Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(22):4156-4165. doi:10.7150/jca.27098 This issue Cite
Review
Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer
1. Department of the 2nd Department of Thoracic Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, P.R. China;
2. Department of the Central Laboratory, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, P.R. China;
3. Department of Pharmacy, Xiangya Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410001, P.R. China;
4. Department of Thoracic Radiotherapy, Key laboratory of Translational Radiation Oncology, Department of Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, P.R. China;
5. Hunan University of Medicine, Huaihua, Hunan 418000, P.R. China.
*These authors contributed equally to this work.
Received 2018-5-5; Accepted 2018-6-24; Published 2018-10-18
Abstract
Lung cancer is the leading cause of cancer-related deaths worldwide. Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts and longer than 200 nucleotides. LncRNAs have been demonstrated to modulate gene expression at transcriptional, post-transcriptional, as well as epigenetic levels in lung cancer. Interestingly, compelling studies have revealed that lncRNAs participated in the EZH2 oncogenic regulatory network. EZH2 plays an important role in the initiation, progression and metastasis of cancer. On one hand, lncRNAs can directly bind to EZH2, recruit EZH2 to the promoter region of genes and repress their expression. On the other hand, lncRNAs can also serve as EZH2 effectors or regulators. In this review, we summarized the types of lncRNA-EZH2 interaction and regulatory network identified till date and discussed their influence on lung cancer. Better understanding regarding the interaction and regulatory network will provide new insights on lncRNA- or EZH2-based therapeutic development in lung cancer.
Keywords: lung cancer, lncRNA, EZH2, regulation, interaction
Introduction
Lung cancer is the most common cause of cancer death, with an estimated 1.6 million deaths in 2012 worldwide [1]. Approximately 85% lung cancers are classified as non-small cell lung cancer (NSCLC) which includes squamous cell carcinoma (SCC), lung adenocarcinoma (LAD), and large cell carcinoma (LCC) histologic subtypes, and the other 15% as small cell lung cancer (SCLC) [2, 3]. Although knowledge regarding lung cancer biology, advances in diagnostic techniques and therapeutic strategies have been improved, the prognosis remains poor with an overall 5-year survival of only 15% [4]. Therefore, there is a strong need to better understand the pathogenesis, early diagnostic biomarkers and therapeutic targets for lung cancer.
Accumulated evidences have demonstrated important roles of long non-coding RNAs (lncRNAs) in various diseases, particularly in cancer. LncRNAs refer to non-protein coding transcripts longer than 200 nucleotides [5, 6]. Although lncRNAs are not translated into proteins, they function to regulate gene transcription at transcriptional level, post-transcriptional level and epigenetic level [7]. The dysregulation of lncRNAs has been demonstrated in various human cancers, including lung cancer, and promoted tumor formation and progression [8, 9]. In this review, we focused on lncRNAs and Enhancer of Zeste Homolog 2 (EZH2) interaction and regulatory network in lung cancer.
Functions and mechanisms of lncRNAs in lung cancer
LncRNAs has been emerged as novel master regulators, playing a major regulatory role in various biological processes, such as cell cycle regulation, proliferation, survival, apoptosis, migration, invasion and chemoresistance [10, 11]. To data, a large number of lncRNAs that play a key role in cell function regulation may be used as potential biomarkers for the diagnosis, treatment and prognosis of various cancers, including lung cancer [12, 13]. For instance, MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1) is shown to be upregulated and linked to clinicopathological features in patients with lung cancer, which may serve as a potential prognostic marker to predict poorer prognosis in patients [14, 15]. HOTAIR (HOX transcript antisenseRNA) exhibited significantly higher expression in lung cancer and its elevated expression was correlated with lymph node metastasis and poor survival rate [16-18]. HOTAIR has been emerged as a key regulator of lung cancer and may be used as a diagnostic and therapeutic potential marker of lung cancer [19, 20].
As known, lncRNA is regulated by mechanisms similar to those of protein-coding genes, such as transcription factor binding, RNA splicing, DNA and histone modifications [21]. Numerous lncRNAs are demonstrated to be mediated by transcription factors, like p53, NF-κB, Oct4 and Sox2 [22, 23]. Jen et al. revealed that expression of NEAT1 and MALAT1 was transcriptionally regulated by Oct4 in lung cancer [24]. LncRNAs can also regulate various key cellular functions in lung cancer, such as gene expression regulation, genomic reprogramming, nuclear cytoplasmic trafficking, nuclear compartmentalization and RNA-splicing [25-27]. H19 promoted cell cycle progression by down-regulating miR-107 in NSCLC cells [28]. SBF2-AS1 could regulate cell cycle through epigenetic inhibition of P21 [29]. LncRNA-HIT (HOXA transcript induced by TGFβ) promoted migration and invasion of NSCLC cells by associating directly with ZEB1[30].
Functions and mechanisms of EZH2 in lung cancer
EZH2 (Enhancer of zeste homolog 2), a 751-amino acid histone-lysine methyltransferase, is located on human chromosome 7q35 [31]. It is the enzymatic subunit of polycomb-repressive complex 2 (PRC2). PRC2 functions as a histone H3 lysine 27 (H3K27) methyltransferase and promotes transcriptional silencing via regulating chromatin structure through posttranslational modification of histones [32, 33]. The PRC2 complex is mainly composed of a trimeric core of SUZ12, EED and EZH1/2 [34]. Sequence analysis showed that EZH family is organized into four homologous domains, where the cysteine-rich region and the SET domain functions in maintaining histone methyl transferase (HMT) activity, and the N-terminal domains H1 and H2 are the protein interaction domains that are required for establishing and maintaining proper PRC2 functions [35, 36].
EZH2 is capable of mono-, di-, and tri-methylation of H3K27 and essential for epigenetic gene silencing [37]. It has been proved to regulate various biological functions and cellular signals in lung cancer. Liu and colleagues reported that knockdown of EZH2 exerted inhibitory effects on proliferation of NSCLC cells, which is achieved through direct binding of EZH2 to the PUMA promoter, thus epigenetically repressing the PUMA expression [38]. Murai et al. revealed that EZH2 promoted SCLC progression by suppressing the TGF-β-Smad-ASCL1 pathway [39]. Another study by Li and colleagues showed that EZH2 inhibited lung cancer cell proliferation through binding to the Nrf2 promoter, where the expression of H3K27me3 was increased and Nrf2 was repressed [40]. Moreover, the EZH2 interaction with lncRNAs could regulate numerous gene expressions at the epigenetic level.
On the other hand, EZH2 expression and activity in cancer cells can be altered at multiple levels. It can be transcriptionally induced by multiple factors, for example, p53 and C-MYC [41, 42], or can be post-transcriptionally regulated through the interaction of miRNAs or lncRNAs [43]. MiR-138 was downregulated in NSCLC tissues and cells, and it can bind to the 3' UTR of EZH2 and suppress the expression levels of EZH2 mRNA and protein [44]. Zhang et al. reported that miR-101 inhibited cell proliferation and invasion in NSCLC cells by directly repressing EZH2 expression [45].
Compelling studies have revealed that lncRNA is a novel player in the EZH2-related cellular biological functions. To summarize and detail the previously findings, we are focusing in the following paragraphs on the up-to-date characterized lncRNAs and EZH2 interaction/regulatory network in lung cancer (Table 1 and Figure 1).
LncRNAs and EZH2 interaction in lung cancer
These lncRNAs are capable of directly binding to EZH2 and epigenetically silencing gene expression.
LncRNAs bind with EZH2 in lung cancer.
| LncRNA | Cancer type | Expression | Bind to | Function mechanism | Associated clinical features | Associated cell process | Ref |
|---|---|---|---|---|---|---|---|
| TUG1 | SCLC | up | EZH2 | represses LIMK2b expression | correlates with clinical stage and shorter survival time | promotes cell growth migration and invasion, increases chemoresistance | [50] |
| TUG1 | LAD | up | EZH2 | represses BAX expression | associates with enhanced tumor size, degree of differentiation, lymph node metastases, distant metastasis and TNM stage. | promotes cell viability and decreases cell apoptosis | [55] |
| TUG1 | NSCLC | down | EZH2, EED | represses CELF1 expression | - | inhibits cell proliferation | [56] |
| TUG1 | NSCLC | down | EZH2 | represses HOXB7 expression | associates with higher TNM stage and tumor size | inhibits cell proliferation | [57] |
| XIST | NSCLC | up | EZH2 | represses KLF2 expression | associates with shorter survival and poorer prognosis | promotes cell proliferation, migration and invasion | [61] |
| SNHG20 | NSCLC | up | EZH2 | represses P21 expression | associates with advanced tumor, TNM stage and tumor size, poorer OS | promotes cell proliferation, migration and inhibits cell apoptosis | [73] |
| LINC01133 | NSCLC | up | EZH2, LSD1 | represses KLF2, P21 and E-cadherin transcription. | associates with poor prognosis and short survival time | increases cell proliferation, migration and invasion, decreases cell apoptosis | [80] |
| LINC00673 | NSCLC | up | EZH2 | silences HOXA5 expression | associates with tumor size, lymph node metastasis, TNM stage | increases cell proliferation | [87] |
| LINC00152 | LAD | up | EZH2 | represses IL24 expression | correlates with advanced TNM stage, larger tumor size, and lymph node metastasis, shorter survival time | promotes cell growth, suppresses cell apoptosis | [98] |
| LINC00511 | NSCLC | up | EZH2 | represses P57 expression | associates with oncogenesis, tumor size, metastasis, and poor prognosis | affected cell proliferation, invasiveness, metastasis, and apoptosis | [102] |
| LINC01207 | LAD | up | EZH2 | represses BAD expression | associates with TNM stage, advanced TNM stage and shorter survival | increases cell proliferation, decreases cell apoptosis | [105] |
| AGAP2-AS1 | NSCLC | up | EZH2, LSD1 | represses KLF2 and LATS2 expression | correlates with poor prognostic outcomes | increases cell proliferation, migration and invasion, and inhibits cell apoptosis. | [108] |
| HOXA11-AS | NSCLC | up | EZH2, DNMT1 | represses miR-200b expression | indicates poor prognosis | promotes cell invasive abilities | [121] |
| FEZF1-AS1 | NSCLC | up | EZH2, LSD1 | Suppresses Wnt/β-catenin signaling | associates with lymph node metastasis, poor differentiation, and advanced TNM stage | promotes cell proliferation, invasion and EMT | [129] |
| FOXF1-AS1 | NSCLC | down | EZH2 | correlates with FOXF1 expression | associates with tumor migration, invasion and metastasis | inhibits cell migration and invasion by regulating EMT | [136] |
TUG1
TUG1 (Taurine-upregulated gene 1) is mapped on chromosome 22q12.2, and has a length of 7100 nt [46]. It was initially detected in a genomic screen for genes upregulated following taurine treatment in developing mouse retinal cells. Later studies demonstrated that TUG1 played an important role in the initiation and progression of malignancies [46]. The expression of TUG1 in human cancer has been shown to overexpress in a variety of cancers, for example, bladder cancer, gastric cancer, osteosarcoma, hepatocellular carcinoma, and colorectal cancer [47]. Recent investigations have indicated that TUG1 could regulate gene expression by binding with EZH2, affecting cell proliferation in human gastric cancer and hepatocellular carcinoma [48, 49].
TUG1 is upregulated in SCLC tissues and cell lines and promoted cell proliferation, migration and invasion, as well as chemoresistance [50]. It affected cell functions through regulation of LIMK2b expression by binding to EZH2. LIMK2b is located at 300kp of TUG1 and is a member of LIMK2, belonging to the LIM kinase family [51]. LIMK2 encodes a kinase that phosphorylates cofilin and then regulates actin dynamics, and is involved in tumor growth, migration and invasion [52-54]. Another study revealed that TUG1 was overexpressed in LAD cells and serum samples and inhibited cell apoptosis through suppressing expression of the pro-apoptotic protein BAX via physically binding with EZH2 [55].
However, TUG1 was significantly decreased in NSCLC tissues compared to surrounding non-tumor lung tissues [56, 57]. The different TUG1 expression on SCLC and NSCLC might be due to its tissue-specific expression patterns of lncRNAs. Knockdown of TUG1 significantly promoted the proliferation of NSCLC cells. TUG1 is induced by p53 and negatively regulates HOXB7 (homeobox B7) by binding to PRC2, and participates in the AKT and MAPK pathway. Lin et al. revealed that TUG1, which is involved in pre-mRNA alternative splicing, RNA editing, RNA decay, and translation was bound to EZH2/EED in NSCLC cells by RIP assay and negatively regulated CELF1 (Elav-like family member 1) [58].
Studies have shown either upregulation or downregulation of TUG1 in lung cancer, suggesting its complex role in cancer biology. More studies are needed to better understand the role of TUG1 in lung cancer.
Involvement of lncRNAs in EZH2 regulatory network. LncRNAs take part in the EZH2 regulatory network to regulate the proliferation, migration, invasion, and EMT of lung cells, and associate with larger tumor size, advanced TNM stage, advanced lymph node metastasis and survival time of lung cancer patients.
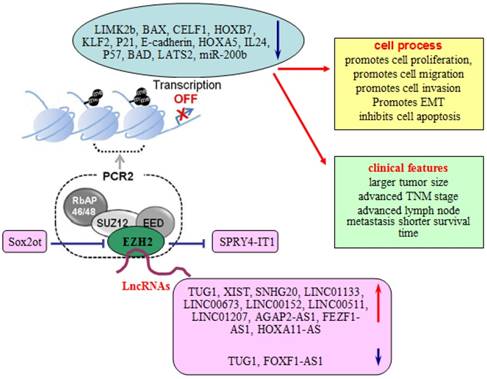
XIST
XIST (X inactivate-specific transcript) is derived from XIST gene and is essential for transcriptional silencing of one X-chromosome during mammalian female development [59, 60]. It is overexpressed in numerous human cancers, including lung cancer, and serves as an oncogene [61-64].
Tantai et.al revealed that XIST was up-regulated in NSCLC tissues and serum, and might serve as a potential diagnostic marker [65, 66]. High expression of XIST might be linked to poorer prognosis and shorter survival in NSCLC patients [61]. XIST silencing inhibited NSCLC cell proliferation, migration and invasion. Mechanistically, RIP and RNA pull-down assays showed that XIST could directly bind to EZH2, and then suppress the transcription of KLF2. KLF2 belongs to the Kruppel-like factor family, which contain Cys2/His2 zinc-finger domains [67]. KLF2 is down-regulated in various cancers, where it inhibits cell proliferation and acts as a tumor suppressor [67]. Thus, XIST might be a potential candidate biomarker and target for treatment of NSCLC, but more studies are needed to better understand the importance of XIST.
SNHG20
SNHG20 (Small nucleolar RNA host gene 20) is mapped on chromosome 17q25.2, and is 2183 nt lncRNA in length [68]. SNHG20 is overexpressed in ovarian cancer, colon cancer and hepatocellular carcinoma, and predicts poor prognosis [69-71]. It promotes cell proliferation, and cell invasion by EMT. Knockdown of SNHG20 suppresses β-catenin expression and inhibits the activity of Wnt/β-catenin signaling [69, 72].
SNHG20 is upregulated in NSCLC tissues and is associated with bigger tumor size, advanced TNM stage, as well as poorer survival rate[73]. SNHG20 functions as an oncogene by promoting NSCLC cell proliferation, migration, and repressing cell apoptosis. Further mechanistic analyses revealed that SNHG20 could interact with EZH2. Moreover, SNHG20 silencing decreases EZH2 by binding to the promoter region of P21 and represses its expression. P21 is a cyclin-dependent kinase (CDK) inhibitor, and functions in multiple cellular processes during cell growth by directly binding to kinases related to G1/S transition [74]. Therefore, SNHG20 plays an important role in NSCLC progression by epigenetically silencing of P21 transcription via binding with EZH2.
LINC01133
LINC01133 is encoded by chromosome 1q23.2 and is a 1154 nt lncRNA long [75]. LINC01133 expression is downregulated in colorectal cancer tissues and inhibits the EMT in colorectal cancer cells by directly interacting with SRSF6, a splicing factor that regulates the proliferation as an oncoprotein [76-78]. However, LINC01133 was found to be statistically overexpressed in osteosarcoma tumor tissues and cell lines and promotes the proliferation, migration and invasion of osteosarcoma cells [79]. It specifically targeted miR-422a, and played a tumor suppressive role in osteosarcoma progression. The differences might be due to the tissue-specific expression patterns of lncRNAs.
Interestingly, LINC01133 is overexpressed in LSCC tissues and predicted poor survival rates, but not in the LAD samples [75]. However, another study revealed that LINC01133 was up-regulated in both NSCLC types and indicated poor prognosis [80]. By performing RNA pulldown assay, the authors found that LINC01133 could directly bind to EZH2, which in turn directly binds to the promoter regions of KLF2, P21 and E-cadherin and represses their transcription. LINC01133 promotes cell proliferation via inhibiting KLF2 and P21, while reduced cell migration and invasion through repressing E-cadherin expression in NSCLC cells.
LINC00673
LINC00673 is located on chromosome 17q25.1, and has a transcript length of 2275 nt [81]. LINC00673 upregulation has been reported in various kinds of cancers, such as pancreatic cancer, gastric cancer and tongue squamous cell carcinoma [82-84]. It is identified as a potential oncogene that promotes cell proliferation and invasion and inhibits cell apoptosis. Furthermore, LINC00673 functions through different mechanisms, including regulation of epigenetic signatures and gene expression. It can directly interact with EZH2 and LSD1 in gastric cancer cells, thereby suppressing KLF2, KLF4 and LATS2 expression levels [82, 83]. It has been reported to negatively regulate miR-205 in hepatocellular carcinoma, and suppress PI3K/AKT signaling in glioma [85].
LINC00673 is found to be overexpressed in NSCLC tissues and correlated with tumor size, lymph node metastasis and TNM stage [86, 87]. LINC00673 increases proliferation, migration and invasion of NSCLC cells. It was found that the oncogenic activity of LINC00673 is partially attributable to its epigenetically inhibition of NCALD (Neurocalcin delta) through binding to LSD1, which could directly bind to the NCALD promoter region [86]. NCALD belongs to the EF-hand calcium-binding protein superfamily, and functions in the regulation of neuronal signal transduction process. It is involved in the pathogenesis of human cancer [88, 89]. In another study, using RIP and RNA-pull down assays, Ma and colleagues revealed that LINC00673 was directly associated with EZH2 in NSCLC cells, and repressesed the expression of HOXA5 [87]. HOXA5 is identified as a tumor suppressor, which functions as a transcription factor and inhibits cancer cell metastasis through cytoskeletal remodeling regulation [90, 91]. Taken together, LINC00673 might be a new diagnostic marker and targeting it might be meaningful for treating patients with NSCLC.
LINC00152
LINC00152, which is 828 nt in length, is located on chromosome 2p11.2 [92]. It was first reported to be overexpressed in gastric tissues and cells and subsequently reported to be involved in cell proliferation, apoptosis, migration, and invasion of cancer cells [93, 94]. Mechanistic investigations revealed that LINC00152 promoted tumor growth through EGFR-mediated PI3/AKT pathway, and it also promoted cell cycle progression by binding to EZH2, thus suppressing p15 and p21 expression in gastric cancer cells [95]. Another study revealed that linc00152 bound to EZH2 and LSD1 epigenetically silenced P16 expression in renal cell carcinoma [96].
The role of LINC00152 in lung cancer has been recently highlighted. Feng et al. reported that LINC00152 was upregulated in lung cancer, and correlated with poor survival [97]. Silencing of LINC00152 inhibited cell proliferation in lung cancer cells through EGFR signaling independent pathway. In consistent with these results, Chen et al. also reported that LINC00152 expression was highly expressed in human LAD tissues and related to tumor progression [98]. Moreover, RIP assays revealed that LINC00152 directly bound to EZH2 and LSD1 in lung cancer cells, and RNA pulldown assays also confirmed the interaction between LINC00152 and EZH2 or LSD1. ChIP assays also showed that LINC00152 could recruit EZH2 to the IL24 promoter region and repressed its transcription by mediating H3K27me3.
LINC00511
LINC00511 (also known as onco-LncRNA-12) was originally found to be elevated and functioned as an oncogene in breast cancer [99, 100]. LINC00511 knockdown showed tumor-suppressive activities via cell proliferation inhibition in breast cancer cells. LINC00511 was also overexpressed in pancreatic ductal adenocarcinoma and exerted oncogene functions through up-regulating VEGFA via acting as a competing endogenous RNA on hsa-miR-29b-3p [101].
A recent study found that LINC00511 was upregulated in NSCLC tumor tissues and correlated with tumor size, TNM stage, and lymph node metastasis [102]. Knockdown of LINC00511 inhibited cell proliferation and metastasis in NSCLC cell lines. Interestingly, further mechanistical study revealed that LINC00511 could directly bind to EZH2 by means of RIP assay, and recruit EZH2 to the promoter region p57, which is an inhibitor of cyclin-dependent kinase, and is deemed to be a tumor-suppressor in numerous types of cancers [103, 104]. Thus, these results showed that LINC00511 is an oncogene in NSCLC.
LINC01207
LINC01207 is an intergenic lncRNA with 3212 nt in length, locates in chromosome 4q32.3, and consists of 3 exons and 2 introns [105]. LINC01207 is found to be significantly up-regualted in LAD but not in LSCC tissues [106]. The higher expression of LINC01207 was associated with advanced TNM stage and poor survival of LAD patients. LINC01207 promotes cell proliferation and inhibits cell apoptosis, while dose not affect cell migration and invasion. Using RIP and ChIP assay, LINC01207 was found to directly bind with EZH2 and mediated H3K27-me3 at the promoter region of Bad, which is an important pro-apoptotic protein of the Bcl-2 family [107]. Furthermore, LINC01207 silencing up-regulates the expression of Bad. However, with only two manuscripts published regarding LINC01207, very little is known about this lncRNA. Further investigation is necessary before its role in cancer can be drawn.
AGAP2-AS1
AGAP2-AS1 (AGAP2 antisense RNA 1) is an antisense lncRNA with 1567 nt in length, and is transcribed from a gene mapped on chromosome 12q14.1[108]. AGAP2-AS1 was found to be upregulated in gastric cancer tissues and cell lines, and its upregulation may be activated partly by SP1, which is a transcription factor [109]. Knockdown of AGAP2-AS1 significantly inhibited gastric cell proliferation, migration and invasion. In addition, AGAP2-AS1 binds with EZH2 and LSD1, and epigenetically suppresses the expression of P21 and E-cadherin in gastric cancer cells [109].
The expression of AGAP2-AS1 was up-regulated in NSCLC tissues, and correlated with tumor stage, lymph nodes metastasis and survival time [108, 110]. AGAP2-AS1 exerts oncogene functions by inducing cell proliferation, migration and inhibiting apoptosis in NSCLC cells. Further RNA IP assays indicated that AGAP2-AS1 could directly bind to EZH2 and LSD1, and then recruited them to LATS2 promoter regions and repressed the transcription of LATS2. LATS2 is a regulator of cellular homeostasis and tumor-suppressor and downregulated in multiple human cancers [111]. Due to its significant correlation with clinical NSCLC progression, AGAP2-AS1 might be a potential biomarker or therapeutic target.
HOXA11-AS
HOXA11-AS is a lncRNA transcribed from the opposite strand of the HOXA11 gene [112]. It is 5100 nt in length, and is mapped on chromosome 7p15.2. HOXA11‑AS is initially discovered in mouse embryonic cDNA library. It is reported that HOXA11-AS takes part in cancer development including glioma, epithelial ovarian cancer, gastric cancer, cervical cancer, colorectal cancer [113-117]. HOXA11-AS can promote cell proliferation and invasion by sponging miR-124, and mediating the expression of Sp1 as a ceRNA of miR-124 [118, 119]. It can also promote cell proliferation through LATS1 expression inhibition via bridging to EZH2 [120].
lncRNA HOXA11-AS was significantly higher in NSCLC tissues compared with adjacent normal tissues, and demonstrated a poor prognosis in NSCLC patients [118, 121]. It is also markedly expressed in NSCLC cells. Knockdown of HOXA11-AS inhibited the proliferation, migration, invasion, as well as the EMT process [121]. Mechanically, RIP assays revealed that HOXA11-AS directly interacts with EZH2 and DNMT1. Subsequently, EZH2 and DNMT1 are recruited to the promoter regions of miR-200b and repressed its expression. MiR-200b is well known to function as a tumor suppressor, for example, miR-200b suppresses migration and invasion in NSCLC cells via targeting FSCN1 [122, 123]. Thus, HOXA11-AS may be a promising candidate for further investigation as therapeutic target for NSCLC therapy.
FEZF1-AS1
LncRNA FEZF1-AS1 (FEZ family zinc finger 1 antisense RNA 1) is located on the opposite strand of gene FEZF1 in chromosome 7, and is 2564 nt long [124]. It is up-regulated in various cancers, including colorectal carcinoma, gastric cancer, stomach adenocarcinoma, pancreatic ductal adenocarcinoma and osteosarcoma [124-128]. Generally, higher expression of FEZF1-AS1 was correlated with larger tumor size, higher clinical stage and poorer survival. Moreover, knockdown of FEZF1-AS1 significantly inhibited cancer cell proliferation, migration and invasiveness.
In accordance with other cancer types, FEZF1-AS1 was overexpressed in NSCLC tissues and correlated with poor differentiation grade, lymph node metastasis, advanced TNM stage and poor prognosis [129, 130]. It exerts oncogenic activity by promoting cell proliferation, migration and invasion, as well as EMT process of NSCLC cells. Using RIP assays, He and colleagues revealed that FEZF1-AS1 could bind to EZH2 and LSD1, which then reduced their binding to the E-cadherin promoter regions. Furthermore, downregulation of FEZF1-AS1 suppressed Wnt/β-catenin signaling in NSCLC, which was significantly associated with tumor metastasis [131].
FOXF1-AS1
FOXF1-AS1 (also referred as FENDRR) is located in chromosome 16q24.1, with 3099 nt in length [132]. FOXF1-AS1 is transcribed from the negative strand of FOXF1 (Forkhead box protein F1), originally identified to be coimmunoprecipitated with SUZ12 in human fetal lung and foot fibroblasts [133]. It was overexpressed with FOXF1 in osteosarcoma tissues and correlated with lung metastasis[134]. However, Xu et al. found that FOXF1-AS1 expression was down-regulated in gastric cancer tissues and correlated with poor prognosis [135].
FOXF1-AS1 was found to be significantly down-regulated in lung cancer tissues and cells, and associated with tumor migration, invasion and metastasis [136]. Moreover, loss of FOXF1-AS1 was also correlated with stem-like properties require EZH2. FOXF1-AS1could demonstrated to bind with EZH2 by means of RPISeq and RIP assay. It was also found that FOXF1-AS1 functions through targeting FOXF1, in consistence with that FOXF1 expression was lower in LAD and LSCC. FOXF1 belongs to the forkhead box family of transcription factors and regulates cell proliferation and function in tumorigenesis by a large number of studies [137]. As the expression of FOXF1-AS1 is not consistent in cancers, it is not easy to accurately define its role in lung cancer with only one signal manuscript published.
LncRNAs as EZH2 effectors/regulators in lung cancer
Similar to protein-coding genes, the transcription of lncRNA was shown to be regulated by some key transcription factors. For example, lncRNA-p21 transcription is promoted by p53 [138], while E2F1 regulated lncRNA ERIC expression[139], and also serve as EZH2 effectors. On the other hand, lncRNAs can also be regulated by EZH2.
Sox2ot (SOX2 overlapping transcript) is located on human chromosome 3q26.3 [140]. Sox2ot is transcribed in the same orientation as Sox2 (sex determining region Y-box 2), and indicated to regulate SOX2 transcription as an important enhancer [141, 142]. SOX2 is a transcription factor of the SRY-related HMG-box family that has been shown to play key roles in many stages of mammalian development. Sox2ot is upregulated and linked with cancer metastasis and poor prognosis in several types of carcinomas, such as gastric cancer, breast cancer, ovarian cancer and esophageal cancer [143-145]. Sox2ot is also overexpressed in lung cancer, especially in LSCCs, indicating poor survival of lung cancer patients [146]. It promoted cancer cell proliferation by inducing G2/M cell cycle arrest. Knockdown of Sox2ot decreased the expression levels of EZH2 mRNA and protein. Meanwhile, enhanced expression of EZH2 reversed the G2/M arrest induced by Sox2ot depletion. This indicated that Sox2ot-mediated lung cancer cell proliferation through regulating EZH2.
SPRY4-IT1 (SPRY4 intronic transcript 1) is mapped on human chromosome 5q31.3, transcribed from the second intron within the SPRY4 gene [147]. It is an inhibitor of the MAPK signaling pathway [148]. SPRY4-IT1 is upregulated in various kinds of tumor tissues and cell lines and identified as an oncogene via promoting tumor progression and metastasis [149]. The expression of SPRY4-IT1 is also upregulated in LAD tissues and cell lines [150]. However, Sun et al. reported that SPRY4-IT1 was downregulated and correlated with advanced pathological stage, lymph node metastasis and poor survival of NSCLC [151]. Overexpression of SPRY4-IT1 is found to inhibit the migration and invasion through its regulation of EMT, while promote apoptosis of NSCLC cells. By performing Chip assays, the authors revealed that EZH2 could directly bind to the promoter region of SPRY4-IT1 and suppress its expression. Moreover, in EZH2-knockdown cells, depletion of SPRY4-IT1 partially reversed the oncogenic phenotype, suggesting that SPRY4-IT1 is involved in the EZH2 oncogenesis.
Conclusion
Numerous lncRNAs have been demonstrated to contribute to cancer cell functions through silencing of tumor suppressors via interaction with EZH2. EZH2 plays an important role in maintaining the integrity of cellular epigenetics, and is highly relevant to human cancer. Moreover, EZH2 expression and activity in cancer cells can be altered at multiple levels, and regulated by lncRNAs. Improving our understanding regarding the role of lncRNA-EZH2 regulatory and interaction network in tumorgenesis helps to diagnose and develop varied therapeutic strategies for lung cancer.
Acknowledgements
This work was supported in part by grants from National Natural Scientific Foundation of China (81802947, 81472595), Health and Family Planning Commission of Hunan Province (B20180545), Natural Science Foundation of Hunan Province (2017JJ2173, 2018JJ3314), Changsha Science and Technology Board (kq1706045).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Chheang S, Brown K. Lung cancer staging: clinical and radiologic perspectives. Semin Intervent Radiol. 2013;30:99-113
3. Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479-85
4. Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175-80
5. Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010-39
6. Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423-37
7. Chen J, Wang R, Zhang K, Chen LB. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18:2425-36
8. Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13-21
9. Cheng N, Li X, Zhao C, Ren S, Chen X, Cai W. et al. Microarray expression profile of long non-coding RNAs in EGFR-TKIs resistance of human non-small cell lung cancer. Oncol Rep. 2015;33:833-9
10. Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crino L, Chiari R. et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33:18
11. Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F. et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586-94
12. Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M. et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908-13
13. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38
14. Wang JZ, Xiang JJ, Wu LG, Bai YS, Chen ZW, Yin XQ. et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17:167
15. Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015;5:e008653
16. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90
17. Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui X, Fewell C. et al. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6:35
18. Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930-6
19. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464
20. Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N. et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319-24
21. Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15-24
22. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223-7
23. Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol. 2014;6:181-91
24. Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104
25. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178-81
26. Cerk S, Schwarzenbacher D, Adiprasito JB, Stotz M, Hutterer GC, Gerger A. et al. Current Status of Long Non-Coding RNAs in Human Breast Cancer. Int J Mol Sci. 2016:17
27. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66
28. Cui J, Mo J, Luo M, Yu Q, Zhou S, Li T. et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400-9
29. Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J. et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75
30. Jia X, Wang Z, Qiu L, Yang Y, Wang Y, Chen Z. et al. Upregulation of LncRNA-HIT promotes migration and invasion of non-small cell lung cancer cells by association with ZEB1. Cancer Med. 2016;5:3555-63
31. Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185-96
32. Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-9
33. Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147-55
34. Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299-313
35. Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL. et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503-18
36. Han Li C, Chen Y. Targeting EZH2 for cancer therapy: progress and perspective. Curr Protein Pept Sci. 2015;16:559-70
37. Wen Y, Cai J, Hou Y, Huang Z, Wang Z. Role of EZH2 in cancer stem cells: from biological insight to a therapeutic target. Oncotarget. 2017;8:37974-90
38. Liu H, Li W, Yu X, Gao F, Duan Z, Ma X. et al. EZH2-mediated Puma gene repression regulates non-small cell lung cancer cell proliferation and cisplatin-induced apoptosis. Oncotarget. 2016;7:56338-54
39. Murai F, Koinuma D, Shinozaki-Ushiku A, Fukayama M, Miyaozono K, Ehata S. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-beta-Smad-ASCL1 pathway. Cell Discov. 2015;1:15026
40. Li Z, Xu L, Tang N, Xu Y, Ye X, Shen S. et al. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS Lett. 2014;588:3000-7
41. Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759-69
42. Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669-83
43. Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E. Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer. 2013;133:267-74
44. Zhang H, Zhao M, Lv Z, Zhang X, Qin X, Wang H. et al. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56-65
45. Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671-8
46. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501-12
47. Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471-5
48. Zhang E, He X, Yin D, Han L, Qiu M, Xu T. et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109
49. Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P. et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165
50. Niu Y, Ma F, Huang W, Fang S, Li M, Wei T. et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16:5
51. Smolich B, Vo M, Buckley S, Plowman G, Papkoff J. Cloning and biochemical characterization of LIMK-2, a protein kinase containing two LIM domains. J Biochem. 1997;121:382-8
52. Acevedo K, Moussi N, Li R, Soo P, Bernard O. LIM kinase 2 is widely expressed in all tissues. J Histochem Cytochem. 2006;54:487-501
53. Scott GA, McClelland LA, Fricke AF, Fender A. Plexin C1, a receptor for semaphorin 7a, inactivates cofilin and is a potential tumor suppressor for melanoma progression. J Invest Dermatol. 2009;129:954-63
54. Suyama E, Wadhwa R, Kawasaki H, Yaguchi T, Kaul SC, Nakajima M. et al. LIM kinase-2 targeting as a possible anti-metastasis therapy. J Gene Med. 2004;6:357-63
55. Liu H, Zhou G, Fu X, Cui H, Pu G, Xiao Y. et al. Long noncoding RNA TUG1 is a diagnostic factor in lung adenocarcinoma and suppresses apoptosis via epigenetic silencing of BAX. Oncotarget. 2017;8:101899-910
56. Lin PC, Huang HD, Chang CC, Chang YS, Yen JC, Lee CC. et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:583
57. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243
58. Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2012;3:104-21
59. Gu S, Xie R, Liu X, Shou J, Gu W, Che X. Long Coding RNA XIST Contributes to Neuronal Apoptosis through the Downregulation of AKT Phosphorylation and Is Negatively Regulated by miR-494 in Rat Spinal Cord Injury. Int J Mol Sci. 2017:18
60. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38-44
61. Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811-7
62. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J. et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75-86
63. Zhou Q, Hu W, Zhu W, Zhang F, Lin-Lin L, Liu C. et al. Long non coding RNA XIST as a prognostic cancer marker - A meta-analysis. Clin Chim Acta. 2018;482:1-7
64. Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J. et al. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer. 2017;8:4106-16
65. Ren C, Li X, Wang T, Wang G, Zhao C, Liang T. et al. Functions and Mechanisms of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2015;25:566-9
66. Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887-95
67. Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088-96
68. Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma. J Cancer. 2016;7:608-17
69. He S, Zhao Y, Wang X, Deng Y, Wan Z, Yao S. et al. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/beta-catenin signaling. Biosci Rep. 2018:38
70. Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655
71. Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857-63
72. Liu J, Liu L, Wan JX, Song Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3beta/ beta-catenin signaling pathway. Oncotarget. 2017;8:80700-8
73. Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B. et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8:e3092
74. Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980-5
75. Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36:7465-71
76. Zhang JH, Li AY, Wei N. Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:2103-7
77. Kong J, Sun W, Li C, Wan L, Wang S, Wu Y. et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476-84
78. Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF. et al. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol. 2013;229:630-9
79. Zeng HF, Qiu HY, Feng FB. Long Noncoding RNA LINC01133 Sponges miR-422a to Aggravate the Tumorigenesis of Human Osteosarcoma. Oncol Res. 2017
80. Zang C, Nie FQ, Wang Q, Sun M, Li W, He J. et al. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016;7:11696-707
81. Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M. et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911-6
82. Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB. et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542-53
83. Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F. et al. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol Ther. 2017;25:1014-26
84. Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-32
85. Zhang LG, Zhou XK, Zhou RJ, Lv HZ, Li WP. Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am J Cancer Res. 2017;7:2536-44
86. Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X. et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558-75
87. Ma C, Wu G, Zhu Q, Liu H, Yao Y, Yuan D. et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. 2017;8:32696-705
88. Couvelard A, Hu J, Steers G, O'Toole D, Sauvanet A, Belghiti J. et al. Identification of potential therapeutic targets by gene-expression profiling in pancreatic endocrine tumors. Gastroenterology. 2006;131:1597-610
89. Isaksson HS, Sorbe B, Nilsson TK. Whole genome expression profiling of blood cells in ovarian cancer patients -prognostic impact of the CYP1B1, MTSS1, NCALD, and NOP14. Oncotarget. 2014;5:4040-9
90. Wang CC, Su KY, Chen HY, Chang SY, Shen CF, Hsieh CH. et al. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS One. 2015;10:e0124191
91. Lee DH, Forscher C, Di Vizio D, Koeffler HP. Induction of p53-independent apoptosis by ectopic expression of HOXA5 in human liposarcomas. Sci Rep. 2015;5:12580
92. Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658-64
93. Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P. et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112-23
94. Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T. et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441-7
95. Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J. et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135
96. Wang Y, Liu J, Bai H, Dang Y, Lv P, Wu S. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7:312-22
97. Feng S, Zhang J, Su W, Bai S, Xiao L, Chen X. et al. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7:2982
98. Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC, Ma HW. et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16:17
99. Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761-72
100. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129
101. Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W. et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655-67
102. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non-small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385
103. Avrahami D, Li C, Yu M, Jiao Y, Zhang J, Naji A. et al. Targeting the cell cycle inhibitor p57Kip2 promotes adult human beta cell replication. J Clin Invest. 2014;124:670-4
104. Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F. et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9:247-61
105. Wang G, Chen H, Liu J. The long noncoding RNA LINC01207 promotes proliferation of lung adenocarcinoma. Am J Cancer Res. 2015;5:3162-73
106. White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429
107. Trecherel E, Godin C, Louandre C, Benchitrit J, Poirot S, Maziere JC. et al. Upregulation of BAD, a pro-apoptotic protein of the BCL2 family, in vascular smooth muscle cells exposed to uremic conditions. Biochem Biophys Res Commun. 2012;417:479-83
108. Li W, Sun M, Zang C, Ma P, He J, Zhang M. et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225
109. Qi F, Liu X, Wu H, Yu X, Wei C, Huang X. et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10:48
110. Fan KJ, Liu Y, Yang B, Tian XD, Li CR, Wang B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21:2392-6
111. Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892-903
112. Chau YM, Pando S, Taylor HS. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. J Clin Endocrinol Metab. 2002;87:2674-80
113. Li T, Xu C, Cai B, Zhang M, Gao F, Gan J. Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer. Oncol Lett. 2016;12:4155-60
114. Richards EJ, Permuth-Wey J, Li Y, Chen YA, Coppola D, Reid BM. et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745-57
115. Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R. et al. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373:251-9
116. Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO, Kim KH. et al. The long noncoding RNA HOXA11 antisense induces tumor progression and stemness maintenance in cervical cancer. Oncotarget. 2016;7:83001-16
117. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D. et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-310
118. Yu W, Peng W, Jiang H, Sha H, Li J. LncRNA HOXA11-AS promotes proliferation and invasion by targeting miR-124 in human non-small cell lung cancer cells. Tumour Biol. 2017;39:1010428317721440
119. Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS Exerts Oncogenic Functions by Repressing p21 and miR-124 in Uveal Melanoma. DNA Cell Biol. 2017;36:837-44
120. Yu J, Hong JF, Kang J, Liao LH, Li CD. Promotion of LncRNA HOXA11-AS on the proliferation of hepatocellular carcinoma by regulating the expression of LATS1. Eur Rev Med Pharmacol Sci. 2017;21:3402-11
121. Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF, Hu CP. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64
122. Cheng YX, Zhang QF, Hong L, Pan F, Huang JL, Li BS. et al. MicroRNA-200b suppresses cell invasion and metastasis by inhibiting the epithelial-mesenchymal transition in cervical carcinoma. Mol Med Rep. 2016;13:3155-60
123. Xiao P, Liu W, Zhou H. miR-200b inhibits migration and invasion in non-small cell lung cancer cells via targeting FSCN1. Mol Med Rep. 2016;14:1835-40
124. Chen N, Guo D, Xu Q, Yang M, Wang D, Peng M. et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7:11271-83
125. Ye H, Zhou Q, Zheng S, Li G, Lin Q, Ye L. et al. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:34
126. Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M. et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39
127. Gu J, Li Y, Fan L, Zhao Q, Tan B, Hua K. et al. Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma. Oncotarget. 2017;8:49201-16
128. Zhou C, Xu J, Lin J, Lin R, Chen K, Kong J. et al. Long non-coding RNA FEZF1-AS1 promotes osteosarcoma progression by regulating miR-4443/NUPR1 axis. Oncol Res. 2018
129. He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother. 2017;95:331-8
130. Liu Z, Zhao P, Han Y, Lu S. LincRNA FEZF1-AS1 is associated with prognosis in lung adenocarcinoma and promotes cell proliferation, migration and invasion. Oncol Res. 2018
131. Lan K, Zhao Y, Fan Y, Ma B, Yang S, Liu Q. et al. Sulfiredoxin May Promote Cervical Cancer Metastasis via Wnt/beta-Catenin Signaling Pathway. Int J Mol Sci. 2017:18
132. Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O. et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23-33
133. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667-72
134. Kun-Peng Z, Chun-Lin Z, Xiao-Long M. Antisense lncRNA FOXF1-AS1 Promotes Migration and Invasion of Osteosarcoma Cells Through the FOXF1/MMP-2/-9 Pathway. Int J Biol Sci. 2017;13:1180-91
135. Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L. et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63
136. Miao L, Huang Z, Zengli Z, Li H, Chen Q, Yao C. et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget. 2016;7:68339-49
137. Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV. et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep. 2017;7:10690
138. Hall JR, Messenger ZJ, Tam HW, Phillips SL, Recio L, Smart RC. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:e1700
139. Feldstein O, Nizri T, Doniger T, Jacob J, Rechavi G, Ginsberg D. The long non-coding RNA ERIC is regulated by E2F and modulates the cellular response to DNA damage. Mol Cancer. 2013;12:131
140. Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D. et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960
141. Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC. et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013-27
142. Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN. et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461-3
143. Shahryari A, Jazi MS, Samaei NM, Mowla SJ. Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front Genet. 2015;6:196
144. Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8:5035-43
145. Han L, Zhang W, Zhang B, Zhan L. Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Exp Ther Med. 2018;15:2182-8
146. Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C. et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380-8
147. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS. et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852-62
148. Leeksma OC, Van Achterberg TA, Tsumura Y, Toshima J, Eldering E, Kroes WG. et al. Human sprouty 4, a new ras antagonist on 5q31, interacts with the dual specificity kinase TESK1. Eur J Biochem. 2002;269:2546-56
149. Li J, Chen Y, Chen Z, He A, Xie H, Zhang Q. et al. SPRY4-IT1: A novel oncogenic long non-coding RNA in human cancers. Tumour Biol. 2017;39:1010428317711406
150. Zhang X, Chi Q, Zhao Z. Up-regulation of long non-coding RNA SPRY4-IT1 promotes tumor cell migration and invasion in lung adenocarcinoma. Oncotarget. 2017;8:51058-65
151. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R. et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298
Author contact
![]() Corresponding author: wwx78com, Tel.: +86-731-8976-2110
Corresponding author: wwx78com, Tel.: +86-731-8976-2110

 Global reach, higher impact
Global reach, higher impact