression was significantly related to depth of tumor (OR=2.92, 95% CI 1.22-7.02), poor histological differentiation (OR=2.72, 95% CI: 1.92-3.86), lymph node metastasis (OR=3.93, 95% CI: 2.65-5.84), distant metastasis (OR=5.34, 95% CI: 2.85-10.02) and tumor stage (OR=2.46, 95% CI 1.42-4.24), but not to tumor size (OR=1.25, 95% CI 0.79-1.96). Altered ZEB1-AS1 expression was found to be an indicator of worse prognosis in OS (HR = 1.94, 95% CI: 1. 66-2.22) among tumor patients.
Conclusions: High ZEB1-AS1expression was associated poor clinical outcome and it can serve as a novel predictive biomarker in various cancers.
Keywords: long non-coding RNA, ZEB1-AS1, prognostic biomarker, cancer
Introduction
In 2018, approximately 1,735,350 new cancer cases and 609,640 cancer deaths were reported in the United States [1]. Despite tremendous efforts to reduce deaths due to cancer, Cancer has been most deadly diseases worldwide. It is thus required for identify more effective biomarkers for cancer prognosis.
Long non-coding RNAs (lncRNAs) have no or little protein-coding potential with more than 200 nt in length [2]. Compared to protein-coding genes, lncRNAs exhibit typically higher degree of spatio-temporal specificity [3]. Collecting evidences suggests that lncRNAs are aberrant expressed in multiple human malignancies and has critical roles in regulating a variety of cellular processes, including cell apoptosis, proliferation and,invasion [4-6]. Moreover, several lncRNAs has been identified as sensitive prognostic biomarkers for cancers, such as TSLNC8 for hepatocellular carcinoma and HOTAIR for glioblastoma multiforme [7, 8]. Zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1) is a noncoding antisense RNA located on chromosome 10 which is in the opposite direction of ZEB1 gene transcription. A number of studies have reported the oncogenic function of ZEB1-AS1 mainly to be mediated through its gene silencing activity of ZEB1. In hepatocellular carcinoma, higher ZEB1-AS1expression is known to be more associated metastasis through ZEB1 expression and poor patient survival [9]. It has also been shown recently that ZEB1-AS1 plays a role in the proliferation and migration of osteosarcoma cells [10]. Moreover, increasing evidence has shown that ZEB1-AS1 is dysregulated in a variety of human cancers, and this dysfunctional expression is associated with the survival and clinicopathologic characteristics in cancer patients. However, due to small sample size and/or methodological limitations, the prognostic significance of ZEB1-AS1 expression in cancer remains to be elucidated. Therefore, it is essential to highlight the important role of ZEB1-AS1 in cancer base on larger samples.
In the present study, we carried out a meta-analysis of all relevant publications to exploit the prognostic effect of ZEB1-AS1 in cancer. Levels of ZEB1-AS1 were closely correlated with increased likelihood of tumor differentiation, depth of invasion, lymph node metastasis, distant metastasis, and tumor stage but not tumor size in cancer. Furthermore, Overall results confirm a correlation between increased ZEB1-AS1 expression and poor overall survival (OS).
Materials and Methods
Search strategy
A literature search was performed in the electronic databases of PubMed and Embase. The latest search was applied on June 13, 2018. Literature search terms were included (“ZEB1-AS1” OR "lncRNA ZEB1-AS1") AND (tumor OR cancer OR neoplasm OR carcinoma). Only English language publications were included in this study.
Study selection criteria
The following criteria for study eligibility were set before collecting articles: (1) associations of ZEB1-AS1 expression levels with clinicopathological features or survival in multiple human tumors were described; or (2) patients were grouped into two groups based on high and low expression levels of ZEB1-AS1 in human tumor tissue. (3) expression levels of ZEB1-AS1 in the patients were measured by qRT-PCR.
The following criteria were used to exclude studies from this meta-analysis: (1) reviews, letters, expert opinions, case reports, commentaries and editorials; (2) studies without original data; (3) duplicate articles; (4) sample cases fewer than 30; (5) non-human studies.
Data extraction and methodological assessment
Two researchers independently evaluated study eligibility. Disagreements were resolved by with a third researcher. The following data were retrieved: first author's surname, publication year, country of origin, tumor type, sample size, ZEB1-AS1 expression detection method, cut-off value, outcome measures, hazard ratio (HR) and its corresponding 95% confident interval (CI), the number of patients with lager tumor size, depth of tumor invasion, histological differentiation, lymph node metastasis, distant metastasis and tumor stage.
Statistical methods
Pooled odds ratios (ORs) or hazard ratios (HRs) with 95 % confidence intervals (CIs) were calculated to determine ZEB1-AS1 as a prognostic biomarker in human cancer. The heterogeneity across the included studies was measured by the Chi squared (χ2)-based Q test [11]. P < 0.1 for the Q test (Ph) and I2 > 50% indicated existence of significant heterogeneity among studies. The fixed effects model was used to analyze the pooled results when the included studies showed heterogeneity [12]. Otherwise, the random effects model was used [13]. Also, the potential publication bias was assessed by the Begg's funnel plot [14]. For the eligible studies that provided both the univariate and multivariate test, the multivariate values were preferred as the multivariate values had higher precision on interpreting covariates. When studies only report Kaplan-Meier curves, the Enguage Digitizer tool (Version 4.1) was used to digitize the survival data from the reported curves. All the P-values were determined by a 2-tailed t test and P<0.05 was representative of a statistically significance. The STATA version 12.0 software (Stata Corporation, College Station, TX) was used to perform all statistical analyses.
Results
Characteristics of the included studies
As shown in Figure 1, a total of fourteen articles were included in the current meta-analysis [9, 15-27]. Additionally, Table 1 summarized the main characteristics of the fourteen studies included in the present meta-analysis. All fourteen studies were conducted in from China, and addressed nine different tumor types: three focused on colorectal cancer, three focused on gastric cancer, one focused on osteosarcoma, one focused on hepatocellular carcinoma, one focused on bladder cancer, one focused on prostate cancer, two focused on glioma, one focused on leukemia and one focused on esophageal squamous cell carcinoma (ESCC). In these included literatures, the level of ZEB1-AS1 was detected in patient's tumor tissue by RT-PCR; and the negative control was adjacent normal tissue. The reference gene of ZEB1-AS1 in these studies was found to be inconsistent, including GAPDH and β-actin.
The flow diagram of the meta-analysis
Association of ZEB1-AS1 expression with tumor size
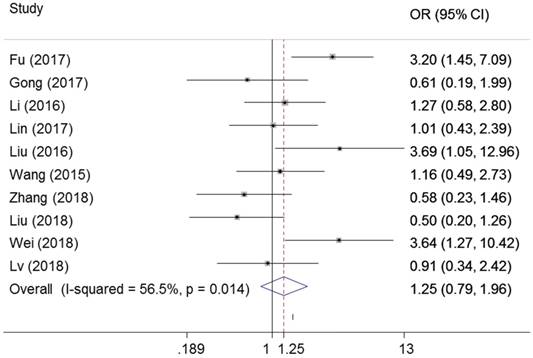
As shown in Figure 2, ten studies examined the association between ZEB1-AS1 and tumor size in six types of cancer. Significant heterogeneity was found among the studies (I²=56.5%, PQ =0.014) and then the random-effects model was applied. No significant connection was found between a high ZEB1-AS1 expression level and high tumor size in cancer patients (OR=1.25, 95%CI 0.79-1.96, P=0.339) (Figure 2).
Association of ZEB1-AS1 expression with depth of invasion
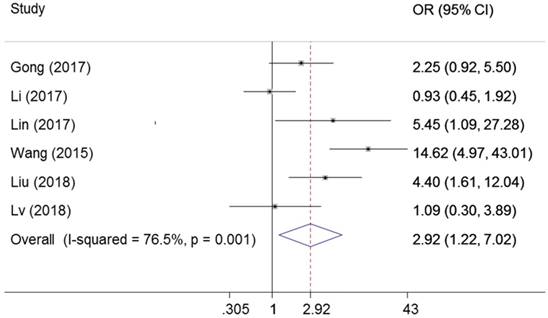
Six studies explored the association between ZEB1-AS1 expression levels with depth of invasion. The random-effects model was used for the significant heterogeneity (I2=76.5%, PQ =0.001). AS presented in Figure 3, the results indicated that the expression of ZEB1-AS1 in cancer was associated with depth of invasion (OR=2.92, 95% CI: 1.22-7.02, P=0.016).
Association of ZEB1-AS1 expression with tumor differentiation
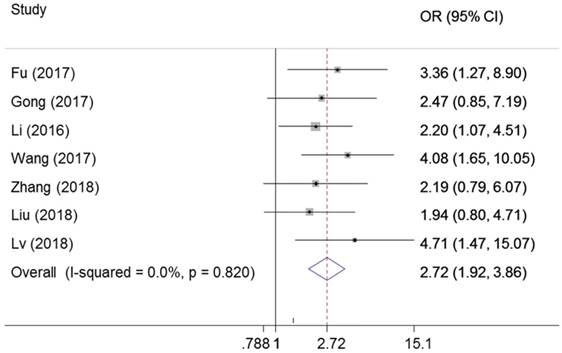
There were seven studies reported the association between ZEB1-AS1 expression levels and tumor difference. The fixed-effects model was adopted due to no significant heterogeneity among the studies (I2=0%, PQ = 0.82). The results of the forest plot demonstrated that patients with higher expression of ZEB1-AS1 in the tumor tissues may have increased probability of poor differentiation (OR = 2.72; 95% CI: 1.92-3.86, P = 0.000) (Figure 4).
Characteristics of studies in this meta-analysis
| ZEB1-AS1 expression | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | |||||||||||||||||||||
| Author | Year | Country | Cancer type | Sample size | Total | LTS | DI | PD | LNM | DM | HTS | Total | LTS | DI | PD | LNM | DM | HTS | Reference gene | Cutoff | HR statistic | OS (95% CI) |
| Fu | 2017 | China | CRC | 108 | 54 | 38 | _ | 47 | _ | 24 | 22 | 54 | 23 | _ | 36 | 8 | 11 | GAPDH | 2.58 | Data in paper | 2.026(1.493-2.748) | |
| Gong | 2017 | China | CRC | 63 | 31 | 6 | 25 | 14 | 24 | _ | 20 | 32 | 9 | 16 | 8 | 13 | _ | 24 | GAPDH | median | Survival curve | 1.77(1.08-2.91) |
| Li | 2016 | China | HCC | 102 | 51 | 23 | _ | 39 | _ | _ | 13 | 51 | 20 | 27 | _ | 11 | β-actin | median | Survival curve | 1.6(1.09-2.36) | ||
| Li | 2017 | China | GC | 124 | 62 | _ | 24 | _ | 46 | 10 | 49 | 62 | _ | 25 | _ | 20 | 0 | 22 | GAPDH | median | Data in paper | 2.363 (1.410-3.962) |
| Lin | 2017 | China | BC | 55 | 37 | 23 | 15 | _ | 2 | _ | 18 | 11 | 2 | _ | 0 | _ | NM | NM | NM | NM | ||
| Liu | 2016 | China | osteosarcoma | 50 | 25 | 12 | _ | _ | 6 | 8 | 25 | 5 | _ | _ | _ | 1 | 1 | GAPDH | median | NM | NM | |
| Lv | 2016 | China | Glioma | 82 | 29 | _ | _ | _ | _ | _ | 21 | 53 | _ | _ | _ | _ | 24 | GAPDH | NM | Data in paper | 1.885(1.068-3.326) | |
| Su | 2017 | China | PC | 114 | 57 | _ | _ | _ | _ | _ | 54 | 57 | _ | _ | _ | _ | _ | 33 | GAPDH | NM | NM | NM |
| Wang | 2017 | China | BL | 30 | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | GAPDH | median | Survival curve | 2.14(1.04-4.38) |
| Wang | 2015 | China | ESCC | 87 | 44 | 19 | 38 | 32 | 24 | _ | 43 | 17 | 13 | 17 | 10 | _ | _ | GAPDH | median | Data in paper | 2.371(1.284-6.115) | |
| Zhang | 2018 | China | GC | 76 | 38 | 13 | _ | 30 | 15 | _ | 25 | 38 | 18 | 24 | 6 | _ | 14 | GAPDH | median | Data in paper | 1.95(1.52-2.49) | |
| Liu | 2018 | China | GC | 75 | 42 | 17 | 33 | 29 | 34 | 31 | 33 | 19 | 15 | 13 | 20 | 12 | GAPDH | 4.5 | Data in paper | 2.28(1.109-4.689) | ||
| Wei | 2018 | China | Glioma | 65 | 34 | 11 | 31 | 23 | GAPDH | median | Data in paper | 2.983(1.189-4.739) | ||||||||||
| Lv | 2018 | China | CRC | 65 | 44 | 19 | 32 | 33 | 36 | 27 | 21 | 17 | 21 | 14 | 7 | 11 | GAPDH | NM | NM | NM | ||
CRC colorectal cancer, HCC hepatocellular carcinoma, GC gastric cancer, BC breast cancer, PC prostate cancer, BL bladder cancer, ESCC esophageal squamouscell carcinoma, LTS large tumor size, DI depth of invasion, PD poor differentiation, LNM lymph node metastasis, DM distant metastasis, HTS high tumor stage, NM not mentioned, HR hazard ratio, OS overall survival.
Forest plot for the association between ZEB1-AS1 expression levels with tumor size
Forest plot for the association between ZEB1-AS1 expression levels with tumor depth
Forest plot for the association between ZEB1-AS1 expression levels with histological differentiation.
Association of ZEB1-AS1 expression with lymph node metastasis
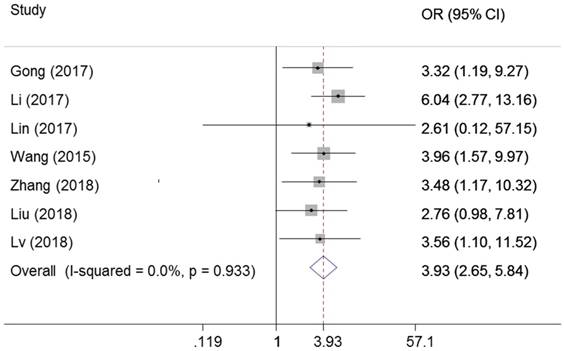
Seven studies examined the correlation of ZEB1-AS1 expression with lymph node metastasis in cancer. There was no heterogeneity among the studies (I2=0%, PQ = 0.933), and the fixed effects model was adopted. We observed significant association between ZEB1-AS1 expression and lymph node metastasis (OR=3.93, 95%CI: 2.65-5.84, P=0.00) (Figure 5).
Association of ZEB1-AS1 expression with distant metastasis
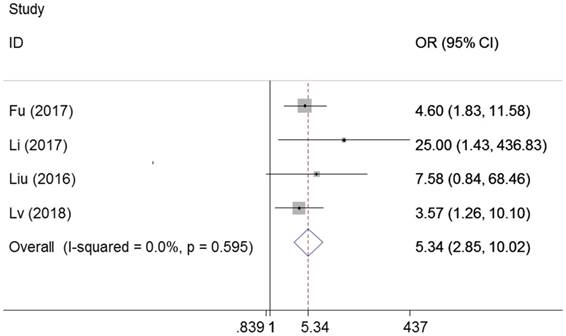
Four studies examined the association between metastasis and ZEB1-AS1 in different cancers, including colorectal cancer, gastric cancer and osteosarcoma. We found high ZEB1-AS1 expression group had a significant elevated distant metastasis incidence compared with low expression group (OR=5.34, 95%CI: 2.85-10.02, P=0.00). There was no heterogeneity among the studies (I2=0%, PQ =0.595) (Figure 6).
Association of ZEB1-AS1 expression with tumor stage
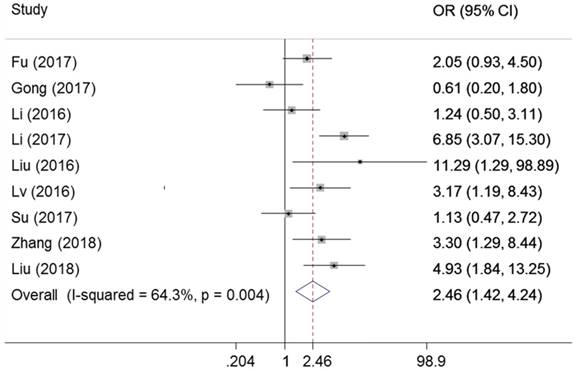
As presented in Figure 7, nine studies declared the association between the ZEB1-AS1 expression levels and tumor stage in cancer. There was significant heterogeneity in these studies, and the random-effects model was used (I2 =64.3%, PQ = 0.004). As a result, patients with a higher expression of ZEB1-AS1 were more liable to developing more advanced tumor stage. (OR=2.46, 95%CI 1.42-4.24, P=0.001)
Forest plot for the association between ZEB1-AS1 expression levels with lymph node metastasis.
Forest plot for the association between ZEB1-AS1 expression levels with distant metastasis.
Association between ZEB1-AS1 expression with survival
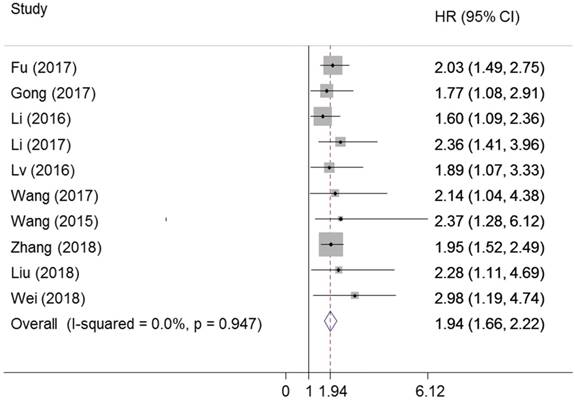
The prognostic value of ZEB1-AS1 in OS was assessed for ten studies with 812 patients. ZEB1-AS1 was significantly associated with OS (pooled HR: 1.94, 95% CI: 1.66-2.22, P=0.00) (Figure 8). There was no significant heterogeneity across the studies (I2=0% , PQ=0.947). This result manifested that a higher expression of ZEB1-AS1 might be correlated with a poorer OS in cancer patients, which demonstrated that ZEB1-AS1 was an independent factor for survival among patients with cancer.
Publication bias
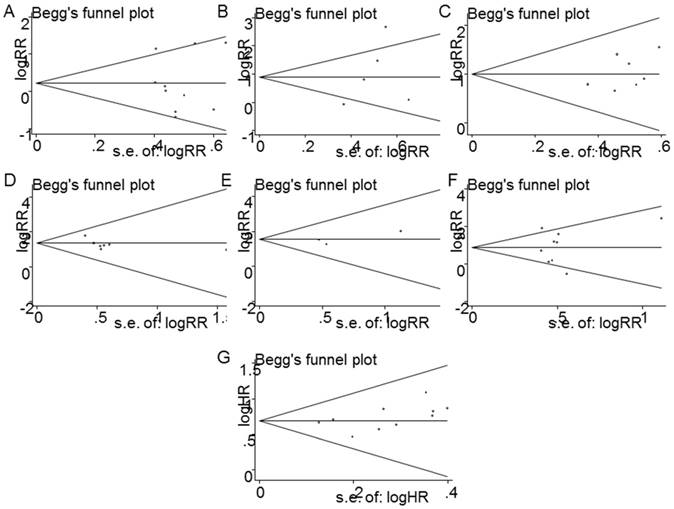
Begg's funnel plot was conducted to access the publication bias in the studies included in this meta-analysis. Figure 9 showed no publication bias for tumor size (P= 0.939) (Figure 9A), depth of invasion (P=0.306) (Figure 9B), differentiation (P= 0.274) (Figure 9C), lymph node metastasis (P=0.209) (Figure 9D), metastasis (P=0.114) (Figure 9E), TNM stage (P= 0.778) (Figure 9F), and OS (P= 0.183) (Figure 9G).
Forest plot for the association between ZEB1-AS1 expression levels with tumor stage.
Forest plot for the association between ZEB1-AS1 expression levels with OS.
Funnel plot analysis of potential publication bias in clinicopathological parameters group and survival. (A) tumor size, (B) depth of tumor, (C) histological grade, (D) lymph node metastasis, (E) distant metastasis, (F) tumor stage (G) OS
Discussion
Recent years, many studies have indicated that lncRNAs play important roles in gene expression and are widely involved in tumor progression and metastasis [28, 29]. It suggested these molecules could improve prognostic value by providing precise and valuable information about tumor phenotypes. ZEB1-AS1 is a novel noncoding antisense transcript in the opposite direction of ZEB1 gene, located on chromosome 10 with approximately 2535 nucleotides in length [21]. In the recent studies, the expression levels of ZEB1-AS1 are found to be abnormal in different types of tumors. Furthermore, it was a key mediator in cancer progression by promoting invasion and metastasis, including colorectal carcinoma, hepatocellular carcinoma, gastric cancer, prostate cancer, osteosarcoma, ESCC and leukemia [30]. Thus, this meta-analysis was performed to evaluate the association between ZEB1-AS1 expression with clinicopathological characteristics and survival in various cancers.
A total of fourteen eligible studies, addressing nine common cancer types, met the selection requirements for inclusion. The meta-analysis results indicated that high ZEB1-AS1 expression is significantly correlated with poor prognosis in patients with different types of cancers. The meta-analysis for the association between increased ZEB1-AS1 expression and clinicopathological parameters was also analyzed in this study, and our results showed that increased ZEB1-AS1 expression was significantly associated with depth of invasion, tumor differentiation, lymph node metastasis, distal metastasis and advanced clinical stage, which may suggest that increased ZEB1-AS1 may be associated with advanced features of cancer. Nevertheless, increased ZEB1-AS1 expression was not found to be associated with tumor size. Notably, the pooled HRs results demonstrated that elevated ZEB1-AS1 expression was positively associated with a shorter OS time. Collectively, these observations provide evidence that ZEB1-AS1 may serve as a potential independent predictive biomarker in cancer patients.
The molecular mechanisms underlying the association between ZEB1-AS1 altered expression and poor clinical outcome in cancer largely unknown. Several articles have suggested ZEB1-AS1 may contribute to migration and invasion of tumors through both positive and negative regulation of gene activity. It is a long known fact that upstream antisense transcription involved in transcriptional regulation of corresponding gene expression [31]. Since ZEB1-AS1 is an antisense transcript in the opposite direction of ZEB1 gene, the target gene expression of ZEB1-AS1 focus on ZEB1 which is a crucial modulator of epithelial-mesenchymal transition (EMT) program. For instance, it was first demonstrated that ZEB1-AS1 has a significantly increased the promoter activity of ZEB1 in hepatocellular carcinoma and gastric cancer [17, 32]. In osteosarcoma, Liu then reported that ZEB1-AS1 could recruit p300 to the ZEB1 promoter region and activate ZEB1 transcription [10]. In addition, Lv showed ZEB1-AS1 promoted invasion by inducing the ZEB1-EMT and regulated the proliferation through cyclin D /CDK2 in glioma [20]. Another underlying mechanism of lncRNAs as oncogenes is to sponge the tumor-suppressive microRNAs. Study from Su et al. showed that ZEB1-AS1 indirectly inhibited the tumor suppressor miR200c, and further regulated the expression of BMI1, the degradation of which was mediated by directly binding of miR200c [21]. In accordance with this finding is another report that the interplay between ZEB1-AS1 and miR-200s contributes to osteosarcoma cell proliferation and migration [33]. Moreover, ZEB1-AS1 was found to promote colorectal cancer cells proliferation and invasion via miR-101/ZEB1 axis [34]. Increasing evidences also showed that ZEB1-AS1 had functional roles on the activation of signaling pathway, such as ZEB1-AS1 IL-11/STAT3 pathway [22]. These evidences indicate that targeting ZEB1-AS1 may affects tumor progression and metastasis and might serve as a novel biomarker for the treatment and prognosis of cancer. Consistent with these mechanisms, we showed in this study that ZEB1-AS1 expression was significantly associated with depth of invasion, tumor differentiation, lymph node metastasis, distal metastasis, advanced clinical stage and a shorter OS. In contrast, increased ZEB1-AS1 expression was not correlated with tumor size. This discrepancy may arise from the heterogeneous molecular profiles embedded in the different types of cancers. Another reason may be that ZEB-AS1 contributes to cancer metastasis but not proliferation in some type of cancer [9]. Therefore, the role of ZEB1-AS1 in other non-studied types of cancer and the detailed mechanisms may be further investigated to confirm the role of ZEB1-AS1.
Some limitations still exist in our meta-analysis worth mentioning. First, the number of research, the type of cancers and the total sample size included was relatively small. Second, some HRs and their corresponding 95%CIs were calculated from survival curves, and may be less reliable than those directly obtained from studies. Third, publication bias may exist, despite the fact that no significant publication bias was observed based on funnel plot analysis. Finally, the cut-off values definition for high ZEB1-AS1 expression and the reference gene was not consistent among the included. Therefore, additional large-size, higher quality and multi-center studies with unified criteria for determining ZEB1-AS1 expression are necessary to validate the results in this study.
Taken together, the results of our meta-analysis indicate that the role ZEB-AS1 in prognosis of patients with different types of cancer. However, multi-center, well designed, larger-sample size, and higher-quality studies may be required to further solidify the clinical value of ZEB1-AS1 as prognostic cancer marker.
Abbreviations
OR: odds ratios; HR: hazard ratios; Cl: confidence interval; OS: overall survival; lncRNAs: long non-coding RNAs; ZEB1-AS1: Zinc finger E-box binding homeobox 1 antisense 1; ESCC: esophageal squamous cell carcinoma; CRC: colorectal cancer; HCC: hepatocellular carcinoma; GC: gastric cancer, BC: breast cancer, PC: prostate cancer, BL: bladder cancer.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 81702279).
Author Contributions
WY and LH contributed to the design of the study. WS, WT, XR collected the data and conducted the statistical analysis. WY contributed to the writing of the manuscript. LH and ZX contributed to the revision of the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. 2018;68:7-30.
2. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs.
3. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155-9
4. Grelet S, Link LA, Howley B. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105-15
5. Zhang S, Zhao BS, Zhou A. et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer cell. 2017;31:591-606.e6
6. Li C, Wang S, Xing Z. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106-19
7. Zhang J, Li Z, Liu L. et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171-87
8. Tan SK, Pastori C, Penas C. et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Molecular cancer. 2018;17:74
9. Li T, Xie J, Shen C. et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575-84
10. Liu C, Lin J. Long noncoding RNA ZEB1-AZS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. American journal of translational research. 2016;8:4095-105
11. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539-58
12. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22:719-48
13. DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemporary clinical trials. 2015;45:139-45
14. Egger M, Smith GD. Bias in location and selection of studies. BMJ (Clinical research ed). 1998;316:61-6
15. Fu J, Cui Y. Long noncoding RNA ZEB1-AS1 expression predicts progression and poor prognosis of colorectal cancer. The International journal of biological markers. 2017;32:e428-e33
16. Gong H, Wen H, Zhu X. et al. High expression of long non-coding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39:1010428317705336
17. Li Y, Wen X, Wang L. et al. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26:527-34
18. Lin J, Zhan Y, Liu Y. et al. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8:24202-12
19. Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. American journal of translational research. 2016;8:4095-105
20. Lv QL, Hu L, Chen SH. et al. A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. International journal of molecular sciences. 2016:17
21. Su W, Xu M, Chen X. et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Molecular cancer. 2017;16:142
22. Wang Q, Du X, Yang M. et al. LncRNA ZEB1-AS1 contributes to STAT3 activation by associating with IL-11 in B-lymphoblastic leukemia. Biotechnology letters. 2017;39:1801-10
23. Wang YL, Bai Y, Yao WJ. et al. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8:11871-6
24. Zhang LL, Zhang LF, Guo XH. et al. Downregulation of miR-335-5p by Long Noncoding RNA ZEB1-AS1 in Gastric Cancer Promotes Tumor Proliferation and Invasion. DNA Cell Biol. 2018;37:46-52
25. Liu XJ, Li SL, Li JS. et al. Long non-coding RNA ZEB1-AS1 is associated with poor prognosis in gastric cancer and promotes cancer cell metastasis. European review for medical and pharmacological sciences. 2018;22:2624-30
26. Wei N, Wei H, Zhang H. Long non-coding RNA ZEB1-AS1 promotes glioma cell proliferation, migration and invasion through regulating miR-577. European review for medical and pharmacological sciences. 2018;22:3085-93
27. Lv SY, Shan TD, Pan XT. et al. The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/beta-catenin signaling. Cell cycle (Georgetown, Tex). 2018:1-10
28. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452-63
29. Wang Z, Yang B, Zhang M. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer cell. 2018;33:706-20.e9
30. Li J, Li Z, Leng K. et al. ZEB1-AS1: A crucial cancer-related long non-coding RNA. Cell Proliferation. 2018:51
31. Wei W, Pelechano V, Jarvelin AI. et al. Functional consequences of bidirectional promoters. Trends in genetics: TIG. 2011;27:267-76
32. Li T, Xie J, Shen C. et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575-84
33. Liu C, Pan C, Cai Y. et al. Interplay Between Long Noncoding RNA ZEB1-AS1 and miR-200s Regulates Osteosarcoma Cell Proliferation and Migration. J Cell Biochem. 2017;118:2250-60
34. Xiong WC, Han N, Wu N. et al. Interplay between long noncoding RNA ZEB1-AS1 and miR-101/ZEB1 axis regulates proliferation and migration of colorectal cancer cells. American journal of translational research. 2018;10:605-17
Author contact
![]() Corresponding authors: Xiaoli Zhu Tel: +86 25 83262821; E-mail: zhuxiaoli62com and Hongbing Liu Tel: +8613852293363; E-mail: 101012112edu.cn or netlhbcom
Corresponding authors: Xiaoli Zhu Tel: +86 25 83262821; E-mail: zhuxiaoli62com and Hongbing Liu Tel: +8613852293363; E-mail: 101012112edu.cn or netlhbcom

 Global reach, higher impact
Global reach, higher impact