3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(20):3755-3764. doi:10.7150/jca.24253 This issue Cite
Research Paper
Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in Caski cells
1. Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, 510515, P.R. China
2. The First School of Clinical Medicine, Southern Medical University, Guangzhou, 510515, P.R. China
3. Department of Gynaecology and Obstetrics, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, P.R. China
Received 2017-12-7; Accepted 2018-4-14; Published 2018-9-27
Abstract
Cervical cancer is one of the most common cancer in female worldwide. The expression of high-risk human papillomavirus E7 oncogene is necessary for the maintenance of malignant phenotypes and transformation. Accumulating studies of this protein has been explored in cervical cancer, however, there are fewer studies on how E7 expression affects the expression of global circular RNA. CircRNA, a promising biomarker and even therapeutic target, has become a star molecular in research after miRNA and long non-coding RNA. Our aim of this study was to investigate the global circRNA levels modulated by HPV E7 expression and identified the potential consequences for mechanism studies. Here we investigated the expression profiles of circRNAs by transfecting E7 siRNA in Caski cells with high-throughput microarray technology. In total, we identified 526 dysregulated circRNAs with fold change ≥2 or≤0.5, and p< 0.05. Among them, 352 were up-regulated and 174 were down-regulated. In addition, 8 selected circRNAs confirmed using qRT-PCR was in line with the results of microarray analysis. Furthermore, bioinformatic analyses indicated that differently expressed circRNAs might implicate in the mTOR signaling pathway, proline metabolism and glutathione metabolism. In conclusion, this study showed the expression profiles of circRNAs regulated by HPV16 E7 in cervical cancer cells and provides novel insights into the new potential candidates for future mechanism studies.
Keywords: cervical cancer, circular RNA, microarray, E7 microRNA
Introduction
Cervical cancer is one of the most common malignant gynecological cancer in the world, with approximately 529,000 new cases and 275,000 deaths annually[1, 2]. Most of these cases occur in developing countries[2]. Therefore, it is significant to explore the mechanisms of tumorigenesis and development and find effective therapeutic strategies for prevention and cure of cervical cancer.
A large number of cervical cancers are persistent cervical squamous or glandular epithelium infection by the oncogenic human papillomaviruses (HPVs). The high-risk types HPV16 and HPV18 are the most common infections and accounted for more than 70% of all the cases[3]. The viral HPV E6 and E7 proteins are of crucial importance for the HPV-associated transforming properties. Moreover, E6 and E7 oncoproteins play important roles in cervical cancer development via dysregulating p53 and pRb pathways, respectively[4]. The E6 induces the degradation of p53 and inhibits p53-dependent signaling, thereby leading to tumorigenesis. E7 oncoprotein binds to pRb tumor-suppressor family[5, 6], and interacts with transcription factor EZH2, a member of the Polycomb Repressive Complex 2 (PRC2) that involves in S phase of cell cycle[7]. Additionally, E7 also can bind the inhibitors of cyclin-dependent kinases (CDK2), p21 and p27 to maintain high CDK2 activity, thus promoting cell cycle progression[8]. Indeed, the expression of HPV E6 and E7 contribute to the tumorigenesis and development as well as the phenotype in cervical cancer.
Circular RNA is a class of novel competing endogenous RNA that lacks the capacity of protein coding[9, 10]. Different from linear RNAs, circRNAs have a special circular covalently closed loop structure, which make them lack of 5' cap or 3' Poly A tail[11, 12]. Although circRNAs had been found for decades in eukaryotic cells, they were considered as transcriptional noise. Recently, with the use of high-throughput microarray screening, circRNAs have been identified in the tumorigenesis and development, which might function as miRNA sponges or decoys, modulate the expression of target genes and so on[13]. Although evidences indicated that circRNAs have a great potential to act as novel significant biomarkers for human cancers, little is known about circRNAs in oncogenesis even more in cervical cancer. For instance, Hongbo Wang and his colleagues performed profiles of long no-coding RNA, circRNA and miRNAs as well as mRNA in HPV 16 cervical cancer patients tissues with RNA sequencing[14]. Evidence also showed that hsa_circ_0018289 is up-regulated in cervical cancer tissues and promotes the tumorigenesis of cervical cancer[15]. However, no research has been reported circRNA related to HPV E7 oncogene both in cervical cancer tissues and cell lines so far.
In the present study, firstly, we investigated the expression profile of circRNAs in HPV16-positive Caski cells, upon silencing of endogenous E7 oncogene expression. We identified 526 aberrant expressed circRNAs, including 352 up-regulated and 174 down-regulated. Secondly, we selected 8 circRNAs and further validated by qRT-PCR, which might be functionally important in HPV16-positve cervical cancer cells. Additionally, we predicted miRNAs targets of circRNAs by TargetScan Human and miRBase. Finally, we investigated the potential canonical pathways to further clarify the mechanism of altered expression of circRNAs via gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Our findings might provide a new useful insight into the crucial question that viral E7 oncogenes can affect the global circRNA composition of HPV-positive cervical cancer cells.
Materials and Methods
Cell culture
The HPV16-positive Caski, SiHa and negative C33A human cervical cancer cell lines were purchased from The Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences, Shanghai, China. The cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, USA), supplemented with 10% fetal bovine serum (Hyclone, Utah, USA) and maintained at 37℃, in a humidified 5% CO2 atmosphere.
Plasmids and siRNA transfection
The full-length E7 human transcript cDNA was amplified by PCR and cloned into the pEGFP-C3 vector to generate an E7 overexpression vector, pEGFP-C3-E7. An empty pEGFP-C3 vector was used as control. The plasmid was purified by TaKaRa MiniBEST Plasmid Purification Kit (TaKaRa, Dalian, China). All constructs were sequenced for confirmation in Sangon Biotechnology Company.
The E7 siRNA and negative control (NC) were purchased from Shanghai GenePharma (Shanghai, China). The siRNA sequences were as follows: NC, 5'-UUCUCCGAACGUGUCACGU-3' (sense); 5'-ACGUGACACGUUCGGAGAA-3' (antisense); E7 siRNA_#1, 5'-AGGAGGAUGAAAUAGAUGG-3' (sense); 5'-CCAUCUAUUUCAUCCUCCU-3' (antisense). E7 siRNA_#2, 5'-GCUUCGGUUGUGCGUACAA-3' (sense); 5'-UUGUACGCACAACCGAAGC-3' (antisense).
Caski, SiHa and C33A cells were seeded in 6-well plates and cultured for 24 h. When growing 70%-80% confluence, Caski and SiHa cells were transfected with a mixture of siRNA and LipofectamineTM 2000 (Invitrogen, Newyork, USA); while C33A cells were transfected with a mixture of plasmid and LipofectamineTM 2000 (Invitrogen, Newyork, USA), according to the manufacturer's instructions. The final concentrations of plasmid and siRNA transfection were 2 μg and 40 μM, respectively. After transfection for 6 h, the mixture medium was replaced with complete medium, and at 48 h post-transfection, the cells were harvested for further experiments.
Total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was used to identify the E7 expression transfected by E7 siRNA and pEGFP-C3-E7 plasmid as well as confirm the differently expressed circRNAs obtained from the microarray data. Total RNA of Caski and SiHa as well as C33A cells were isolated with TRIzolTM reagent (TaKaRa, Dalian, China) following the manufacturer's protocol. The concentration of RNA was detected by the spectrophotometer of NanoDropTM ND-1000 (Applied Biosystems, CA, USA). Total RNA (≤500 ng) was reverse transcribed into cDNA using the PrimeScriptTMⅡ 1st Strand cDNA Synthesis kit (TaKaRa, Dalian, China). qRT-PCR was performed using the SYBR Premix Ex TaqTM Ⅱ Kit (TaKaRa, Dalian, China). The primers used for amplification were shown in Table 1. GAPDH was used as internal control. Each experiment was repeated in triplicate. All assays were performed using the ABI 7500 system (Applied Biosystems, CA, USA). The results were calculated by the method of 2-ΔΔCt.
The list of primers was used in our study
| Name | Primer sequence ( 5' - 3' ) |
|---|---|
| GAPDH | F: TCTCCTCTGACTTCAACAGC |
| R: GAAATGAGCTTGACAAAGTC | |
| E7 | F: CATGGAGATACACCTACATTGC |
| R: CACAACCGAAGCGTAGAGTC | |
| hsa_circ_0048867 | F: CCGAGAGCATGGTTGTCTTT |
| R: GTAGCAGCTTGTGGTTGACG | |
| hsa_circ_0038475 | F: TTCTTACATGCCACCATCCA |
| R: CCAATCAAAGCCATTCTTGC | |
| hsa_circ_0035918 | F: ACCGGACGATTCAGTTTCAC |
| R: ATTTCTTCCATGGTGCCAAA | |
| hsa_circ_0056353 | F: CCAGGAATGGGAATACCAGA |
| R: ACAYAYCCTCCGTGGTCTGC | |
| hsa_circ_0026527 | F: AGGAAAGCTGGCAAAAAGGA |
| R: CCGATACCCATCCCGATAC | |
| hsa_circ_0037213 | F: GACCTCAGCCCTGCTCACT |
| R: CTGCCGGCATTTCAGGAG | |
| hsa_circ_0051620 | F: ACCTGCTGTCACTCCAGAGG |
| R: ATGAAACGGCTGATGTGCTT | |
| hsa_circ_0052602 | F: TGCTGCCTCTACGTTCAATG |
| R: CAAAGGCAACTCTCCAGGCC |
Western blotting
After 72 h transient transfection, total proteins were isolated using RIPA lysis buffer (1g/mL leupeptin and 1g/mL PMSF; Beyotime, Guangzhou, China) following to the manufacturer's instructions. The lysate was centrifuged for 15 min at 12,000 rpm at 4℃. Protein samples (approximately 30 μg each) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel and transferred onto poly-vinylidene fluoride membranes (Millipore, Billerica, MA). Primary antibodies included mouse monoclonal anti-HPV16 E7 (Abcam, Cambridge, MA), β-actin (Neobioscience, Shenzhen, China). β-actin was used as internal control to normalize the relative levels of protein.
CircRNA microarray analysis
After 48 h transfection with E7 siRNA #2, total RNA of Caski was extracted using TRIZOL Reagent (Life technologies, Carlsbad, USA) following the manufacturer's instructions and checked for a RIN number to inspect RNA integrity by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, USA). Qualified total RNA was further purified by RNeasy mini kit (Qiagen, Hamburg, Germany) and RNase-Free DNase Set (Qiagen, Hamburg, Germany). Using RNase R to remove the linear RNAs and enrich the circRNAs. The enriched circRNAs were amplified and transcribed into fluorescent complementary RNA (cRNA) by Low Input Quick Amp WT Labeling Kit (Agilent technologies, Santa Clara, USA), following the manufacturer's instructions. Labeled cRNA were purified by RNeasy mini kit (Qiagen, Hamburg, Germany). In the experiment, each slide was hybridized with 1.65 μg Cy3-labeled cRNA using Gene Expression Hybridization Kit (Agilent technologies, Santa Clara, USA), according to the manufacturer's instructions. After 17 h hybridization, slides were washed in staining dishes with Gene Expression Wash Buffer Kit (Agilent technologies, Santa Clara, USA) followed the manufacturer's instructions. After washing, slides were scanned by Agilent Microarray Scanner (Agilent technologies, Santa Clara, USA) with default settings. Dye channel: Green, Scan resolution = 3 μm, PMT 100%, 20 bit. Data were extracted with Feature Extraction software 10.7 (Agilent technologies, Santa Clara, USA). Raw data were normalized by algorithm, limma packages in R.
Bioinformatics analysis
Using Agilent Feature Extraction software to analyze the acquired array images, and then using R software package to perform quantile normalization and subsequent data processing. Differently expressed circRNAs were performed by volcano plot filtering. Hierarchical clustering was shown the distinguishable expression patterns of circRNAs. Fold changes≥2, and P<0.05 were selected and considered as significantly differential expression between two groups.
miRNA target prediction and functional analysis
circRNAs screened from profile data interacted with mRNAs via combining with targeted miRNAs. The miRNAs prediction of dysregulated circRNAs were performed using the TargetScan Human 7.1 (http://www.targetscan.org/vert_7.1/) and miRBase release 20.0 (http://www.mirbase.org/). The top five targeted miRNAs interacted with circRNAs were selected for further analysis. Gene ontology (GO) and Kyoto encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the website: http://amp.pharm.mssm.edu/Enrichr/. DAVID is an integrated data program to analyze gene lists.
Statistical analysis
All experiments were performed at least three times. All data were expressed as mean ± S.E.M. Statistical analysis was performed using GraphPad Prism 5.0 (Graph Pad Software Inc., San Diego, CA, USA) and SPSS version 20.0 (IBM, San Jose, CA, USA). Paired t-test and student's t-test was used for data analysis. P<0.05 was considered statistically significant different.
Results
E7 siRNA transfection inhibits the expression of E7 oncogene
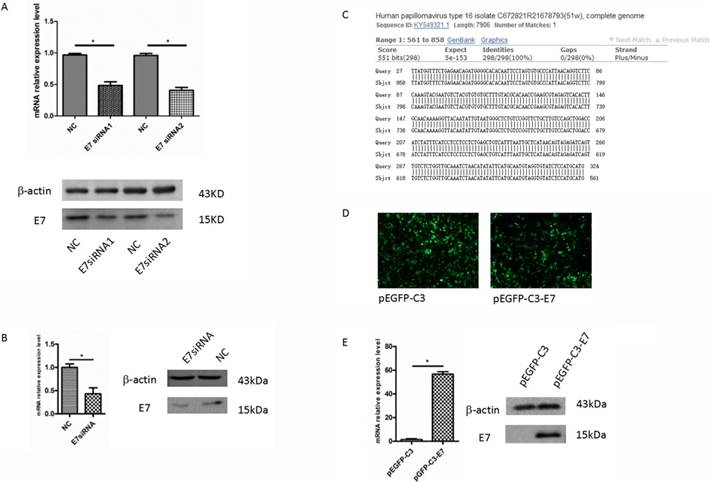
In Caski cells, after 48 h transfection with E7 siRNA1 and E7 siRNA2, E7 mRNA expression was determined by qRT-PCR, and the result showed that the level of E7 mRNA was significantly decreased as compared with that in negative control (P<0.05, Figure 1A). Western blot analysis revealed that E7 protein in siRNA group was significantly decreased, compared to that in the negative control (Figure 1A). The result was consistent with the qRT-PCR , which suggested that E7 siRNA could significantly repress the expression of E7 in Caski cells. Thus, we used these Caski cells to perform microarray analysis for the further study.
E7 overexpression in C33A cell line
The over-expression vector, pEGFP-C3-E7, was sequenced for confirmation in Sangon Biotechnology Company. The BLAST analysis showed that the original sequence was identical to the constructed sequence (Figure 1C). pEGFP-C3-E7 was transfected into HPV negative cervical cancer cell line C33A. After 48 h transfection, total RNA was isolated and E7 mRNA level was measured by qRT-PCR. The green fluorescent protein effect can directly reflect an effective transfection in C33A cells (Figure 1D). The qRT-PCR result showed that pEGFP-C3-E7 vector significantly up-regulated the E7 mRNA expression in C33A cells, as compared with that in negative control. Western blot analysis also showed that E7 protein was significantly up-regulated in pEGFP-C3-E7 group (Figure 1E), which indicated that pEGFP-C3-E7 vector can significantly increase the expression of E7.
Overview of circRNAs profile
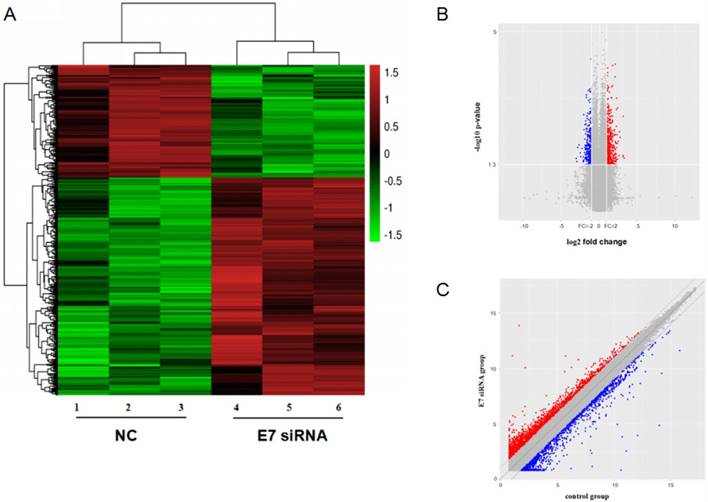
To investigate the expression of circRNAs, we transfected HPV16 positive Caski cells with E7 siRNA and performed the circRNAs microarray to collect the deregulated circRNAs. The hierarchical clustering showed that the relative levels of circRNAs expression were distinguishable.
The expression of E7 in cervical cancer cells. (A). The expression of E7 in Caski cells were significantly decreased by E7 siRNA compared with the negative control. (B). The expression of E7 in SiHa cells were significantly decreased by E7 siRNA compared with the negative control. (C). pEGFP-C3-E7 vectors were sequenced for confirmation in Sangon Biotechnology Company and the results were analyzed by BLAST. (D). The effects of pEGFP-C3/ pEGFP-C3-E7 vector in C33A cells. (E). pEGFP-C3-E7 vectors was significantly up-regulated the E7 expression level compared with the pEGFP-C3 in C33A cells. All data were expressed as mean ± S.E.M for three replicate determination. Student's t-test was used for data analysis, P<0.05 was considered statistically significant difference.
The scatter plot of circRNAs expression in microarray revealed significant variation between E7 siRNA group and NC group. And dysregulated expressed circRNAs also observed between two groups in volcano plot. There were 526 dysregulated expressed circRNAs between two groups (Figure 2). Among these circRNAs, 352 circRNAs were found to be up-regulated more than 2-fold change in Caski cells transfected with E7 siRNA as compared to that in negative control. In contrast, 174 circRNAs were identified to be down-regulated less than 0.5-fold change in E7 siRNA transfected cells.
Validation of circRNA expression
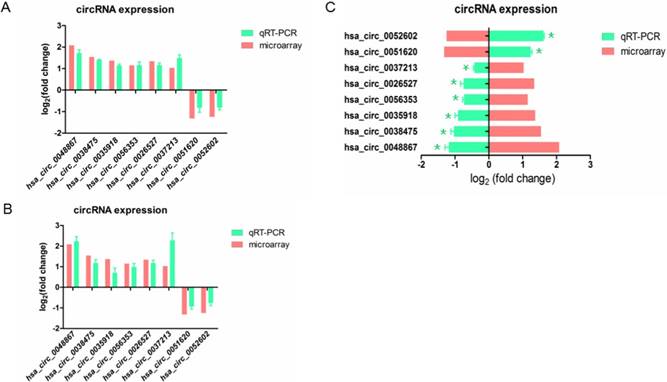
In order to confirm the result of microarray, we selected 8 differently expressed circRNAs including 6 up-regulated and 2 down-regulated circRNAs, and validated their expression by qRT-PCR.
To examine whether these aberrant circRNAs have similar change in other HPV16 positive cell lines, we knockdown E7 in SiHa cell (Figure 1B). The result revealed that when knockdown E7 in Caski or SiHa cells, hsa_circ_0048867, hsa_circ_0035918, hsa_circ_0038475, hsa_circ_0056353, hsa_circ_0026527, hsa_circ_0037213 were up-regulated, while hsa_circ_0051620, hsa_circ_0052602 were down-regulated (Figure 3A, 3B), showing no significantly differential expression of candidate circRNAs compared to microarray analysis.
We also transfected E7 over-expressed vector in C33A cell, we observed that hsa_circ_0048867, hsa_circ_0035918, hsa_circ_0038475, hsa_circ_0056353, hsa_circ_0026527, hsa_circ_0037213 were down-regulated, while hsa_circ_0051620, hsa_circ_0052602 were up-regulated (Figure 3C), the result was significantly opposite to the microarray analysis. In brief, the expressions of these 8 circRNAs validated by qRT-PCR were in accordance with the result of microarray analysis.
miRNA prediction that bind to circRNA
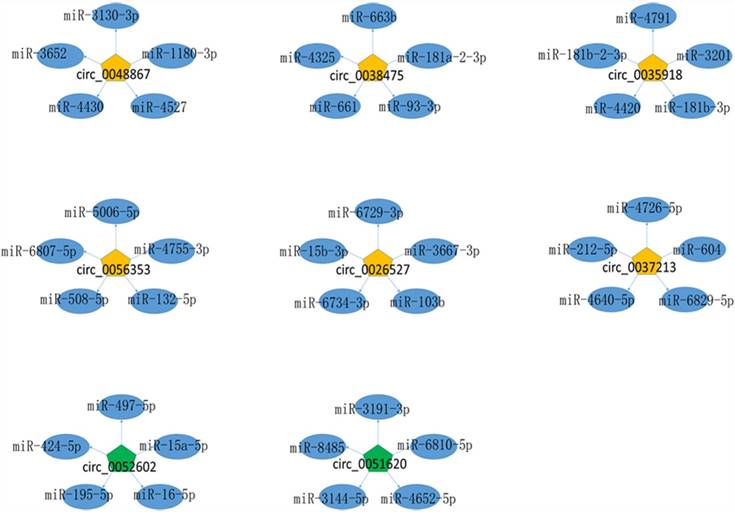
Given that circRNAs can function as miRNA sponges, we predicted the targeted miRNAs of dysregulated expressed circRNAs for the further study. The summary of 8 circRNAs validated by qRT-PCR were showed in Table 2 and the top five predicted target miRNAs/circRNA interaction networks were established and showed in Figure 4.
circRNA expression profile in HPV 16 E7 silencing cervical cancer Caski cells. (A). hierarchical clustering showed distinguishable circRNA expression between E7 siRNA group and NC group. Red represented up-regulated circRNA expression with high fold change, and green represented down-regulated circRNA expression with low fold change. (B). scatter plot revealed significant variation between two groups. (C). volcano plot showed differentially expressed circRNAs.
Validation of 8 differentially expressed circRNAs by qRT-PCR. (A). The expression of circRNAs transfected with E7siRNA in Caski cells were validated by qRT-PCR, compared with the microarray. (B). The expression of circRNAs transfected with E7siRNA in SiHa cells were validated by qRT-PCR, compared with the microarray. The Y-axis column represents the mean of fold change (log2 transformed) measured from microarray analysis and qRT-PCR. (C). The expression of circRNAs transfected with pEGFP-C3-E7 vector in C33A cells were validated by qRT-PCR, compared with the microarray. The X-axis column represents the mean of fold change (log2 transformed) measured from microarray analysis and qRT-PCR. P<0.05 was considered statistically significant difference.
The miRNA-circRNA interaction network. Yellow pentagon: up-regulated differentially expressed circRNAs; green pentagon: down-regulated differentially expressed circRNAs; blue ellipse: the top five binding miRNAs.
8 differently expressed circRNAs validated by qRT-PCR.
| circRNA | P value | Fold change (abs) | Regulation | circ_chrom | Gene symbol |
|---|---|---|---|---|---|
| hsa_circ_0048867 | 0.02581 | 4.249988 | up | chr19 | C3 |
| hsa_circ_0038475 | 0.04357 | 2.594571 | up | chr16 | UQCRC2 |
| hsa_circ_0035918 | 0.04347 | 2.903045 | up | chr15 | PTPLAD1 |
| hsa_circ_0056353 | 0.04594 | 2.233130 | up | chr2 | WDR33 |
| hsa_circ_0026527 | 0.00588 | 2.525893 | up | chr12 | EIF4B |
| hsa_circ_0037213 | 0.03586 | 2.053445 | up | chr16 | FAM195A |
| hsa_circ_0051620 | 0.03245 | 0.401975 | down | chr19 | SLC1A5 |
| hsa_circ_0052602 | 0.03571 | 0.421226 | down | chr2 | ODC1 |
Fold change (abs): the absolute ratio (not logarithm) of normalized intensities between two conditions; Formula: if fold change>1, fold change (abs) = fold change, fold change<1, fold change (abs) =1/fold change.
GO and pathway analysis of differently expressed gene of circRNAs
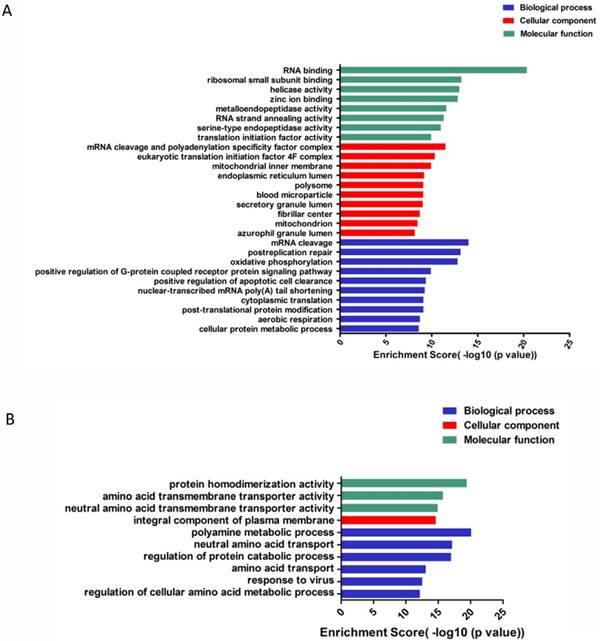
To go further investigation of circRNA, we applied GO analysis to predict and clarify the function of circRNAs. circRNAs were enriched to perform differences in biological processes, cellular components and molecular function. For up-regulated circRNAs, the top 3 enriched GO terms in biological processes included mRNA cleavage, post-replication repair, oxidative phosphorylation; cellular components included mRNA cleavage and polyadenylation specificity factor complex, eukaryotic translation initiation factor 4F complex, mitochondrial inner membrane; and molecular function included RNA binding, ribosomal small subunit binding and helicase activity (Figure 5A). For the down-regulated circRNAs, the top enriched GO terms included polyamine metabolic process, regulation of protein catabolic process, neutral amino acid transport; integral component of plasma membrane; amino acid transmembrane and neutral amino acid transmembrane transporter activity (Figure 5B).
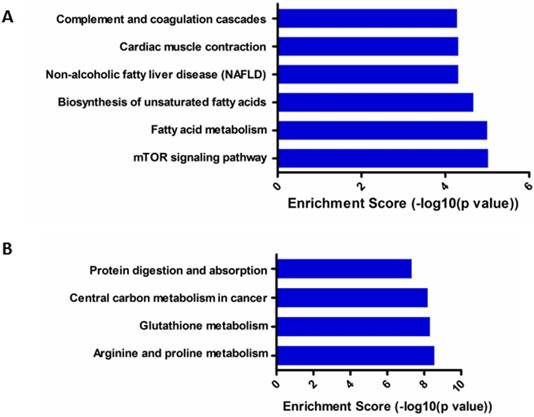
KEGG pathway analysis showed that six top predicted pathways associated with up-regulated circRNA, including mTOR signaling pathway, fatty acid metabolism and so on (Figure 6A); while four top predicted pathways associated with down-regulated circRNA, including arginine and proline metabolism, glutathione metabolism, central carbon metabolism in cancer and so on (Figure 6B).
Discussion
In recent years, emerging evidences have shown that lncRNAs or miRNAs play crucial roles in cervical carcinogenesis, implicated with cell migration and invasion, cell proliferation, cell cycle progression and apoptosis, and other biological processes[16, 17]. In view of the role of lncRNA or miRNA in the development of cancer, we hypothesis that circRNA also plays an important role in tumorigenesis.
circRNAs have been discovered decades ago and natural widely present in organisms. It is characterized by covalently closed loop resulting from 3'- and 5'- ends back-spliced[18, 19]. Thus, circRNAs have higher stable structure and conservation to resist the digestion of RNase R than linear RNA, which make them enrich in cytoplasm. Due to their stability, abundance and tissue specificity, circRNAs play crucial roles in modulating cellular function. For instance, evidence has shown that when interfering with circ-Foxo3, the cell cycle and proliferation capacity were facilitated, while overexpression of circ-Foxo3, the cell cycle process was inhibited, which suggested that Foxo3 gene encoding the circular RNA circ-Foxo3 was closely related to cell cycle process[20]. Down-regulation of hsa_circ_0001895 was associated with cell differentiation and tissue carcinoembryonic antigen expression in gastric cancer[21]. Knockdown the expression of hsa_circ_0000069 can suppress cell proliferation, invasion and migration in colorectal cancer[22].
Cervical cancer is one of the most common cause of death from cancer in female worldwide with its high mortality and morbidity[23]. HPV infection, especially HPV16 or HPV18, is the major cause for the development of cervical cancer. Also, the E6 and E7 proteins encoded by HPV are almost always involved in the transformation of cervical epithelial cells, contributing to the pathogenesis of cervical cancer. Considering the E7 protein is a causative agent of cervical carcinoma, this study determined whether it alters the expression profiles of circRNAs. Here we are the first to comprehensively delineate the expression profiles of circRNAs regulated by HPV16 E7 in cervical cancer cells and to provide potential candidates for the future mechanism studies. The results showed that there were 526 significantly differentially expressed circRNAs, including 352 up-regulated and 174 down-regulated, suggesting that some host circRNAs that display critical roles in cervical cancer may be regulated specifically by viral oncoprotein E7. Furthermore, the 8 selected circRNAs (6 up-regulated and 2 down-regulated) were validated to be consistent with the results of microarray experiment by qRT-PCR analysis, which indicated that the microarray data was relative stable.
GO enrichment of differentially expressed gene of circRNAs. (A). GO enrichment for up-regulated ciRNAs. (B). GO enrichment for down-regulated ciRNAs.
KEGG pathway enrichment analysis of differentially expressed gene of circRNAs. (A). KEGG pathway analysis of up-regulated circRNAs; (B). KEGG pathway analysis of down-regulated circRNAs.
CircRNA can function as sponges interacting with miRNA or competing endogenous RNA (ceRNA)[24, 25]. Wei et al. showed that circLMO7 suppress cell differentiation and facilitates cell survival by sponging miR-378a-3p in bovine myoblasts[26]. Also, circBIRC6 promotes pluripotency maintenance by sponging miR-34a and miR-145 in hESCs[27]. In our study, the miRNA-circRNA network revealed potential associations between circRNA and miRNA, which is of great importance in further investigation on the functions of dysregulated circRNAs and their targets. The up-regulated circRNAs, hsa_circ_0048867, hsa_circ_0038475, hsa_circ_0035918, hsa_circ_0056353, hsa_circ_0026527, hsa_circ_0037213, interacted with hsa-miR-1180-3p, hsa-miR-181b-3p, hsa-miR-181a-2-3p, hsa-miR-132-5p, hsa-miR-15b-3p, hsa-miR-212-5p, respectively. Down-regulated circRNAs, hsa_circ_0051620, hsa_circ_0052602 bind to hsa-miR-3144-5p and hsa-miR-497-5p, respectively. Moreover, hsa-miR-1180-3p, hsa-miR-181b-3p, hsa-miR-181a-2-3p, hsa-miR-132-5p, hsa-miR-15b-3p, hsa-miR-212-5p, hsa-miR-3144-5p and hsa-miR-497-5p have been reported in the pathogenesis of cervical cancer[28-30]. Whether these miRNAs direct to sponge circRNAs or target to E7 to regulate their expression leading to the tumorigenesis is an important part of our study in the future.
Recently, study on circRNAs of cervical cancer is rarely limited. Wang et al. and Gao et al. performed profiles of circRNA in tissues[14, 15], but did not mention the role of E7 in the regulation of cervical cancer. In our study, GO and KEGG pathway analyses were used to classify the aberrant circRNAs with E7 siRNA transfected. GO analysis showed that up-regulated circRNAs was most involved in molecular function acting as RNA binding. These data indicated that several circRNAs might serve as ceRNA to modulate the development and progression of cervical cancer. In addition, KEGG pathway analysis was involved in the pathway of mTOR signaling pathway, arginine and proline metabolism and glutathione metabolism and so on. Among them, mTOR signaling pathway is now widely accepted that plays a vital role in human cervical cancer[31, 32]. These findings provide a novel strategy for the treatment of cervical cancer based on E7.
In summary, using circRNA array analysis, we first identified a subset of host circRNAs that are regulated by HPV16 E7, which showed direct evidence that the viral oncogenes have the potential to influence the abundance of multiple circRNAs likely contributing to the growth of HPV-positive cancer cells. Additionally, differential expression circRNAs were verified by qRT-PCR in knockdown and over-expression E7 cell respectively. Bioinformatic analyses predicted the potential binding miRNAs and the functions of candidate circRNAs in the development and progression of cervical cancer. In brief, our study is potentially of critical significance in uncovering the roles of circRNAs in cervical cancer, and provides novel insights into the new potential biomarkers for diagnostic and therapeutic of cervical cancer.
Acknowledgements
This study was supported by the grants from the National Natural Science Foundation of China (81672588, 81372154) and the Teaching and Research Award Program for Outstanding Young Teachers in Higher Education institutions of Guangdong Province, China (2014) and the National Undergraduate Training Program for Innovation and Entrepreneurship (201712121035, 201712121033).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jonson AL, Rogers LM, Ramakrishnan S. et al. Gene silencing with siRNA targeting E6/E7 as a therapeutic intervention in a mouse model of cervical cancer. Gynecol Oncol. 2008;111:356-64
2. Cao S, Liu W, Li F. et al. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776-83
3. Ghittoni R, Accardi R, Hasan U. et al. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes. 2010;40:1-13
4. Burroni E, Bisanzi S, Sani C. et al. Codon 72 polymorphism of p53 and HPV type 16 E6 variants as risk factors for patients with squamous epithelial lesion of the uterine cervix. J Med Virol. 2013;85:83-90
5. Honegger A, Schilling D, Sponagel J. et al. Dependence of Intracellular and Exosomal microRNAs on Viral E6/E7 Oncogene Expression in HPV-positive Tumor Cells. PLoS Pathog. 2015;11:e1004712
6. Yamato K, Yamada T, Kizaki M. et al. New highly potent and specific E6 and E7 siRNAs for treatment of HPV16 positive cervical cancer. Cancer Gene Ther. 2008;15:140-53
7. Sharma S, Mandal P, Sadhukhan T. et al. Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci Rep. 2015;5:11724
8. Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550-60
9. Greene J, Baird AM, Brady L. et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38
10. Zhao Z, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514-21
11. Meng S, Zhou H, Feng Z. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94
12. Liang HF, Zhang XZ, Liu BG. et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566-76
13. Zou M, Huang C, Li X. et al. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. 2017;8:81825-37
14. Wang H, Zhao Y, Chen M. et al. Identification of Novel Long Non-coding and Circular RNAs in Human Papillomavirus-Mediated Cervical Cancer. Front Microbiol. 2017;8:1720
15. Gao YL, Zhang MY, Xu B. et al. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8:86625-33
16. Kim HJ, Lee DW, Yim GW. et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521-30
17. Gómez-Gómez Y, Organista-Nava J, Gariglio P. Deregulation of the miRNAs Expression in Cervical Cancer: Human Papillomavirus Implications. Biomed Res Int. 2013;2013:407052
18. Li H, Hao X, Wang H. et al. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell Physiol Biochem. 2016;40:1334-44
19. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167-76
20. Huang S, Yang B, Chen BJ. et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;9:401-7
21. Shao Y, Chen L, Lu R. et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumor Biol. 2017;39:1-6
22. Guo JN, Li J, Zhu CL. et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451-8
23. Iempridee T. Long non-coding RNA H19 enhances cell proliferation and anchorage-independent growth of cervical cancer cell lines. Exp Biol Med. 2016;242:184-93
24. Shi L, Yan P, Liang Y. et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171
25. He R, Liu P, Xie X. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145
26. Wei X, Li H, Yang J. et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8:e3153
27. Yu CY, Li TC, Wu YY. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149
28. Harden ME, Prasad N, Griffiths A. et al. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. MBio. 2017;8:e02170-16
29. Wang X, Wang HK, Li Y. et al. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci U S A. 2014;111:4262-7
30. Pereira PM, Marques JP, Soares AR. et al. MicroRNA Expression Variability in Human Cervical Tissues. PLoS One. 2010;5:e11780
31. Oh KJ, Kalinina A, Park NH. et al. Deregulation of eIF4E: 4E-BP1 in Differentiated Human Papillomavirus-Containing Cells Leads to High Levels of Expression of the E7 Oncoprotein. J Virol. 2006;80:7079-88
32. Ueno S, Sudo T, Oka N. et al. Absence of Human Papillomavirus Infection and Activation of PI3K-AKT Pathway in Cervical Clear Cell Carcinoma. Int J Gynecol Cancer. 2013;23:1084-91
Author contact
![]() Corresponding author: Jue-Yu Zhou, e-mail: zhoujueyucom
Corresponding author: Jue-Yu Zhou, e-mail: zhoujueyucom

 Global reach, higher impact
Global reach, higher impact