3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(20):3765-3775. doi:10.7150/jca.25576 This issue Cite
Review
MicroRNA-34 family in breast cancer: from research to therapeutic potential
1. Key Laboratory of Epigenetics and Oncology, the Research Center for Preclinical Medicine, Southwest Medical University, Luzhou, Sichuan 646000, P.R. China.
2. Department of Biochemistry and Molecular Medicine, the George Washington University, Washington, DC 20052, USA.
Received 2018-2-15; Accepted 2018-8-3; Published 2018-9-28
Abstract
MicroRNA (miRNA)-34 family (miR-34s), including miR-34a/b/c, is the most well studied non-coding RNAs that regulate gene expression post-transcriptionally. The miR-34s mediates the tumor suppressor function of p53 in the pathogenesis of breast cancer by targeting different oncogenes. This review focuses on the anti-oncogenic regulation of the miR-34s, emphasizing the major signaling pathways that are involved in the modulation of miR-34s in breast cancer. Moreover, it highlights how epigenetic modification by the p53/miR-34s axis regulates the proliferation, invasiveness, chemoresistance, and sternness of breast cancer. A better understanding of the molecular mechanisms of miR-34s will open new opportunities for the development of novel therapeutic strategies and define a new approach in identifying potential biomarkers for early diagnosis of breast cancer.
Keywords: MicroRNA-34, Breast cancer, Tumor suppressor miRNA, Diagnosis, Treatment.
Introduction
According to the American Cancer Society (http://www.cancer.org), breast cancer is the second-leading cause of mortality in female and the most frequently diagnosed cancer in the United States, with estimated 252,710 new cases and 40,610 expected breast cancer deaths every year [1]. Among these cases, only 20 percent of these cases were diagnosed at early stage when the cancer is still localized and treatable. Breast cancer is a heterogeneous disease that can be classified into 25 subtypes based on distinct histological and gene expression profiles [2, 3]. The etiology of breast cancer is still unknown and no potential prognostic biomarkers could predict the survival rate of breast cancer. This highlights the importance of early diagnosis to improve therapy and molecular diagnostics of breast cancer [4, 5]. To date, many research centers recognized that several miRNAs play critical roles in breast cancer initiation, progression, and metastasis; thus they are attractive targets for therapy supplementing traditional treatments, such as surgery, chemotherapy, and radiotherapy.
MicroRNAs (miRNAs) are in a class of endogenous, small, non-coding RNAs 21~23 nucleotides (nt) in length. MiRNAs are involved in various human cancers and can either modulate as oncogenic miRNAs (oncomiRs) or tumor suppressor miRNAs. In the cancer cells, most tumor suppressor genes are inhibited by the upregulation of oncomiRs or expression of proto-oncogenes [6-8]. The functional balance between oncomiRs and tumor suppressor miRNAs play critical roles in tumor proliferation, differentiation, angiogenesis, invasion, metastasis, and treatment outcome [9]. The microRNA-34/499 (miR-34/499) super family was mainly established with the discovery of microRNA-34 (miR-34) family and miR-499 family as a small single-stranded miRNA [10, 11]. The miR-34family consists of three closely related members; miR-34b, -34c, and -34a, is the most well studied tumor suppressor miRNAs [12]. The miR-449 cluster, with highly conserved miR-449a, -449b, and -449c, contains secondary structures and sequences similar to the miR-34 family [13]. Both miR-34 and miR-449 family were categorized as one family of miRNAs because they share the same seed sequence and mRNA targets [12]; for the purpose of this review, miRNA-34/499 super family will be called as miR-34. Corresponding to the tumor-suppressive role of miR-449, expression of the miR-34 family is down-regulated in a wide range of cancers, including lung [14, 15], multiple myeloma [16], kidney [17, 18], gastric [19], breast [20, 21], colorectal [22], hepatocellular [23], prostate [24], and ovarian [25, 26].
This review summarizes the epigenetic mechanisms of miR-34 family members in regulating of proliferation, apoptosis, invasion, and metastasis of breast cancer cells. Furthermore, we will try to explore the possible biomarker roles of miR-34 family for diagnosis, prognosis, and therapeutic targets of breast cancer.
Expression, biogenesis, and structure of miR-34 family
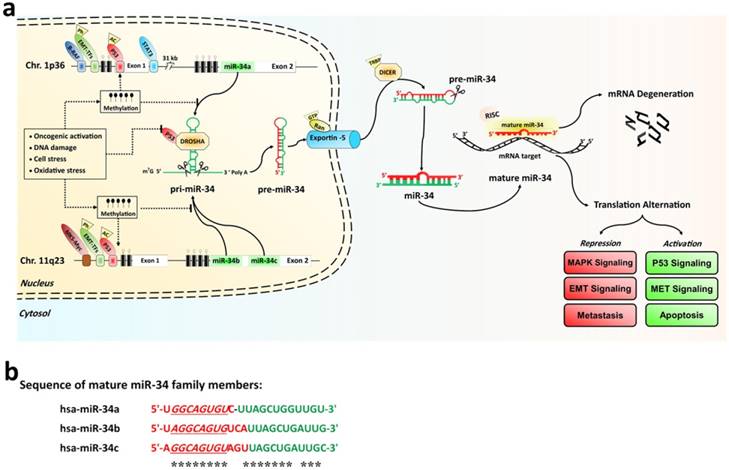
The miR-34 family, which contains three members, is encoded by two genes located on chromosomes 1 and 11 [27]. Within the human genome, miR-34b/c shares a common primary seed sequence located at one transcription unit on chromosome 11q23.1, whereas miR-34a is encoded in the second exon of a transcript located on chromosome 1p36.22. The mature miR-34a shares 86% identity (19/22 nt) with miR-34b and 82% identity (18/22 nt) with miR-34c, respectively. The position 2-9 adjacent at the 5' end (8 nt) is considered the “seed region” for all three members (See the Fig. 1a) [27-29].
Among these members, miR-34a expression levels are higher than miR-34b/c, in most human cells, except for lung tissue in humans and brain tissue in mice. In lung tissues, miR-34b/c is usually expressed instead. The biogenesis of miR-34s like other miRNAs, is a multistep process. MiR-34 family encoding genes are initially transcribed by RNA polymerase II or III as long hairpin molecule (pri-miRNA) in the nucleus. The pri-miRNA is processed by the RNase III DROSHA into a stem-loop-structured miRNA precursor molecule (pre-miRNA) (≈70 nt length) [30]. Then, Pre-miR-34s are transported to the cytoplasm by active exportin-5 nucleus transporter. The cytoplasmic biogenesis process of pre-miR-34s into mature miR-34 is mediated by another human RNase III (DICER); resulting in a 20-23 bp RNA duplex consisting of the mature miRNA and its anti-sense strand (miRNA*) [30]. Finally, one strand of the mature form of duplex stand is incorporated into an RNA-induced silencing complex (RISC), while the other is degraded. If the binding sites on the 3'-UTRs or 5'-UTRs of target mRNAs and miR-34 are fully complementary, it may lead to mRNA degradation and inhibit target gene expression. Conversely, miR-34 family can suppress translation or transcriptional activation if only partially complementary sequences are present in its target genes [28, 31, 32]. The sequence alignment of the mature miR-34a, miR-34b, and miR-34c molecules were compared in the Fig. 1b. The expression of all miR-34s genesis is tightly controlled at the transcriptional and posttranscriptional levels by the p53 tumor suppressor, ETS domain-containing protein Elk-1, signal transducer, activator of transcription 3' (STAT3), CpG island methylation, and EMT-inducing transcription factors (EMT-TFs, such as zinc-finger E-box-binding (ZEB) and basic helix-loo-helix (bHLH) families) [27]. It is well established that p53 is an important inducers of miR-34s expression, which binds to the promoter regions of both miR-34a and miR-34b/c [27, 29, 33]. Hypermethylation of the CpG islands of miR-34 promoter directly induced miR-34s silencing [34]. Furthermore, any DNA and/or cellular stress damage led to the silencing of the miR-34s expression by the activation of the p53 network (Fig. 1a).
Functions of miR-34 family in normal and cancer cells
The miR-34 family regulates vital biological processes such as cell development, metabolism and differentiation [35, 36]. For example, during normal human bronchial epithelial cell differentiation and embryonic central nervous system development, miR-34s are upregulated [35, 37]. Specifically, miR-34a transcriptionally regulated lineage selection and B-cell development in murine bone marrow [29] and affected critical developmental checkpoints during hematopoiesis [38]; miR-34b/c were involved in the differentiation of male primordial germ cells into spermatozoa process, as well as in the maintenance of embryonic stem cells in an undifferentiated state [39].
In the cancer cells, miR-34a was the first class of the miR-34 family in neuroblastoma cancer cells recognized as a potential tumor suppressor miRNA through integration in TP53 network [27, 33, 40]. All three components of the p53 tumor suppressor, p53, p63, and p73, are directly and indirectly coordinated in the activation of miR-34s family [29, 41]. It had been shown that induction of miR-34s by p53 triggered apoptosis and cell cycle arrest in a wide range of hematological and solid malignant cells. In this regard, the miR-34a functions at the core of tumorigenic processes known as "apoptomiR" [40].
a) The miR-34 family biogenesis, regulators, and their functions. Genomic structures of the human miR-34a and miR-34b/c loci. The human RNase III enzymes are distinctive by the octagonal box. CpG island miR-34s promoter hypermethylation induces miR-34s silencing that is dominant over its transactivation by oncogenic activations, DNA damage, and any cellular stress damage. b) Sequence alignment of the mature miR-34a, miR-34b, and miR-34c molecules. The seed sequences and identical nucleotides are show in underline and asterisks indicate markers, respectively. White and green boxes represent exons and miR-34 family hairpins on the genome, respectively. CpG islands are represented by hatched box. Methylations and un-methylations are marked by the black and white cycle, respectively. Red triangle and rhombus indicate the posttranscriptional modifications, respectively.
Ectopic expression of the miR-34s can antagonize many different oncogenic pathways in cancer cells, including cell cycle control, proliferation, metastasis, and apoptotic pathways [21, 40]. MiR-34s were significantly downregulated in many human breast cancer cells by affecting numerous oncogenes and cancer pathways. MiR-34a levels were more than three folds lower in triple negative breast cancer cells (TNBCs) and mesenchymal breast cancer cell lines in comparison to normal Her-2+ cell lines [20, 27, 33]. This downregulation was related with p53 mutation, loss of heterozygosity, and hypermethylation of the neighboring CpG islands [42, 43]. Interestingly, point mutations in miR-34s genes were found in most breast cancer cells, leading to down-regulated expression of the miR-34s in these cancer cells. Remarkably, most mutations were found in the binging site of the p53 protein and CpG rich genomic region [44-46].
The target genes of miR-34s in breast cancer
The tumor suppressors of miR-34 members control an analogous set of target genes with more than 82% homology [21, 40]. Table 1 sorts the main, direct miR-34s targets in breast cancer identified by bioinformatics methods and cellular experiments in human breast cancer tissue and cell lines. The miR-34s regulate their targets via binding of seed-sequence (7 nt) located in their 5′- or 3′-UTR of the target mRNA. Although many genes are identified as targets of miR-34s by target prediction tools (Target Scan [http://www.targetscan.org; release 5.1), PicTar [http://pictar.mdc-berlin.de), and miRanda [http://www.microrna.org)) [47], a few of them were confirmed by in vitro and in vivo study (Fig. 2) [29, 40]. The luciferase reporter assay, biotinylated miRNA, comparative genomics, and hybridization are fundamentally different methods that many researchers used to identify miR-34s targeted genes and their tumor suppressive roles in different cancer cells [27]. Among the different high-throughput technologies, microarray, RNA-sequencing, and next-generation sequencing are now first array approaches in prediction of miRNA targets [48-51]. Technically advanced and well-stablished microarray platforms can now be evaluated by distance bioinformatics tools. As sorts in Table 1, most of the identified targets encoded factors involved in G1 cell cycle progression, apoptosis, proliferations, and invasion of breast cancer cells [52, 53], which are similarly illustrated in the Fig. 2.
Main current miR-34s targets in breast cancer.
| Cellular Process | Target gene | MiR-34s member | mRNA target/hsa-miR-34a alignment * | Biological effect | Validated methods | Cell lines | SMS | Ref. |
|---|---|---|---|---|---|---|---|---|
| Metastasis /Wnt Signaling | AXL | a | 5'-GGAUCCAAGCUAAGCACUGCCA-3' || ||||| 3'-UGUUGGUCGAUUCUGUGACGGU -5' | Inhibition of proliferation, migration, and invasion | Luc. reporter (Mut), WB, qPCR | BRC | Y | [54, 55] |
| Far-1 | a/c | 5'-UAACCCCUUCCAGAUCACUGCCA-3' || ||| | | 3'-CGUUAGUCGAUUGAUGUGACGGA-5' | Inhibition of invasion and metastasis | Luc. reporter, WB, qPCR | BRC | Y | [56] | |
| WNT1 | a/b/c | 5'-UGGAAUCUGACAUAAGCUGAUUG-3' | ||| | | | 3'-UCCCGUCAUAUGAACGACUAAC-5' | Inhibition of Wnt signaling | Luc. reporter, WB, qPCR | BRC | Y | [57] | |
| WNT3 | a/b/c | 5'-CUGGGAACCGCCCUCCUGAUUAA-3' || ||| | 3'-UCCCGUCAUAUGAACGACUAAC-5' | Inhibition of Wnt signaling | Luc. reporter, WB, qPCR | BRC | Y | [57] | |
| ZEB1 | a | 5'-AGAGGUUAAAGGAAGCUGAUUA-3' | |||| | | 3'-UCCCGUCAUAUGAACGACUAAC-5' | Inhibition of EMT and invasion | Luc. reporter (Mut), WB, qPCR | BRC/BC | Y | [20] | |
| SNAI1 | a/b/c | 5'-CAGAGCUGGGAUGCUGCU-3' || ||||| ||| | | 3'-UGUUCGAUUCUGUGACGGU-5' | Inhibition of EMT | Luc. reporter (Mut), WB, qPCR | BRC/BC | Y | [58] | |
| LMTK3 | a | 5'-TGTGGATGACGGCGCCACTGCCA-3' ||||||||| 3'-UGUUGUCGAUUCUGUGACGGU-5' | Inhibition of proliferation and invasion | Luc. reporter (Mut), WB, qPCR | BC | Y | [53] | |
| TWIST1 | a | 5'-AUUUUUUAUUUCAUUCUGAUUAU-3' | | | ||| | | | 3'-UCCCGUCAUAUGAAC-GACUAAC-5' | Inhibition of EMT and invasion | Luc. reporter (Mut), WB, qPCR | BRC/BC | Y | [20] | |
| Apoptosis/P53 Pathway | Bcl-2 | a | 5'-UCGAAUCAGCUAUUUACUGCCA-3' | || |||| 3'-UGUUGGUCGAUUCUGUGACGGU-5'd | Inhibition of proliferation and apoptosis | WB, qPCR | BRC/BC | N | [53, 59, 60] |
| FasR | a | 5'-AGGGUCUUCUGACCUCUGAUUAG-3' |||||| 3'-UCCCGUCAUAUGAACGACUAAC-5' | Inhibition of proliferation and G1 -arrest, | Luc. reporter, WB, qPCR | BRC | Y | [61] | |
| SIRT1 | a | 5'-CCAGCUAGGACCAUUACUGCCA-3' |||||| | 3'-UGUUGGUCGAUUCUGUGACGGU-5' | G1/S-arrest and apoptosis, Inhibition of proliferation | WB, qPCR | BRC/BC | N | [60, 62] | |
| Cancer Stemness | CD24 | a | 5'- AGUAAAUCUUUUACAACUGCCU-3' |||| | | 3'- UGUUGGUCGAUUCUGUGACGGU-5' | Inhibition of differentiation and positive regulation of p53-cell cycle | Luc. reporter, WB, qPCR | BRC | Y | [63] |
| NOTCH1 | a | 5'-AUUUUACACAGAAACACUGCCU-3' | | || | || 3'- UGUUGGUCGAUUCUGUGACGGU-5' | Apoptosis and inhibition of EMT, proliferation, and invasion | Luc. reporter (Mut), WB, qPCR | BRC/BC | Y | [20, 64, 65] | |
| NOTCH4 | c | 5'-GUCCCCAUAAUAAAGCUGAUUU-3' |||| | | | 3'-UCCCGUCAUAUGAACGACUAAC-5' | Inhibition of Notch signaling and senescence | Luc. reporter (Mut), WB | BRC | N | [66] | |
| HDAC1 | a | 5'-AAGUGAGCCAAGAAACACUGCCU-3' ||| | | || 3'- UGUUGGUCGAUUCUGUGACGGU-5' | Inhibition of tumor progression and cell proliferation | Luc. reporter (Mut), WB, qPCR | BRC | Y | [53, 67, 68] | |
| HDAC7 | a | 5'-CUGGGACCCCUCGGCCACUGCCC-3' ||||| | | 3'-UGUUGGUCGGAUUCUGUGACGGU-5' | Inhibition of tumor progression and cell proliferation | Luc. reporter (Mut), WB | BRC | Y | [67] | |
| Cell Cycle | CDK6 | a/b | 5'-UAUAACUACAUAUUGACUGCCA-3' ||| ||| | 3'-UGUUGGUCGAUUCUGUGACGGU-5' | G1-arrest,inhibition of invasion and metastasis | Luc. reporter, WB, qPCR | BRC/BC | Y | [69] |
| MDM4 | a | 5'-AGAUUUUUUUUACUCACUGCCA-3' ||| ||| | 3'-UGUUGGUCGAUUCUGUGACGGU-5' | Positive regulation of p53-cell cycle | Luc. reporter (Mut), WB, qPCR | BRC | Y | [52, 53] | |
| LMTK3 | a | 5'- GUGGAUGACGGCGCCACUGCCA-3' ||| ||| | 3'- UGUUGGUCGAUUCUGUGACGGU-5' | G1-arrest and inhibition of proliferation | Luc. reporter, WB | BRC | Y | [53] | |
| Oncogenic Transcription | MET | b/c | 5'-UCCAAUGGUUUUUUCACUGCCU-3' ||| ||| | 3'-UGUUGGUCGAUUCUGUGACGGU-5' | G1-arrest,inhibition of proliferation and invasion | Luc. reporter (Mut), WB, qPCR | BRC | Y | [70] |
| Msi1 | a | 5'-GGCCAAGGCCCACCCACUGCCA-3' ||| ||| | 3'-UGUUGGUCGAUUCUGUGACGGU-5' | G1/S-arrest and inhibition of proliferation | Luc. reporter (Mut), WB, qPCR | BRC | Y | [71] | |
| Src | a | 5'-GAGGACGUGUUACCCACUGCCA-3' ||| ||| | 3'- UGUUGGUCGAUUCUGUGACGGU-5' | Inhibition of progression and regulation of p53-cell cycle | Luc. reporter (Mut), WB, qPCR | BRC | Y | [63] |
* The alignments between the hsa-miR-34 family and the 3'-UTR of validated targets are reported above. The miR-34s species shown to bind or regulate the respective targets are listed in the third column. Overall, the alignment with miR-34a and miR-34b are as shown and the miR-34s seed region is highlighted by gray shading and bold letters. Vertical lines between both sequences indicate perfect Watson-Crick base pairs. The indicated, putative miR-34-binding sites were recognized, using bioinformatics analysis by the following target prediction tools: TargetScan (http://www.targetscan.org; release 5.1), PicTar (http://pictar.mdc-berlin.de) and miRanda (http://www.microrna.org). This table shows the publication identified a seed-matching sequence (SMS) in the 3'-UTR.
Abbreviation: AXL, AXL receptor tyrosine kinase; Fra-1, Fos-related antigen 1; IL-6R, Interleukin 6 receptor; WNT1,3, Wingless-related MMTV integration site member 1,3; ZEB1, Zinc finger binding protein 1; SNAI1, Snail family transcriptional repressor 1; TWIST1, Twist family BHLH transcription factor 1; Bcl-2, B-cell leukemia/lymphoma 2; FasR, FAS receptor; SIRT1, Sirtuin 1, Silent information regulator 1; MDM4, MDM4 as P53 regulator; C-MYC, C-myc myelocytomatosis viral oncogene homolog; CD24, Cluster of differentiation 24; NOTCH 2,4, Notch homolog 2,4; CDK6, Cyclin-dependent kinase 6; LMTK3, Lemur tyrosine kinase 3; E2F3, E2F transcription factor 3; Met, Met proto-oncogene; Msi1, Musashi RNA binding protein 1; SRC, SRC Proto-Oncogene; BRC, Breast cancer cell line; BR, Human breast cancer sample; SMS, seed-matching sequence; luc. reporter, Luciferase reporter assay; mut, Mutagenesis of the SMS in the 3'-UTR-reporter construct; qPCR, Quantitative real-time PCR; WB, Western blotting analysis. Y, Yes; N, No.
Cellular outcomes associated with miR-34s-induced gene silencing in cancer cells. The increased levels of any members of miR-34s re-enforced the cancer-related pathways in response to cell cycle arrest, proliferation, metastatic, stemness, apoptosis, and chemo drug resistance in breast cancer.
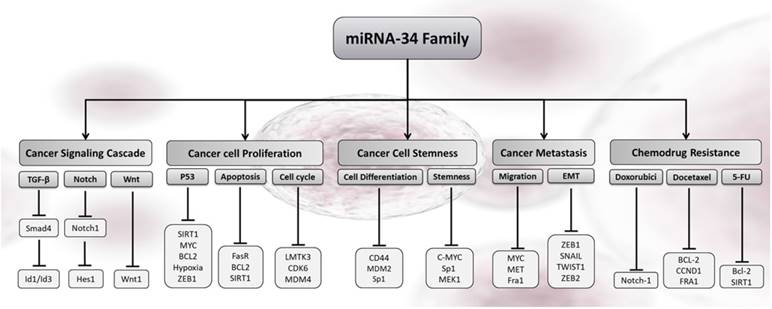
Molecular mechanisms of miR-34s in breast cancer
The tumor suppressor and inducing of apoptosis
In the last decade, miR-34s emerged as critical regulators of apoptosis by multitudes of signaling pathways [42]. MiR34s, as regulation of multiple genes, is responsible for cancer cell death and G1/S cell cycle arrest in response to any oncogenic triggers at the posttranscriptional level [27, 29]. As an ApoptomiR, re-introducing mimics of miR-34s inhibits numerous cancer cell types and initiates the apoptotic pathways [28, 33]. In the view of the large tumor suppressor deregulated targets, we sought to narrow down the number of targets related to apoptosis. It is generally accepted that miR-34a has directly trigged p53-induced apoptosis in breast cancer. Pro-apoptotic functions of miR-34a in breast cancer interplayed with anti-apoptotic proteins such as Bcl-xL and Bcl-2 (Table 1) [29, 53, 59]. Remarkably, Gou et al. confirmed that miR-34a is a main member of miR-34 family that triggers p53-induced apoptosis by directly repressing Bcl-2 in the TNBC and MCF-7 cell lines [29, 72]. Similarly, SIRT1 and NAD+-dependent protein deacetylase class is another miR-34a target in p53/miR-34-induced apoptosis [56, 60, 62]. In detail, repression of SIRT1 by miR-34a in breast cancer cells leads to reduction of p53 deacetylation. Deacetyled-p53 more actively induces G1/S cell cycle arrest or apoptosis. Therefore, SIRT1, and p53/miR-34a forms a positive feedback axis to inhibit proliferation that inducts tumor suppression process by triggering of the p53-meditated apoptosis network [60, 62]. Furthermore, FasR can be extracellular anti-apoptotic targets of miR-34a, presumably resulting from a reduction of cancer cell proliferations and blocked apoptotic signaling (Fig. 2) [61]. As shown in the Fig. 2, the miR-34s family interferes with apoptotic properties across a broad spectrum of pro-apoptotic and oncogenes regulators (TP53, NOTCH1, and SMAD4). The upstream apoptotic signaling, such as Wnt and TGF-β cascade, are involved in the miR-34-apoptotic related pathways in metastatic breast cancer (MBC) [20, 56, 64-66, 73]. Consequently, overexpression of the pro-oncogenes and particular miR-34s target proteins revealed regulation of p53-apoptosis in breast cancer patients [27]. Increasing progress in cancer biology research has found several cell cycle related gene targets of miR-34s, such as LMTK3, MDM4, CDK6 [52, 53, 69]. These evidences suggested that important role of ApoptomiRs may have parts in maintaining balance between cell cycle and apoptosis [74, 75]. Generally, miR-34a functions as an ApoptomiR in breast cancer and reported for other cancers. However, the detailed roles of miR-34b/c in breast cancer need to be confirmed by more substantial and comprehensive research.
The Reduction of chemoresistance
Breast cancer patients who do not respond to chemotherapy usually have a low expression of miR-34s. Ectopic miR-34a expression reduces the resistance to chemo drugs such as doxorubicin, docetaxel, Adriamycin, and 5- fluorouracil (5-FU) [60, 64, 65, 76]. MiR-34a was considerably upregulated in HDAC1/7-depleted breast cancer cells. MiR-34a-HDAC1/HDAC7-HSP70 K246 crosstalk is identified as a novel molecular signature predictive of therapy resistance [77]. The targeting of histone deacetylases (HDACs) with miR-34s is a potential anti-cancer therapy, resulting in the reduction of the chemo resistance of breast cancer and increase in the chemo drug efflux [53, 67, 68]. Also, miR-34a level significantly is reduced by methylation of the promoter region of the miR-34a gene in radio sensitivities of breast cancer cells. In detail, the MDA-MB-231 cell line (with low miR-34a level) is significantly more sensitive to radiation than normal human mammary epithelial cells (HMECs, with high miR-34a level). This approach attended earlier reports that had suggested targeted-specific genes silencing after chemotherapy in breast cancer patients [56, 60, 64, 65, 76]. For example, NOTCH1, Bcl-2, CCND1, FRA1, and SIRT1 are the targets of miR-34a in a combination treatment of miR-34a with 5-FU and/or docetaxel (Fig. 2). Remarkably, restoration of miR-34a sensitized MCF-7 breast cancer cells to 5-FU and docetaxel suggested this can be a useful therapy approach for chemo drug resistant breast cancer [56, 60]. The miR-34 family roles in radiation-induced cell death in breast cancer cells are still unknown. Hence, further research is needed to complete the clinical usage of miR-34s as therapeutic targets to overcome chemoresistance in breast cancer.
Inhibition of proliferation, invasion, and metastasis
MBC is an end-stage, deadly aggravation of breast cancer with complex mechanisms, including local invasion, transport, extravasations, and colonization. EMT-TFs (such as zinc-finger E-box-binding family, such as TWIST1, SLUG1, SNAIL1 and ZEB1) are key factors that accelerate the progression of non-invasive to invasive breast cancer (IBC) by several transcriptional repressors of Vimentin, E-cadherin [67, 78-80]. Epigenetically, miR-34s/EMT-TFs axis plays inhibition roles of MBC migration and invasive at in vivo and in vitro levels [42, 81-83]. MiR-34s are often down-regulated in lymph node metastases of breast cancer cells as comparted normal breast by binding to EMT-TFs in TNBC cells. Overexpression of miR-34a significantly inhibits EMT-TFs, like TWIST1, and ZEB1 in TNBC [20, 81]. Ultimately, the above findings strongly propose why miR-34a/EMT-TFs contributes to MBC progression and identification of novel therapeutic targets of stage ІІ/ІІІ of breast cancer progress. Incredibly, migration and invasive target crucial genes, such as MYC, MET, and Fra1 involved mainly in the apoptotic related pathways in MBC [29, 56, 80, 84]. MiR-34a silenced c-SRC and attenuated tumor growth and invasion in TNBC in vitro and in vivo [85]. Meanwhile, the published evidence is limited and further studies are needed for illustrating the exact cellular functions of miR-34-induced gene silencing.
Regulation of the breast cancer stem cells
Breast cancer stem cell (BCSCs) are the potential stem cells of new tumor forms characterized by cell surface markers ESA+, CD44+, and CD24- [86, 87]. Delivery of miR-34s had significant improvement in patient outcomes with successful targeting of CD44+/CD24low BCSCs [80, 88]. MiR-34a targets CD44, Sp1, and MDM2 as a cancer cell differentiation marker, resulting in impaired tumor growth and decreased metastases in mouse models of BCSCs [66]. Consistently, ectopic expression of miR-34c in BCSCs is mediated by CpG islands-methylation in the promoter region of miR-34c gene, which reduces DNA binding activities of Sp1 [66, 87]. Yu et al. showed that the expression of miR-34c is lower in CD44+ BCSCs when compared to CD44 non-stem cells, resulting in impaired tumor growth and decreased metastases in breast cancer [66]. From this evidence, miR-34c's ability to target BCSCs proposes that they may have significant therapeutic potential, due to therapeutic metastasis and cancer relapse with elimination of all BCSC populations.
Diagnostic accuracy and survival potential of miR-34s
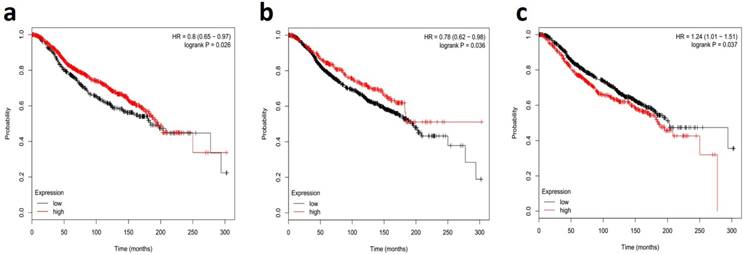
MiR-34s members are down-modulated in metastatic lymph node and IBC tissue samples with a high aggregative index, proposing that a deficiency of miR-34s is related with a poor prognosis. In this regard, the recent comprehensive systematic review and meta-analysis study showed the miR-34 family as a potential diagnostic biomarker in radiation-induced breast cancer patients [89]. The literature review indicated that serum and plasma levels of miR-34a were related with histological grades of breast cancer. However, other studies described no significant association between serum miR-34a expression and clinicopathological features, such as lymph node metastasis and hormone receptors [90, 91]. Nonetheless, our systematic review and meta-analysis confirmed the diagnostic value of miR-34a in detecting of breast cancer with 85.50% sensitivity and 70.50% specificity [92, 93]. Furthermore, reported documents have evidences for miR-34a as an accurate diagnostic biomarker in tissue-based samples of breast cancer [67, 79]. Interestingly, miR-34a, more deep-rooted than the other two miR-34 family members, is a new class of non-invasive urine-based biomarker for diagnosis of breast cancer, with averagely 61.0% sensitivity and 79.7% specificity [90, 91, 94]. This creates great interest in the early-noninvasive, breast cancer detection capabilities as a novel target for tumor suppression. Taken together, these results comprehensively showed that miR-34a could be a promising and novel non-invasive biomarker in the early diagnosing of patients with breast cancer. It is clear that profiling all members of miR-34s need early stage diagnosis and the determination of therapeutic prognosis of breast cancer [95-97]. The Fig. 3 shows the survival correlation of all members of miR-34s in human breast cancer, according the online dataset [98, 99]. This figure shows the lower expression of miR-34a/b were associated with poor outcomes in breast cancer, verifying its role as tumor suppressor biomarkers (Fig. 3A and 3B, respectively). Downregulation of miR-34c was associated with poor outcomes in breast cancer patients with short overall survival (Fig. 3C). Undoubtedly, well-designed large-scale, matched case-control studies are required to find intervention points of the miR-34s in different types of breast cancer [93, 100, 101].
Therapeutic promising of miR-34s
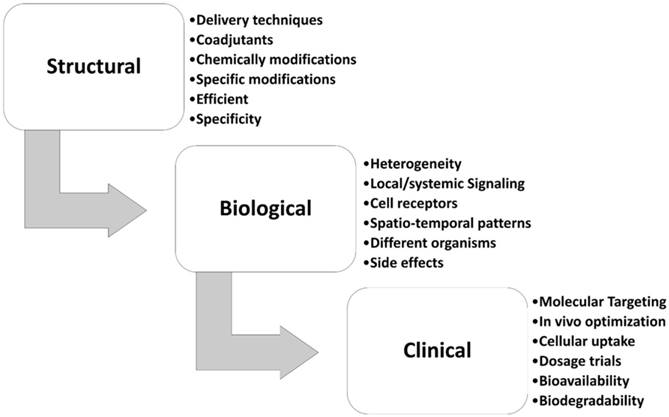
MiR-34s are the first class of tumor suppressor miRNA mimic therapy. Replacing synthetic miR-34s mimics revealed an experimental strategy for the treatment of solid tumors and hematological malignancies [27, 28, 102]. The miR-34a successfully passed the phase I/II of multicenter clinical trial (MRX34, NCT01829971) for patients with unrespectable primary liver cancer, lymphoma, and lung cancer [103]. Also, the MRX34, a mimic of naturally occurring miR-34a encapsulated in liposomal nanoparticle formulation, is in a phase ІІ clinical trial of liver cancer [104]. Retroviral expression vectors are cloned individual human miR-34 family into a derivative of the murine stem cell virus that mostly expresses luciferase or green fluorescent protein [28]. Besides many in vivo and in vitro studies, low efficacy in delivery, non-specific biodistribution, low specific, poorly cellular uptake, and high side effects are the main challenge in miRNA biased therapy of breast cancer [24, 105-108]. Fig. 4 summarizes the steps from the research to the clinical application that should be considered for miR-34s therapeutic approach in breast cancer, in term of biological, structural, and clinical settings. The chemical modifications, nano-delivery, and/or co-delivery of miR-34s mimic's therapy are the most used research approach for the bio-pharmacists research centers (Fig. 4). In this regard, Bader et al systematically compared the administration of the treatment by intravenously injecting miR-34a delivery in mouse models of lymphoma, melanoma, breast, prostate, non-small cell lung, and pancreatic cancers [102]. They found that xenograft breast cancer mice had a 38% repression of tumor growth in comparing with the control group [102]. Hui et al. showed that orally administration of flavonoid compound 3, 6-dihydroxyflavone reduces MNU-induced breast carcinogenesis, with overexpression of miR-34a associated in rats' model [109]. Xie et al. indicated that nanoparticle delivery of miR-34a eradicated long-term-cultured BCSCs by targeting C22ORF28 [87].
Survival correlations of miR-34 family in breast human cancer datasets. A dataset kaplan-meier survival analysis for the relationship between survival time and global expression profiling of miR-34a (a), miR-34b (b), and miR-34c (c) signature in high-risk ER+ breast cancer from patients receiving adjuvant Tamoxifen mono-therapy was performed by using online data set tools of MIRUMIR [98] and Kaplan-Meier [99].
Different challenging areas of miR-34s therapeutic approach in breast cancer. This figure show different stepwise settings for miR-34a therapy in breast cancer.
Co-delivery with chemo drugs
It is well established that co-delivery of miR-34s with the chemo drugs had interesting synergetic effects upon breast cancer, where it could use as a biomarker for chemotherapeutic response. In one comprehensive study, Frères et al. recognized that of 188 circulating miRNAs assessed in the plasma of 25 breast cancer patients, miR-34a is significantly increased after neoadjuvant chemotherapy [110]. This finding introduces miR-34a as a biomarker for chemotherapeutic response [102]. Tissue-specific delivery and cellular uptake are another challenge of miR-34a-based therapy. To achieve sustained target inhibition of tumors in response to oligonucleotides, nanotechnology-based formulations of miRNA are the one of the best options for researchers [111]. In an interesting report, Deng et al. claimed that co-delivery of miR-34a with doxorubicin in hyaluronic acid chitosan nanoparticles significantly increased the anti-tumor effects of doxorubicin in the TNBCs [112]. Misso G et al. discussed the substantial benefits of a new therapeutic concept based on nanotechnology delivery of miRNA mimics in their review [40]. Lipid-based formulation of nano-carriers, like hyaluronic acid and chitosan, are most investigated systems for miR34a-based therapeutic delivery, indicated in a study where Wang et al. deigned against the TNBC samples [113]. In this regard, Zhang et al. introduced the core-shell nanocarrier co-loading with docetaxel and miR-34a as a new nano-platform for the combination of insoluble drugs. This report shows that a combination of miR-34a and docetaxel achieves synergistic therapeutic effects in MBC treatment, due to the highly permeable endothelium of the capillaries and the possibility of passive accumulation of the drug in breast cancer tissues. The co-delivery of miR-34a and docetaxel in nano-carriers suppressed the apoptosis pathways and tumor cell migration by targeting of Bcl-2 in the 4T1 breast cancer cells [59]. In line with this work, the deigning of miR-34a can sensitize a panel of breast cancer cell lines with another breast cancer compounds, indicating that the new formulations of miR-34a is more suitable for therapy of patients with MBC.
Co-delivery with natural compounds
Co-delivery of mir-34s with the natural compounds from dietary sources has a valuable experimental strategy for treatment of solid tumors like breast carcinoma. Co-delivery of miR-34a with Thymoquinone (TQ), a potential small molecular component of Nigella sativa, enhanced inhibition of breast cancer metastasis in vitro [20]. For the first time, this report had shown that the co-delivery of miR-34a+TQ was able to inactivate the downstream of the EMT signaling pathway by directly targeting TWIST1 and ZEB1 [111, 114-116]. In total, re-expression of miR-34s and replacement therapy using miR-34s mimics strongly inhibited cell proliferation, cell cycle progression, self-renewal, EMT, and invasion in breast cancer cell lines [31]. All findings noted in above, making miR-34a a promising therapeutic agent for patients with these diseases [27, 57].
Further directions and changes
Despite extensive studies on detection and therapy of breast cancer, lack of a proper diagnosis and treatment accuracy is major problem for medical researchers. Novel methods should be proposed where miR-34s-based cancer therapy is designed to target more than one miRNA in breast cancer, which depends on the availability of a clinically relevant delivery system [84, 117]. Co-delivery with the common chemo drugs, natural component/chemical-modifying drugs, and mimic therapy of miR-34s such as DNA methylation inhibitors and HDAC inhibitors have shown clinical promises for breast cancer therapy (Fig. 4). However, there are some questions that need to be addressed. MiR-34s are therapeutic targets acting through the entire gene regulatory networks and complex's regulatory cascades. We should consider potential side effects for clinical applications using miR-34s-based drugs. Indeed, further studies are necessary to develop new miR-34s-based drugs that specifically affect the CpG islands promoter region of miR-34s only to reduce the side effects. Therefore, the major challenges and assessment to tackle are: (i) efficacy in appropriate of delivery of miR-34s at in vivo or in vitro models of breast cancer, (ii) miRNA biodistribution, and (iii) preliminary biosafety. The potential usefulness of miR-34s-based therapy in breast cancer needs more research to find epigenetically mechanisms of this noncoding RNA in diagnostic/therapeutic tools of breast cancer.
Conclusions
This review highlights roles of miR-34 family members in tumorigenesis, apoptosis, metastasis, invasion, and chemoresistance of breast cancer by regulating of numerous proto-oncogenes. It is relevant to mention that some miR-34 members, like miR-34a and miR-34c, have independent protective effects on prognosis of breast cancer patients, which are the therapeutic candidates of breast cancer patients in the future. In addition, the possibility of nano-delivery of miR-34s will be useful in targeting of breast cancer cells. Meanwhile, laboratory investigations should continue for better understanding of molecular mechanisms of miR-34s to develop more convenient diagnostics, prognostics, and treatment of breast cancer.
Abbreviations
PR, Progesterone receptor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; miR-34s, MicroRNAs-34; Bcl-2, B-cell lymphoma 2; miRNAs, microRNAs; nt, nucleotides; UTR, untranslated region; mRNAs, messenger RNAs; TNBC, triple negative breast cancer cell; MBC, metastatic breast cancer; oncomiRs, oncogenic miRNAs; RISC, RNA-induced silencing complex; STAT3, signal transducer and activator of transcription 3; SMS, seed-matching sequence; BCSCs, breast cancer stem cell; EMT-TFs, EMT-inducing transcription factors; ZEB, zinc-finger E-box-binding; BCSCs, breast cancer stem cells; HDACs, histone deacetylases; IBC, invasive breast cancer; TQ, Thymoquinone.
Acknowledgements
We gratefully acknowledge all authors and coworkers for their contributions. We sincerely apologize that we cannot cite all contributions due to space limits. We gratefully thank Shangyi Fu for editing the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81672887).
Competing Interests
The authors have declared that no competing interest exists.
References
1. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA: a cancer journal for clinicians. 2017;67(6):439-448
2. Umemura S, Sakamoto G, Sasano H, Tsuda H, Akiyama F, Kurosumi M, Tokuda Y, Watanabe T, Toi M, Hasegawa T. et al. Evaluation of HER2 status: for the treatment of metastatic breast cancers by humanized anti-HER2 Monoclonal antibody (trastuzumab) (Pathological committee for optimal use of trastuzumab). Breast cancer. 2001;8(4):316-320
3. Hurvitz SA, Lalla D, Crosby RD, Mathias SD. Use of the metastatic breast cancer progression (MBC-P) questionnaire to assess the value of progression-free survival for women with metastatic breast cancer. Breast cancer research and treatment. 2013;142(3):603-609
4. Wang M, Ji S, Shao G, Zhang J, Zhao K, Wang Z, Wu A. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clinical & translational oncology. 2017 doi: 10.1007/s12094-017-1805-0
5. Fu S, Cheng J, Wei C, Yang L, Xiao X, Zhang D, Stewart MD, Fu J. Development of diagnostic SCAR markers for genomic DNA amplifications in breast carcinoma by DNA cloning of high-GC RAMP-PCR fragments. Oncotarget. 2017;8(27):43866-43877
6. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature reviews Genetics. 2007;8(2):93-103
7. Tahiri A, Aure MR, Kristensen VN. MicroRNA Networks in Breast Cancer Cells. Methods in molecular biology. 2018;1711:55-81
8. Koleckova M, Janikova M, Kolar Z. MicroRNAs in triple-negative breast cancer. Neoplasma. 2018;65(1):1-13
9. Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochimica et biophysica acta. 2014;1845(2):255-265
10. Yong-Ming H, Ai-Jun J, Xiao-Yue X, Jian-Wei L, Chen Y, Ye C. miR-449a: a potential therapeutic agent for cancer. Anti-cancer drugs. 2017;28(10):1067-1078
11. Mercey O, Popa A, Cavard A, Paquet A, Chevalier B, Pons N, Magnone V, Zangari J, Brest P, Zaragosi LE. et al. Characterizing isomiR variants within the microRNA-34/449 family. FEBS letters. 2017;591(5):693-705
12. Maroof H, Salajegheh A, Smith RA, Lam AK. Role of microRNA-34 family in cancer with particular reference to cancer angiogenesis. Experimental and molecular pathology. 2014;97(2):298-304
13. Ma YY, Tao HQ. Microribonucleic acids and gastric cancer. Cancer science. 2012;103(4):620-625
14. Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB. et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current biology. 2007;17(15):1298-1307
15. Zhao K, Cheng J, Chen B, Liu Q, Xu D, Zhang Y. Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. Journal of thoracic disease. 2017;9(10):3735-3746
16. Yamazaki H, Chijiwa T, Inoue Y, Abe Y, Suemizu H, Kawai K, Wakui M, Furukawa D, Mukai M, Kuwao S. et al. Overexpression of the miR-34 family suppresses invasive growth of malignant melanoma with the wild-type p53 gene. Experimental and therapeutic medicine. 2012;3(5):793-796
17. Beresneva EV, Rykov SV, Hodyrev DS, Pronina IV, Ermilova VD, Kazubskaia TP, Braga EA, Loginov VI. Methylation profile of group of miRNA genes in clear cell renal cell carcinoma; involvement in cancer progression. Genetika. 2013;49(3):366-375
18. Fujino T, Yokosuka A, Higurashi H, Yokokawa R, Sakurai R, Harashima W, Miki Y, Fujiwara Y, Mimaki Y, Hayakawa M. AU-1 from Agavaceae plants causes transient increase in p21/Cip1 expression in renal adenocarcinoma ACHN cells in an miR-34-dependent manner. Journal of natural medicines. 2017;71(1):36-43
19. Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC cancer. 2008;8:266
20. Imani S, Wei C, Cheng J, Khan MA, Fu S, Yang L, Tania M, Zhang X, Xiao X, Zhang X. et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8(13):21362-21379
21. Kaboli PJ, Rahmat A, Ismail P, Ling KH. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacological research. 2015;97:104-121
22. Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer research. 2008;68(11):4123-4132
23. Liu CJ, Ma XW, Zhang XJ, Shen SQ. pri-miR-34b/c rs4938723 polymorphism is associated with hepatocellular carcinoma risk: a case-control study in a Chinese population. International journal of molecular epidemiology and genetics. 2017;8(1):1-7
24. Wang L, Yu J, Xu J, Zheng C, Li X, Du J. The analysis of microRNA-34 family expression in human cancer studies comparing cancer tissues with corresponding pericarcinous tissues. Gene. 2015;554(1):1-8
25. Zhang Q, He XJ, Ma LP, Li N, Yang J, Cheng YX, Cui H. [Expression and significance of microRNAs in the p53 pathway in ovarian cancer cells and serous ovarian cancer tissues]. Zhonghua zhong liu za zhi [Chinese journal of oncology]. 2011;33(12):885-890
26. Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343-349
27. Hermeking H. The miR-34 family in cancer and apoptosis. Cell death and differentiation. 2010;17(2):193-199
28. Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget. 2014;5(4):872-881
29. Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. Journal of molecular cell biology. 2014;6(3):214-230
30. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends in cell biology. 2007;17(3):118-126
31. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-233
32. Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Molecular and cellular pharmacology. 2011;3(3):83-92
33. Maroof H, Salajegheh A, Smith RA, Lam AK. MicroRNA-34 family, mechanisms of action in cancer: a review. Current cancer drug targets. 2014;14(8):737-751
34. Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell cycle. 2008;7(16):2591-2600
35. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9(10):1274-1281
36. Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome biology. 2004;5(9):R68
37. Tarantino C, Paolella G, Cozzuto L, Minopoli G, Pastore L, Parisi S, Russo T. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB journl: official publication of the Federation of American Societies for Experimental Biology. 2010;24(9):3255-3263
38. Rao DS, O'Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48-59
39. Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14(3):432-444
40. Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P. et al. Mir-34: a new weapon against cancer? Molecular therapy Nucleic acids. 2014;3:e194
41. Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell cycle. 2009;8(5):712-715
42. Adams BD, Parsons C, Slack FJ. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert opinion on therapeutic targets. 2016;20(6):737-753
43. Wong KY, Yu L, Chim CS. DNA methylation of tumor suppressor miRNA genes: a lesson from the miR-34 family. Epigenomics. 2011;3(1):83-92
44. Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, Fiegl H, Rossler J, Wiedemair A, Concin N. et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC cancer. 2016;16:102
45. Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, Bu P, Vogel H, Jablons DM, Keller AC. et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes & development. 2014;28(5):438-450
46. Javeri A, Ghaffarpour M, Taha MF, Houshmand M. Downregulation of miR-34a in breast tumors is not associated with either p53 mutations or promoter hypermethylation while it correlates with metastasis. Medical oncology. 2013;30(1):413
47. Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for Sequence-Based miRNA Target Prediction: What to Choose?. International journal of molecular sciences. 2016:17 (12)
48. Wirth H, Cakir MV, Hopp L, Binder H. Analysis of microRNA expression using machine learning. Methods in molecular biology. 2014;1107:257-278
49. Cirillo D, Livi CM, Agostini F, Tartaglia GG. Discovery of protein-RNA networks. Molecular bioSystems. 2014;10(7):1632-1642
50. Walsh CJ, Hu P, Batt J, Dos Santos CC. Discovering MicroRNA-Regulatory Modules in Multi-Dimensional Cancer Genomic Data: A Survey of Computational Methods. Cancer informatics. 2016;15(Suppl 2):25-42
51. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nature reviews Genetics. 2012;13(5):358-369
52. Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PloS one. 2012;7(8):e42034
53. Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, Lv Z, Cong X. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA and cell biology. 2013;32(12):699-707
54. Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30(25):2888-2899
55. Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast cancer research and treatment. 2011;130(2):663-679
56. Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32(36):4294-4303
57. Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM. et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Science signaling. 2011;4(197):ra71
58. Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER. et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011;195(3):417-433
59. Zhang L, Yang X, Lv Y, Xin X, Qin C, Han X, Yang L, He W, Yin L. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Scientific reports. 2017;7:46186
60. Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clinical and experimental medicine. 2013;13(2):109-117
61. Hau A, Ceppi P, Peter ME. CD95 is part of a let-7/p53/miR-34 regulatory network. PloS one. 2012;7(11):e49636
62. Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang L, Fan S, Fan P, Hou Z, Li J. et al. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget. 2015;6(12):10432-10444
63. Muppala S, Mudduluru G, Leupold JH, Buergy D, Sleeman JP, Allgayer H. CD24 induces expression of the oncomir miR-21 via Src, and CD24 and Src are both post-transcriptionally downregulated by the tumor suppressor miR-34a. PloS one. 2013;8(3):e59563
64. Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H. et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer research. 2014;74(24):7573-7582
65. Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, Tang JH. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Archives of medical research. 2012;43(7):514-521
66. Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X, Liu Y, He Y, Park EY, Zhang H. et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. The Journal of biological chemistry. 2012;287(1):465-473
67. Wushou A, Hou J, Zhao YJ, Shao ZM. Twist-1 up-regulation in carcinoma correlates to poor survival. International journal of molecular sciences. 2014;15(12):21621-21630
68. Zhao J, Lammers P, Torrance CJ, Bader AG. TP53-independent Function of miR-34a via HDAC1 and p21CIP1/WAF1. Molecular therapy. 2013;21(9):1678-1686
69. Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ. et al. A microRNA DNA methylation signature for human cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13556-13561
70. Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer research. 2008;68(24):10128-10136
71. Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, Penalva LO. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA biology. 2011;8(5):817-828
72. Zhang DG, Zheng JN, Pei DS. P53/microRNA-34-induced metabolic regulation: new opportunities in anticancer therapy. Molecular cancer. 2014;13:115
73. Werner TV, Hart M, Nickels R, Kim YJ, Menger MD, Bohle RM, Keller A, Ludwig N, Meese E. MiR-34a-3p alters proliferation and apoptosis of meningioma cells in vitro and is directly targeting SMAD4, FRAT1 and BCL2. Aging. 2017;9(3):932-954
74. Achari C, Winslow S, Ceder Y, Larsson C. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC cancer. 2014;14:538
75. Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecular cell. 2007;26(5):731-743
76. O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast cancer research. 2010;12(2):201
77. Wu MY, Fu J, Xiao X, Wu J, Wu RC. MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer letters. 2014;354(2):311-319
78. Bao B, Azmi AS, Ali S, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness. Biochimica et biophysica acta. 2012;1826(2):272-296
79. Imani S, Hosseinifard H, Cheng J, Wei C, Fu J. Prognostic Value of EMT-inducing Transcription Factors (EMT-TFs) in Metastatic Breast Cancer: A Systematic Review and Meta-analysis. Scientific reports. 2016;6:28587
80. Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell cycle. 2011;10(24):4256-4271
81. Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. International journal of molecular sciences. 2012;13(10):13414-13437
82. Si W, Li Y, Shao H, Hu R, Wang W, Zhang K, Yang Q. MiR-34a Inhibits Breast Cancer Proliferation and Progression by Targeting Wnt1 in Wnt/beta-Catenin Signaling Pathway. The American journal of the medical sciences. 2016;352(2):191-199
83. Mansoori B, Mohammadi A, Shirjang S, Baghbani E, Baradaran B. Micro RNA 34a and Let-7a Expression in Human Breast Cancers is Associated with Apoptotic Expression Genes. Asian Pacific journal of cancer prevention: APJCP. 2016;17(4):1887-1890
84. Wang R, Ma J, Wu Q, Xia J, Miele L, Sarkar FH, Wang Z. Functional role of miR-34 family in human cancer. Current drug targets. 2013;14(10):1185-1191
85. Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, Rimm DL, Gyorffy B, Santarpia L, Pusztai L, Saltzman WM, Slack FJ. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer research. 2016;76(4):927-939
86. Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28(25):2419-2424
87. Lin X, Chen W, Wei F, Zhou BP, Hung MC, Xie X. Nanoparticle Delivery of miR-34a Eradicates Long-term-cultured Breast Cancer Stem Cells via Targeting C22ORF28 Directly. Theranostics. 2017;7(19):4805-4824
88. Siemens H, Jackstadt R, Kaller M, Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4(9):1399-1415
89. John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Simone CB 2nd, Falduto MT, Magnuson SR, Coleman CN. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiation research. 2012;178(3):105-117
90. Erbes T, Hirschfeld M, Rücker G, Jaeger M, Boas J, Iborra S, Mayer S, Gitsch G, Stickeler E. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC cancer. 2015;15(1):1
91. Mishra S, Srivastava AK, Suman S, Kumar V, Shukla Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer letters. 2015;369(1):67-75
92. Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clinical chemistry. 2013;59(10):1489-1496
93. Imani S, Zhang X, Hosseinifard H, Fu S, Fu J. The diagnostic role of microRNA-34a in breast cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(14):23177-23187
94. Agarwal S, Hanna J, Sherman M, Figueroa J, Rimm D. Quantitative assessment of miR34a as an independent prognostic marker in breast cancer. British journal of cancer. 2015;112(1):61-68
95. Lee JY, Kong G. Roles and epigenetic regulation of epithelial-mesenchymal transition and its transcription factors in cancer initiation and progression. Cellular and molecular life sciences. 2016;73(24):4643-4660
96. Sun L, Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cellular and molecular life sciences. 2016;73(23):4493-4515
97. Peng F, Xiong L, Tang H, Peng C, Chen J. Regulation of epithelial-mesenchymal transition through microRNAs: clinical and biological significance of microRNAs in breast cancer. Tumour biology. 2016;37(11):14463-14477
98. Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell death and differentiation. 2013;20(2):367
99. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123(3):725-731
100. Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. Journal of surgical oncology. 2012;106(8):947-952
101. Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochimica et biophysica acta. 2010;1806(2):200-207
102. Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in genetics. 2012;3:120
103. Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investigational new drugs. 2017;35(2):180-188
104. Van Roosbroeck K, Calin GA. Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. Advances in cancer research. 2017;135:119-149
105. Casey MC, Sweeney KJ, Brown JA, Kerin MJ. Exploring circulating micro-RNA in the neoadjuvant treatment of breast cancer. International journal of cancer. 2016;139(1):12-22
106. Zeng Y, Zhou Z, Fan M, Gong T, Zhang Z, Sun X. PEGylated Cationic Vectors Containing a Protease-Sensitive Peptide as a miRNA Delivery System for Treating Breast Cancer. Molecular pharmaceutics. 2017;14(1):81-92
107. Ji Y, Han Z, Shao L, Zhao Y. Evaluation of in vivo antitumor effects of low-frequency ultrasound-mediated miRNA-133a microbubble delivery in breast cancer. Cancer medicine. 2016;5(9):2534-2543
108. Shu D, Li H, Shu Y, Xiong G, Carson WE 3rd, Haque F, Xu R, Guo P. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS nano. 2015;9(10):9731-9740
109. Hui C, Yujie F, Lijia Y, Long Y, Hongxia X, Yong Z, Jundong Z, Qianyong Z, Mantian M. MicroRNA-34a and microRNA-21 play roles in the chemopreventive effects of 3,6-dihydroxyflavone on 1-methyl-1-nitrosourea-induced breast carcinogenesis. Breast cancer research. 2012;14(3):R80
110. Freres P, Wenric S, Boukerroucha M, Fasquelle C, Thiry J, Bovy N, Struman I, Geurts P, Collignon J, Schroeder H. et al. Circulating microRNA-based screening tool for breast cancer. Oncotarget. 2016;7(5):5416-5428
111. Wang J, Yang M, Li Y, Han B. The Role of MicroRNAs in the Chemoresistance of Breast Cancer. Drug development research. 2015;76(7):368-374
112. Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y. et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35(14):4333-4344
113. Wang S, Cao M, Deng X, Xiao X, Yin Z, Hu Q, Zhou Z, Zhang F, Zhang R, Wu Y. et al. Degradable hyaluronic acid/protamine sulfate interpolyelectrolyte complexes as miRNA-delivery nanocapsules for triple-negative breast cancer therapy. Advanced healthcare materials. 2015;4(2):281-290
114. Yang J, Kuang XR, Lv PT, Yan XX. Thymoquinone inhibits proliferation and invasion of human nonsmall-cell lung cancer cells via ERK pathway. Tumour biology. 2015;36(1):259-269
115. Abdelfadil E, Cheng YH, Bau DT, Ting WJ, Chen LM, Hsu HH, Lin YM, Chen RJ, Tsai FJ, Tsai CH. et al. Thymoquinone induces apoptosis in oral cancer cells through p38beta inhibition. The American journal of Chinese medicine. 2013;41(3):683-696
116. Peng L, Liu A, Shen Y, Xu HZ, Yang SZ, Ying XZ, Liao W, Liu HX, Lin ZQ, Chen QY. et al. Antitumor and anti-angiogenesis effects of thymoquinone on osteosarcoma through the NF-kappaB pathway. Oncology reports. 2013;29(2):571-578
117. Huang RY, Wong MK, Tan TZ, Kuay KT, Ng AH, Chung VY, Chu YS, Matsumura N, Lai HC, Lee YF. et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell death & disease. 2013;4:e915
118. Voorhoeve PM, Agami R. Classifying microRNAs in cancer: the good, the bad and the ugly. Biochimica et biophysica acta. 2007;1775(2):274-282
Author contact
![]() Corresponding author: Tel-fax: +86-830-3160283. E-mail: fujunjiangedu.cn and fujunjiangcom.
Corresponding author: Tel-fax: +86-830-3160283. E-mail: fujunjiangedu.cn and fujunjiangcom.

 Global reach, higher impact
Global reach, higher impact