Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(21):3991-3999. doi:10.7150/jca.27697 This issue Cite
Research Paper
Serum level of co-expressed hub miRNAs as diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma
1. Department of Oncology, Beijing Friendship Hospital, Capital Medical University, Beijing, 100050, China.
2. Desautels Faculty of Management, McGill University, Montreal, Quebec, H3A 1G5, Canada.
3. Schulich School of Music, McGill University, Montreal, Quebec, H3A 1E3, Canada.
4. Laboratory of Biorheology and Medical Ultrasonics, University of Montreal Hospital Research Center, Montreal, Quebec, H2X0A9, Canada.
5. State Key Laboratory of Radiation Medicine and Protection, School of Radiation Medicine and Protection, Soochow University, Suzhou 215123, China.
6. Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Capital Medical University, Beijing, 100069, China.
7. Department of Mathematics, Nanjing University, 210023, China.
8. Department of Pathology, Research Institute of McGill University Health Center, Montreal, Quebec, H4A 3J1, Canada
Received 2018-6-6; Accepted 2018-7-29; Published 2018-10-11
Abstract
Background: Sensitive and specific non-invasive biomarkers are urgently needed in order to improve the survival of patients with pancreatic ductal adenocarcinoma (PDAC), which is the fourth leading cause of cancer-related death. We aim to identify serum hub miRNAs as potential diagnostic and prognostic biomarkers for PDAC.
Methods: A total of 2578 serum miRNA expression data from 88 PDAC patients and 19 healthy subjects were downloaded from the Gene Expression Omnibus database. Weighted gene co-expression network analysis (WGCNA) was constructed and significant modules were extracted from the network by WGCNA R package. Network modules and hub miRNAs closely related to PDAC were identified. The prognostic value of hub miRNAs was assessed by Kaplan-Meier overall survival analysis.
Results: Two modules strongly associated with PDAC were identified by WGCNA, which were labeled as turquoise and brown respectively. Within each module, twenty hub miRNAs were found. At the functional level, turquoise module was mainly associated with tumorigenesis pathways such as P53 and WNT signaling pathway, while the brown module was mostly related to the pathways of cancer such as RNA transport and MAPK signaling pathway. Utilizing overall survival analyses, five “real” miRNAs were able to stratify PDAC patients into low-risk and high-risk groups.
Conclusions: The association of specific Hub miRNAs with the development of pancreatic cancer was established by WGCNA analysis. Five miRNAs (mir-16-2-3p, mir-890, mir-3201, mir-602, and mir-877) were identified as potential diagnostic and prognostic biomarkers for PDAC.
Keywords: Hub miRNAs, miRNAs, pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) contributes to nearly 80% pancreatic cancer PDAC which is the fourth leading cause of cancer-related death. Compared with the steady improvement in survival for most other cancer types, little advancement have been made in PDAC, which remains a 3% 5-year survival rate [1]. By 2030, PDAC will likely surpass colorectal and breast cancers to become the second leading cause of tumor-related death in both men and women [2]. Due to the deep anatomical location of the human pancreas, tumor-specific symptoms such as abdominal mass, jaundice, and weight loss typically emerge only after the tumor has reached an advanced stage, and is either unresectable or has already metastasized to the liver or other organs [3]. Up to now, CA19-9 is the only clinically available FDA approved serum biomarker for PDAC. However, due to its low sensitivity and specificity, CA19-9 is primarily used for monitoring tumor recurrence after surgery rather than being used as a primary diagnostic and prognostic biomarker [4-7]. In order to improve the survival of patients with pancreatic cancer, sensitive and specific non-invasive diagnostic and prognostic biomarkers are urgently needed.
MiRNAs are small (20-24 nucleotides) non-encoding RNAs that regulate gene expression at the transcriptional or post-transcriptional levels [8]. The circulating miRNAs bind to targeted messenger RNA (mRNAs) and generally inhibit translation or reduce the stability of those mRNAs [9]. MiRNAs could regulate a broad range of biological processes, like cell-cycle, stress response, cell differentiation, apoptosis and invasion. Aberration of expression and functions of miRNAs has been reported to be greatly involved in tumorigenesis and progression [10-12]. Recent studies showed that miRNA-143 can promote cell apoptosis and regulate the process of pancreatic cancer by inhibiting the migration and invasion of pancreatic cancer cells. Eight miRNAs (miR-1301, miR-598, miR-1180, miR-155, miR-496, miR-203, miR-193b, miR-135b) were reported as independent predictors for the survival of patients with PDAC [13,14]. Therefore, miRNAs are considered to hold the potential as novel diagnostic biomarkers and therapeutic targets of cancers including PDAC.
Weighted gene co-expression network analysis (WGCNA) is an effective modular biological analysis methodology that could help us to identify and screen co-expressed modules and key biomarkers, such as miRNAs and lncRNAs (long non-coding RNAs) [15-18]. WGCNA can group miRNAs into a module or network due to their similar expression in association with a given cancer type. In this study, we applied WGCNA methodology to build co-expressed modules and to identify hub miRNAs potentially involved in the pathogenesis and malignant behavior of pancreatic cancer. We attempted to stratify PDAC patients into low-risk and high-risk groups using a specific group of miRNAs identified from this analysis. Data derived from this analysis is supposed to provide statistically convincing evidence that certain hub miRNAs have the potential to become clinically applicable diagnostic and prognostic biomarkers for PDAC.
2 Materials and methods
2.1 MiRNA expression data collection
Microarray dataset of the serum samples of pancreatic cancer patients was downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Dataset GSE85589 was used to construct co-expression networks and identify hub genes in this study. It contained miRNA expression data of serum samples obtained from 19 healthy subjects and 88 PDAC patients. The platform was GPL19117, Affymetrix Multispecies miRNA-4 Array.
The expression data GSE85589 which had been normalized using Robust Multi-Array Average (RMA)[19] was downloaded. Then, probe sets were mapped to gene symbols according to the probe annotation files of the GPL10558 platform. Afterwards, the gene expression values were log2 transformed. Reshape2 and ggplot2 were R-based open-source software packages, which conducted a box plot and correlation plot of normalized samples data for pancreatic cancers (PC) and normal subjects (N) (Supplementary Fig 1). Then, miRNA probe sets were mapped to gene-symbols according to the probe annotation file of the GPL19117 platform. If multiple probes mapped to a same gene-symbol, the biggest value of all probes that mapped to the same gene symbol was chosen to represent this particular gene.
2.2 Weighted gene co-expression networks and their modules
WGCNA package was a freely accessible R software (R 3.4.3), for the construction of weighted gene co-expression networks [11]. Under “WGCNA” environment of R software, by means of goodSamplesGenes function, miRNA data matrix was checked whether they were the good samples and good genes. Then, we applied the pickSoftThreshold function to assess whether the topology of the networks was scale-free, as is required for WGCNA analysis. β was a softthresholding-power parameter that could strengthen strong correlations and penalize weak correlations between genes. Here, the power of β = 1 (scale free R2 =0.89), which was selected to ensure a scale free network (Fig 1). Then, a hierarchical clustering tree was constructed with different branches of the tree representing different gene modules [20]. Three different ways can be selected to construct the network and identify modules according to different needs. In this study, the one-step function was used for network construction and detection of consensus modules. (Fig 1)
2.3 Identification of clinical significant modules and hub miRNAs and functional annotation
To identify clinically significant modules, the module eigengene (ME) was calculated for representing and summarizing each module. This measure can be considered to be representative of gene expression profiles and can be used to capture the maximal amount of variation in the module. To quantify module-trait associations, given that we had a summary profile (ME) for each module, we correlated ME with external traits and searched for the most significant associations. This calculation was referred to as the Module-Trait Relationships [21]. For inter-modular analysis, we evaluated the Gene Significance (GS) and Module Membership (MM), the latter is also known as eigengene-based connectivity (kME). GS is the absolute value of the correlation between a specific gene and a clinical trait. Using the GS and ME, we can identify key modules that have a high significance for a clinical trait and important module membership [21].
Checking scale free topology (up) and Clustering dendrograms and modules identified by WGCNA (down)
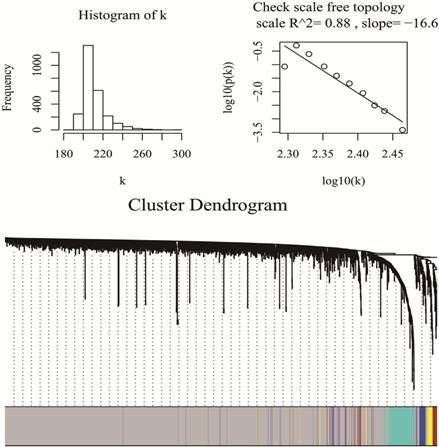
To identify hub miRNAs, the functions of R software module Eigengenes and signedKME were used to identify highly connected intra-modular miRNAs, also called hub miRNAs. In particular, the moduleEigengenes function calculates the eigengene of a module, which is a virtual gene representing the gene expression profile of the entire module. The signedKME function measures the distance of the expression profile of a gene to that of the module eigengene and, thus, quantifies how close a gene is to a module, i.e., it measures the module membership of a gene. Within a module, the genes with the highest module membership scores are considered as hub genes of that module.
2.4 Functional annotation of hub miRNAs
In analogy to previous studies [18, 22], we selected the 20 most connected hub miRNAs for each key module. The miRNet web tool [23] was used to identify the biological pathways, processes, molecular functions and cellular components statistically enriched for the corresponding miRNA target genes. In particular, this tool identifies the enriched KEGG and REACTOME pathways and Gene Ontology (GO) terms based on experimentally confirmed miRNA targets. Since this tool exploits validated miRNA targets, it ensures a higher reliability than tools based on predicted miRNA targets.
2.5 Kaplan-Meier Survival analyses and efficacy evaluation
In order to evaluate the identified hub miRNAs as prognostic PDAC biomarkers, we conducted survival analyses using the SurvMicro web method [24] Survival analyses were performed on an independent PDAC dataset present in The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) containing miRNA expression and survival data derived from 54 PDAC patients (Supplementary Table 1). The SurvMicro tool divides samples into high-risk and low-risk groups through the median of the prognostic index obtained via a Cox Regression model. After this, it generates the hazard ratios (HR) with relative confidence intervals (CI) and p-values. We aim to find the “real” hub miRNAs associated with the prognosis of PDAC.
3. Results
3.1 Screening the miRNA modules
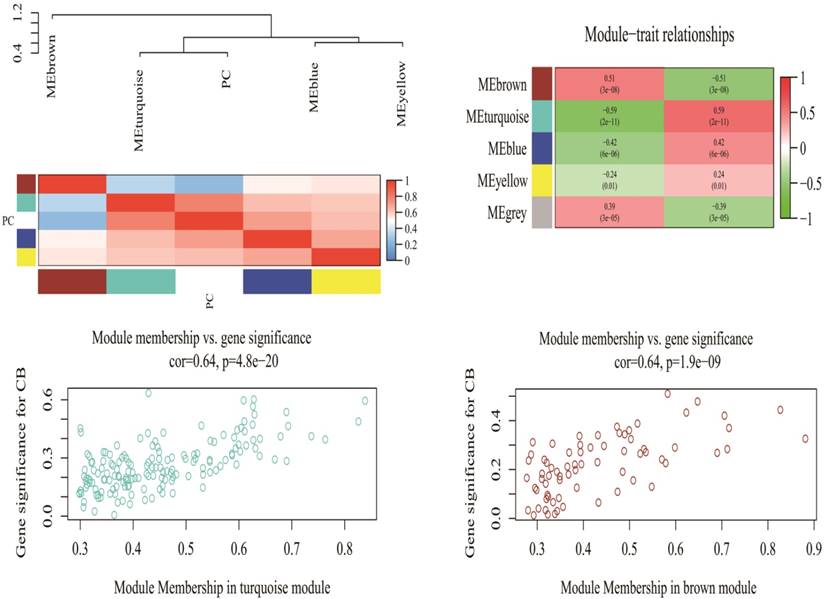
A total of 2578 miRNAs were collected after data preprocessing. The selected miRNAs in each category were plotted in their corresponding expression heat-maps (Supplementary Fig 2). WGCNA analysis of the sequencing data from serum miRNA of PDAC patients and healthy subjects revealed a total of five functional modules (i.e., turquoise, brown, blue, yellow and grey module) (Fig 1). In line with previous reported approach, the grey module belongs to the special WGCNA category that has no association with the disease being analyzed [12]. As shown in the Matrix of Module-Trait Relationships (Fig 2), the module represented by a turquoise color showed a positive association (r = 0.59; p = 1e-22) with PDAC, whereas, the brown module had a negative association with PDAC (r = -0.51; p = 3e-08).Using the GS and MM measures, compared with blue and yellow modules, the turquoise and brown modules showed a significantly higher difference between PDAC group and normal group (Turquoise: r = 0.64; p = 4.8e-20; Brown: r = 0.64; p = 1.9e-09). These data justified the selection of the turquoise and brown modules as key miRNA modules for PDAC. (Fig 2)
3.2 Detection of hub miRNAs and their functional annotations
The signedKME and module Eigengenes functions in “WGCNA” package were applied to determine the highly-connected intramodular miRNAs, i.e hub miRNAs. In particular, the moduleEigengene function calculates the eigengene of one module, which was as a virtual miRNA representing the expression profile of the entire module.
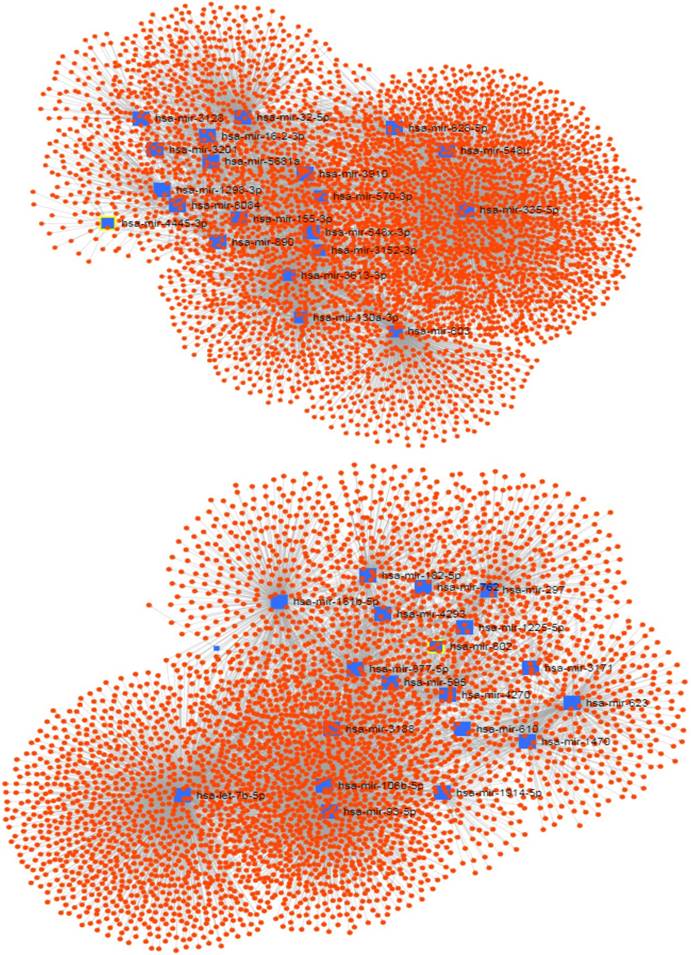
The signedKME function measured the module membership of each miRNA in a special module. The miRNAs with the highest module membership scores are considered as hub miRNAs of that module. Since the turquoise and brown modules were found to be highly associated with the PDAC serum miRNA network, we set out to determine the hub miRNAs that are represented by selecting miRNAs with the highest module membership scores. It is conceivable that these miRNAs may play important roles in the pathogenesis of pancreatic cancer. The top 20 hub miRNAs identified for each module in the PDAC network are listed in Table 1, and their network of with their experimentally validated target genes are shown in Figure 3.
Matrix of Module-Trait Relationships (MTRs) and p-values for pancreatic cancer (up). Scatter plot of Gene Significance (GS) for the cancer condition versus Module Membership (kME) for the tumor-related module (down). PC stands for pancreatic cancer. Each row corresponds to a module eigengene, and each column corresponds to PC. Each cell contains a corresponding correlation and p-value in brackets.
20 Hub miRNAs identified in the PDAC network and Functional annotation of hub miRNAs and enriched KEGG pathways
| Module | Top 20 microRNAs | KEGG | Reactame | GO:BP | GO:MF | GO:CC |
|---|---|---|---|---|---|---|
| Turquoise | miR-3201 miR-8084 miR-3128miR-3910 miR-548x-3p miR-1298-3pmiR-32-5p miR-4445-3p miR-5681amiR-890 miR-335-5p miR-603miR-3152-3p miR-16-2-3p miR-3613-3p miR-155-3p miR-548u miR-628-5p miR-130a-3p miR-570-3p | Pancreatic cancer,Pathways in cancer,MAPK signaling pathway,ErbB signaling pathway,Wnt signaling pathway,Prostate cancer,colorectal cancer,Cytokine-cytokine receptor interaction,T cell receptor signaling pathway | Cytokine Signaling in Immune system,Transmembrane transport of small molecules,Axon guidance, Signalling by NGF,Signaling by PDGF,Downstream signal transduction,NGF signalling via TRKA from the plasma membrane,(Signalling by NGF,FGFR1-4,and ERBB2,ERBB4),Developmental Biology,Generic Transcription Pathway | regulation of anatomical structure morphogenesis,regulation of cell proliferation | regulation of transcription DNA-dependent,enzyme binding,zinc ion binding | nucleoplasm |
| Brown | miR-7b-5p miR-93-5p miR-181b-5pmiR-182-5p miR-106b-5p miR-595miR-602 miR-610 miR-623miR-297 miR-877-5p miR-1225-5pmiR-1469 miR-1470 miR-1914-5pmiR-762 miR-3171 miR-3188miR-4293 miR-4270 | Pathways in cancer,Prostate cancer,Insulin signaling pathway,RNA transport,Non-small cell lung cancer,Pancreatic cancer,MAPK signaling pathway,Regulation of actin cytoskeleton | Gene Expression,Cell Cycle,Immune System,Signalling by NGF,Signaling by FGFR1-4,Generic Transcription Pathway,Signaling by the B Cell Receptor (BCR) | cellular macromolecule catabolic process,cell division | nucleotide binding,transition metal ion binding,purine nucleotide binding | cytosol,nuclear lumen |
In order to functionally annotate the hub miRNAs, we conducted a functional enrichment analysis limited to experimentally confirmed miRNA target genes. By means of the miRNet web tool, the hub miRNAs in each module was analyzed to identify enriched KEGG and REACTOME pathways and Gene Ontology (GO) terms (Table 1). According to KEGG and REACTOME and GO function analyses, the turquoise module was mainly enriched in cancer-related terms such as MAPK and WNT signaling, etc, while the brown module was mainly related to the functions including cancer-associated pathways, such as RNA transport.
3.3 Hub miRNAs validation and further distinction
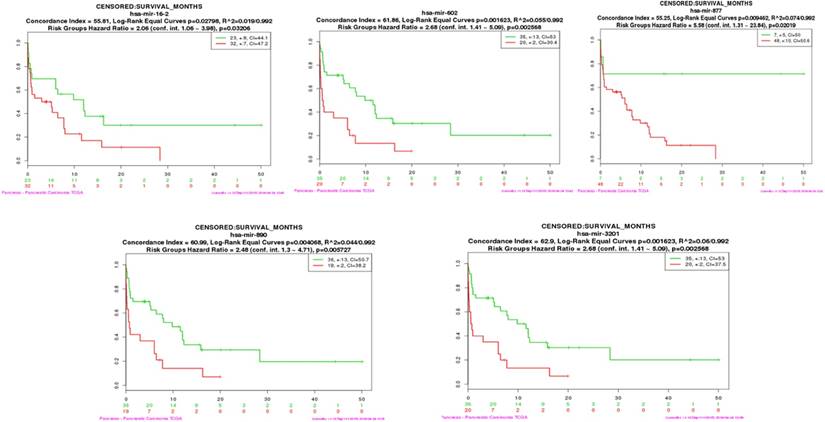
To determine whether the serum candidate miRNAs identified in this study could be used as prognostic biomarkers, we assessed if hub miRNAs in the turquoise and brown modules were connected with the overall survival (OS) of pancreatic cancer patients [18]. We performed both multi-miRNA (top 20 hub miRNAs) survival analyses of PDAC patients in each module by using the SurvMicro tool. By applying the top 20 hub miRNAs as input for the SurvMicro tool, we found that the miRNA profiles in all modules and single miRNA successfully stratified PDAC patients into high-risk and low-risk groups as revealed by Kaplan-Meier survival plots (table 2). Specifically, the OS time in the high-risk group patients was found at least two-fold shorter compared with those of patients in the low-risk group (HR 6.93 [95% CI 3.03-15.89] p = 4.687e-06 for the turquoise module, HR 3.80 [95% CI 1.94-7.46] p = 9.923e-05 for the brown module) (supplementary Fig 3). In addition, we set out to identify “real” miRNAs that were most significantly associated with PDAC survival outcomes by judging whether the P value of each miRNA was less than 0.05. An increased expression of three miRNAs in turquoise module (mir-16-2-3p (HR 2.05 [95% CI 1.08-3.89], p=0.032), mir-890 (HR 2.48 [95% CI 1.30-4.71], p=0.006), and mir-3201 (HR 2.68 [95% CI 1.41-5.09], p=0.003) and two miRNAs in brown module ( mir-602 (HR 2.68 [95% CI 1.41-5.09], p=0.003), and mir-877 (HR 5.58 [95% CI 1.31-23.84], p=0.020) were found to be associated with a significantly better survival (Fig 4 and Table 2).
Discussion
Since its introduction in 2005, WGCNA methodology has been proven to be an effective tool for describing the correlation patterns among genes across microarray samples [25]. WGCNA is particularly useful in finding modules of highly correlated genes and summarizing many intramodular hub genes that can be extrapolated for the diagnosis and prognosis of cancers. WGCNA involves several sequential steps. Firstly, the absolute values of the connection of paired miRNAs were calculated for the co-expression network. Secondly, an adjacency matrix was built to define the miRNAs strength with each other. A soft threshold parameter was under the weighted network. Topological overlap (TOM) was used to cluster miRNAs into different network modules that combined the adjacency of two direct miRNAs and the connection strengths with other indirect miRNAs. The miRNAs in one module could be characterized with the module eigengene (ME). To find the key modules related to clinical traits, the correlations were calculated between MEs and those clinical traits. The correlation between miRNAs and MEs was applied to distinguish hub miRNAs [21]. In this study, we applied this methodology to build co-expressed modules and identify hub miRNAs potentially involved in the tumorigenesis of pancreatic cancer. Among those identified hub miRNAs, we further determined “real” miRNAs that could predict the behavior of PDAC.
The network of hub miRNAs of the turquoise (up) and brown (down) modules with their experimentally validated target genes.
The survival analysis plot of miR-16-2-3p, miR-602, miR-877-5p, miR-890, and miR-3201
Single hub miRNA survival analysis tested by TCGA PDAC miRNAs data
| Module | Hub miRNA | HR | 95%CI | P value |
|---|---|---|---|---|
| Turquoise | miR-16-2-3p | 2.06 | 1.06-3.98 | 0.032 |
| miR-3201 | 2.68 | 1.41-5.09 | 0.003 | |
| miR-890 | 2.48 | 1.30-4.71 | 0.006 | |
| Brown | miR-877-5p | 5.58 | 1.31-23.84 | 0.02 |
| miR-602 | 2.68 | 1.41-5.09 | 0.003 |
Similar work by Siân Jones [26], et al has identified 12 partially overlapping signaling pathways that are genetically altered in the great majority of pancreatic cancer. By means of WGCNA, we found that 20 hub miRNAs were closely associated with, MAPK, P53 and WNT signaling pathways, which are included (WNT and MAPK signaling) or greatly involved (p53 in cell cycle, invasion, DNA damage and cell apoptosis signaling) in the pathways mentioned above. More concretely, target genes of those miRNAs includes TP53BP2 (tumor protein p53 binding protein 2), dkk3 (WNT signaling pathway inhibitor dickkopf-3), and WISP2 (WNT1 inducible signaling pathway protein 2) [27]. We also identified another set of 20 hub miRNAs that were closely associated with the functions of PDAC and RNA transport, whose targeted genes include TP53INP2 (tumor protein p53 inducible nuclear protein). Our data suggest miRNAs are key regulators of these core tumorigenic and functional pathways in PDAC.
High serum levels of certain miRNAs might indicate the presence of an underlying malignancy. Mahito Miyamae reported that high level of six circulating oncogenic miRNAs (miR-615-5p, -744, -575, -557, -675, and -550a) could be useful biomarkers for PDAC screening, monitoring recurrence after surgery, and predicting the prognosis of patients with PDAC [28]. Schultz et al reported that the area under the curve (AUC) was 0.91 (95% CI, 0.87-0.94) when using whole blood level of 10 miRNAs (miR-26b, miR-34a, miR-122, miR-126, miR-145, miR-150, miR-223, miR-505, miR-636, miR-885-5p) to diagnose PDAC [29]. Moreover, the increased miR-16 and miR-196a combined with CA19-9 have been shown to be more effective to distinguish pancreatic cancer from normal individuals [30]. Brand et al found that the combination of CA19-9, ICAM-1 and OPG could discriminate PDAC patients from health controls with the 78% sensitivity and 94% specificity [31]. In the present study, we identified a list of specific Hub miRNAs that are associated with PDAC tumorigenesis and a list of hub miRNAs that are associated with the development of pancreatic cancer. The association of circulating miRNAs with PDAC tumorigenesis and progression indicates that circulating hub miRNAs could not only be a diagnostic biomarker, but also predict the malignant behavior of the tumor.
To further study the prognostic value of circulating hub miRNAs, we identified 5 “real” miRNAs (mir-16-2-3p, mir-890, mir-3201, mir-602, and has-mir-877) that could stratify PDAC patients into high and low risk groups. miR-15b/16-2 cluster could modulate the expression of cell cycle-regulating genes, CCND2, CyclinD2 and IGF1R, and was deeply involved in proliferation and anti-apoptotic pathways of different kind of malignancies such as leukemia and colorectal cancer [32]. Mir-15b was also suggested to promote epithelial-mesenchymal transition (EMT) and metastasis in pancreatic cancers by targeting smurf2 [33]. MiR-16-2-3p has been reported in several studies as a diagnostic biomarker for hepatocellular carcinoma with a high sensitivity and specificity [34, 35]. Mir-890 belongs to a mir-888 genomic cluster located on human X chromosome Xq27.3 that specifically targets RBL1, KLF5, SMAD4, and TIMP2 and functions as pro-oncogenic factors that increased prostate tumor growth in vivo [36], whose function, however, has not been stratified in other cancer types [37]. The function of miR-3201 has been still controversial. MiR-3201 expression was significantly reduced in recurrent epithelial ovarian cancer (EOC) [38], indicative of a suppressive role during cancer recurrence. Other study showed high expression of mir-3201 in melanoma patients [39]. However, in the current study, serum level of mir-3201, which was suggested to be involved in several of cancer-promoting pathways (table 1), predicts a poor prognosis of PDAC patients. The discrepancies between these studies might arise from the cancer types of different tissue origin, or the sample type, with the former being cancerous tissues and the latter being serum. Mir-602 plays a pro-carcinogenic role in HBV-related hepatocarcinoma by inhibiting RASSF1A [40], and with other three miRNAs, was selected to identify the overall survival of glioblastoma multiforme [41]. It can also regulates the expression of sonic hedghog pathway in chondrocytes [42], which was also one of the core signalings during the tumorigenesis of PDAC [26]; miR‑877 has been shown to suppress the proliferation and migration of (RCC) cells by modulating the eEF2K/eEF2 signaling pathway [43]. The binding site of miR-877-3p was on the promoter site of cancer suppressor gene p16 which alters frequently in tumors. MiR-877-3p targets the p16 promoter and are related to increased risk of a wide range of cancers such as bladder cancer, renal cell carcinoma cells as well as pancreatic ductal adenocarcinoma [44-46]. It is conceivable that these 5 miRNAs involved in the tumorigeneis and mailgnant function of PDAC via completely different mechanisms. The conbination of these 5 miRNAs can be a reliable biomarker for predicting the behavior of PDAC.
In summary, using WGCNA technology we identified 20 hub miRNAs that are associated with tumorigenesis pathways and 20 hub miRNAs that are associated with functional pathways in tumors. Among the 40 hub miRNAs, 5 “real” miRNAs were able to stratify the PDAC patients into high and low risk groups. Our data indicate certain hub miRNAs can function as diagnostic and prognostic biomarkers for PDAC.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the Beijing Li Huanying Medical Foundation.
Author Contributions
Q.S., and W.W.Q. had access to all the data included in the study and are responsible for the completeness of the data and the accuracy of our analysis. D.Y.W. helped to design the study, Q.S., T.W. and C.G.Z. contributed to the statistical analysis and the revision of this manuscript. C.Z. and Z.H.G. approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68(1):7-30
2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913-2921
3. Eckel F, Brunner T, Jelic S, Group ObotEGW. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2011;22(suppl 6):vi40-vi44
4. Poruk KE, Firpo MA, Scaife CL, Adler DG, Emerson LL, Boucher KM, Mulvihill SJ. Serum osteopontin and tissue inhibitor of metalloproteinase 1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma. Pancreas. 2013;42(2):193-197
5. Ducreux M, Cuhna AS, Caramella C, Hollebecque A, Burtin P, Goere D, Seufferlein T, Haustermans K, Van Laethem JL, Conroy T. et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v56-68
6. Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. European journal of surgical oncology: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2007;33(3):266-270
7. Duffy MJ, Sturgeon C, Lamerz R, Haglund C, Holubec VL, Klapdor R, Nicolini A, Topolcan O, Heinemann V. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann Oncol. 2010;21(3):441-447
8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-297
9. Filip A. MiRNA-new mechanisms of gene expression control. Postepy biochemii. 2007;53(4):413-419
10. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1608-1613
11. Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116(24):5637-5649
12. Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu H, Ye Z, Li LC. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS letters. 2014;588(24):4654-4664
13. Dou D, Yang S, Lin Y, Zhang J. An eight-miRNA signature expression-based risk scoring system for prediction of survival in pancreatic adenocarcinoma. Cancer Biomark. 2018;22:3233
14. Xu B, Liu J, Xiang X, Liu S, Zhong P, Xie F, Mou T, Lai L. Expression of miRNA-143 in Pancreatic Cancer and Its Clinical Significance. Cancer Biother Radiopharm. 2018;13:1089
15. Esposti DD, Hernandez-Vargas H, Voegele C, Fernandez-Jimenez N, Forey N, Bancel B, Le Calvez-Kelm F, McKay J, Merle P, Herceg Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget. 2016;7(22):31862-31877
16. Giulietti M, Occhipinti G, Principato G, Piva F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cellular oncology (Dordrecht). 2017;40(2):181-192
17. Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther. 2017;10:4493-4506
18. Giulietti M, Occhipinti G, Principato G, Piva F. Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development. Cellular oncology (Dordrecht). 2016;39(4):379-388
19. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics (Oxford, England). 2004;20(3):307-315
20. Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. Journal of statistical software. 2012:46 (11)
21. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559
22. Vauleon E, Tony A, Hamlat A, Etcheverry A, Chiforeanu DC, Menei P, Mosser J, Quillien V, Aubry M. Immune genes are associated with human glioblastoma pathology and patient survival. BMC medical genomics. 2012;5:41
23. Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic acids research. 2016;44(W1):W135-141
24. Aguirre-Gamboa R, Trevino V. SurvMicro: assessment of miRNA-based prognostic signatures for cancer clinical outcomes by multivariate survival analysis. Bioinformatics (Oxford, England). 2014;30(11):1630-1632
25. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4:Article17
26. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY). 2008;321(5897):1801-1806
27. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015:4
28. Miyamae M, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H, Shiozaki A. et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. British journal of cancer. 2015;113(10):1467-1476
29. Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Hollander NH. et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. Jama. 2014;311(4):392-404
30. Li Y, Sarkar FH. MicroRNA Targeted Therapeutic Approach for Pancreatic Cancer. International journal of biological sciences. 2016;12(3):326-337
31. Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM. et al. Serum biomarker panels for the detection of pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(4):805-816
32. Lovat F, Fassan M, Gasparini P, Rizzotto L, Cascione L, Pizzi M, Vicentini C, Balatti V, Palmieri D, Costinean S. et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(37):11636-11641
33. Zhang WL, Zhang JH, Wu XZ, Yan T, Lv W. miR-15b promotes epithelial-mesenchymal transition by inhibiting SMURF2 in pancreatic cancer. International journal of oncology. 2015;47(3):1043-1053
34. Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, Wei Q, Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine. 2017;96(2):e5642
35. El-Tawdi AH, Matboli M, Shehata HH, Tash F, El-Khazragy N, Azazy Ael S, Abdel-Rahman O. Evaluation of Circulatory RNA-Based Biomarker Panel in Hepatocellular Carcinoma. Molecular diagnosis & therapy. 2016;20(3):265-277
36. Hasegawa T, Glavich GJ, Pahuski M, Short A, Semmes OJ, Yang L, Galkin V, Drake R, Esquela-Kerscher A. Characterization and Evidence of the miR-888 Cluster as a Novel Cancer Network in Prostate. Molecular cancer research: MCR. 2018;16(4):669-681
37. Hatano K, Kumar B, Zhang Y. et al. a functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015;43(8):4075-86
38. Chong GO, Jeon HS, Han HS, Son JW, Lee YH, Hong DG, Lee YS, Cho YL. Differential MicroRNA Expression Profiles in Primary and Recurrent Epithelial Ovarian Cancer. Anticancer research. 2015;35(5):2611-2617
39. Margue C, Reinsbach S, Philippidou D, Beaume N. et al. Comparison of a healthy miRNome with melanoma patient miRNomes: are microRNAs suitable serum biomarkers for cancer? Oncotarget. 2015;6(14):12110-27
40. Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer biology & therapy. 2010;9(10):803-808
41. Li R, Gao K, Luo H, Wang X, Shi Y, Dong Q, Luan W, You Y. Identification of intrinsic subtype-specific prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer Res. 2014;19(33):9
42. Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis & rheumatology (Hoboken, NJ). 2015;67(2):423-434
43. Shi Q, Xu X, Liu Q, Luo F, Shi J, He X. MicroRNA-877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncology letters. 2016;11(2):1474-1480
44. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. International journal of cancer. 2012;130(8):1715-1725
45. Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Experimental cell research. 2001;264(1):42-55
46. Liggett WHJr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16(3):1197-1206
Author contact
![]() Corresponding authors: Chen-guang Zhang, Ting Wu & Zu-hua Gao. (Tel +86-13146303355; email: chzhangedu.cn, tingwuedu.cn and zu-hua.gaoca)
Corresponding authors: Chen-guang Zhang, Ting Wu & Zu-hua Gao. (Tel +86-13146303355; email: chzhangedu.cn, tingwuedu.cn and zu-hua.gaoca)

 Global reach, higher impact
Global reach, higher impact