Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(22):4187-4196. doi:10.7150/jca.27939 This issue Cite
Research Paper
MicroRNA-376c-3p Facilitates Human Hepatocellular Carcinoma Progression via Repressing AT-Rich Interaction Domain 2
1. Department of Infectious Diseases, the Second Affiliated Hospital of Xi'an Jiaotong University, 157 Xiwu Road, Xi'an 710004, China
2. Department of General Surgery, the First Affiliated Hospital of Xi'an Medical University, 48 Fenghao West Road, Xi'an 710077, China
Received 2018-6-17; Accepted 2018-8-2; Published 2018-10-18
Abstract
Hepatocellular carcinoma (HCC), accounting for approximately 90% of liver cancer, is the most lethal malignant tumors in the world. Large amount of evidence indicate that microRNAs (miRNAs) contribute to the tumorigenesis and progression of HCC. Among them, miR-376c-3p was recently identified as a tumor-related miRNA and is up-regulated in HBV-related HCC. But, the clinical significance of miR-376c-3p and its biological function in HCC progression are still unclear. Here, we confirmed that miR-376c-3p expression level in HCC was markedly higher than that in noncancerous tissues. Up-regulation of miR-376c-3p was detected in four different HCC cell lines. High miR-376c-3p expression correlated with poor prognostic features, such as large tumor size and venous infiltration. Follow-up data indicated that high miR-376c-3p level evidently correlated with poor clinical outcomes of HCC patients. Moreover, knockdown of miR-376c-3p repressed HCC cell growth, migration and invasion in vitro. miR-376c-3p overexpression facilitated these malignant behaviors of Bel-7402 cells. Mechanistically, miR-376c-3p posttranscriptionally repressed ARID2 expression by directly interacting with its 3'-UTR. Furthermore, an obvious negative correlation between miR-376c-3p and ARID2 mRNA expression in HCC tissues was confirmed. Notably, miR-376c-3p knockdown suppressed HCC growth and metastasis in nude mice. Gain-of-function experiments showed that ARID2 inhibited cell growth and mobility of Hep3B cells. Subsequently, ARID2 knockdown rescued miR-376c-3p silencing attenuated Hep3B cell proliferation and mobility. Our results suggest that miR-376c-3p exerts an oncogenic role in HCC progression.
Keywords: miR-376c-3p, hepatocellular carcinoma, ARID2, proliferation, invasion
Introduction
Hepatocellular carcinoma (HCC) treatment is a worldwide problem that still cannot be solved despite the great advances in diagnosis and treatment in the past decade. Surgical resection is the only and best option for the early stage HCC[1]. However, most patients are usually diagnosed as advanced HCC, which accompanies with intrahepatic and distant metastases[1]. The clinical outcomes of HCC patients is still poor resulted from postoperative recurrence and metastasis[2-4]. Thus, it is urgent to investigate the molecular mechanism underlying HCC initiation and progression to develop new treatment approaches.
MicroRNAs (miRNAs) are potential regulators of tumorigenesis and progression[5]. miRNAs directly bind to 3'-UTR of target mRNA via complementary pairing sequences[6], which induces degradation of the target mRNA and/or leads to translational silence[6]. During the hepatocarcinogenesis, it has been demonstrated that miRNAs function as either tumor promoters or suppressors[7-9]. Recent years, abnormal expression of miRNA-376c is determined in several types of human cancer including ovarian cancer[10], osteosarcoma[11], head and neck squamous cell carcinoma (HNSCC)[12], colorectal cancer (CRC)[13] and oral squamous cell carcinoma (OSCC)[14]. Furthermore, deregulation of serum miR-376c is recognized as a novel prognostic as well as diagnostic marker in gliomas[15], colon cancer[16], breast cancers[17-19] and gastric cancer[20, 21]. According to previous studies, miR-376c functions as either oncogenic or tumor suppressive miRNA , which depends on cancer types. For instance, miRNA-376c upregulation contributes to cell proliferation, survival and poorly response to chemotherapy, and inhibits apoptosis by repressing activin receptor-like kinase 7 (ALK7) in ovarian cancer[10]. In CRC, miR-376c-3p facilitates the growth of cancer cells by suppressing long noncoding RNA (lncRNA) LINC00152[13]. While, miR-376c overexpression restricts cell growth and migration in melanoma[22]. Restoration of miR-376c-3p expression suppresses migratory and invasive abilities of HNSCC cells by repressing runt related transcription factor 2 (RUNX2)[12]. A recent study reports that miR-376c-3p is up-regulated in HBV-related HCC, suggesting an oncogenic role of miR-376c-3p in HCC[23]. Yet, the biological function of miR-376c-3p and its downstream target are still unclear in HCC progression.
In this research, our data revealed that miR-376c-3p was over-expressed in HCC. Knockdown of miR-376c-3p suppressed cell cycle progression, proliferation, migration and invasion of HCC cells in vitro, and inhibited HCC growth and metastasis in vivo. Moreover, we verified that miR-376c-3p exhibited its oncogenic role by directly targeting AT-rich interaction domain 2 (ARID2) in HCC cells. Together, these data suggest that miR-376-3p is a new posttranscriptional regulator of ARID2 and may play a critical role in HCC progression.
Materials and Methods
Patients and tissue samples
Sixty cases of HCC and matched normal liver samples were collected from the Second Affiliated Hospital of Xi'an Jiaotong University. HCC patients who received radiofrequency ablation, chemotherapy, immunotherapy or sorafenib before surgery were excluded. After surgical resection, the specimens were immediately frozen in liquid nitrogen and subsequently stored at -80°C for further analyses. Written informed consent was signed by enrolled HCC patients. The study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi'an Jiaotong University.
Cell culture
HCC cell lines including Hep3B, Bel-7402, HepG2 and SMMC-7721, and the normal hepatocyte cell line LO2 were bought from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HCC cells were grown in the DMEM (Gibco-BRL, Grand Island, NY, USA) added with 10% fetal bovine serum (FBS, Gibco-BRL) and antibiotics (Sigma, St-Louis, MO, USA) in a humidified incubator containing 5% CO2 at 37°C.
Cell transfection and reagent
Vectors mediated precursor miR-376c-3p (HmiR0227-MR04) and miRNA inhibitors against miR-376c-3p (HmiR-AN0470-AM02), and their matching control vectors (CmiR0001-MR04 and CmiR-AN0001-AM02) were purchased from GeneCopoeia Inc. (Guangzhou, China). For ARID2 overexpression, Homo sapiens ARID2 cDNA was obtained from GeneCopoeia Inc and inserted into the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA, USA). Small interfering RNA (siRNA) used for ARID2 silencing and non-targeting (NT) siRNA were brought from GenePharma (Shanghai, China). HCC cell transfection was carried out using the Lipofectamine 2000 Reagent (Invitrogen). In transwell assay, the proliferation of HCC cells were blocked by treating with mitomycin C (10 μg/ml, Sigma-Aldrich) for 2h.
Cell proliferation and flow cytometry assay
HCC cells (5 × 103 per well) were seeded in a 96-well plate and cell proliferation was detected using a cell counting kit-8 (CCK-8) kit (Dojindo, Tokyo, Honshu, Japan). The results were measured the absorbance at 450 nm using a Multiskan Ex microtitre plate reader (Labsystems, Helsinki, Finland) at 0 h, 24 h, 48 h, and 72 h after seeding. To analyze cell cycle progression, HCC cells were incubated with propidium iodide (PI, Sigma) and RNAase (40710ES03, Shanghai Qcbio Science & Technology Co., Ltd., Shanghai, China). Cell cycle distribution was tested using flow cytometer equipped with Cell Quest software (FACSCalibur, BD Biosciences, San Jose, CA, USA).
Colony formation assay
Transected HCC cells (1 × 103 cells each well) were added into a 6-well plate and cultured in the incubator. After two weeks incubation, the formed cell colonies were fixed with 4% paraformaldehyde for 30 min and subsequently stained with 0.5% crystal violet (meilunbio, Dalian, China) for 30 min at room temperature.
Transwell assay
Transwell Cell Migration Chambers and BioCoat Matrigel Invasion Chambers from Corning Costar (Corning, NY, USA) were used for cell migration and invasion assays. Transfected HCC cells (5 × 104) were seeded in the upper chamber of the Transwell and 600 μl DMEM supplemented with 10% FBS was added into the lower chamber. Then the chambers were incubated for 24h at 37°C, migrated or invaded cells in the lower surface of the membrane were fixed 4% paraformaldehyde and stained with 1% crystal violet, and photographed using a Nikon E600 microscopy (Nikon Instrument Inc., Tokyo, Japan). Five fields of each well were randomly selected for quantification.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen) and was reverse transcribed into cDNA using a TakaRa PrimeScript™ RT kit (Takara, Dalian, China). miR-376c-3p expression level was quantified by TaqMan miRNA assays (Applied Biosystems, Foster City, CA, USA). qRT-PCR of ARID2 mRNA was carried out using SYBR Green Premix PCR Master Mix (Roche, Mannheim, Germany) in a StepOnePlus real-time PCR system (Applied Biosystems). 2-ΔΔCt method was used to calculate the levels of miR-376c-3p and ARID2 mRNA, which are normalized to U6 and GAPDH, respectively. All primers for qRT-PCR were listed in Table 2.
Primers for qRT-PCR
| Gene name | Primer sequences (5' to 3') | |
|---|---|---|
| miR-376c-3p | Forward | AACATAGAGGAAATTCCACG |
| ARID2 | Forward | GCAGACAGCCGAGTACATCTT |
| Reverse | CCTATGCCTTCGTCCTTGTCC | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward | TGCACCACCAACTGCTTAGC |
| Reverse | GGCATGGACTGTGGTCATGAG |
Western blot analysis
Xenograft tissues and HCC cells were lysed with RIPA lysis buffer (Beyotime, Beijing, China) on ice. A Enhanced BCA Protein Assay kit (Beyotime) was used for measuring protein concentration. The protein samples were separated by 10% SDS-PAGE and subsequently transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes then were incubated at 4°C overnight with specific primary antibody against ARID2 (ab166850, Abcam, Cambridge, UK), p-AKT (Ser473, #4060, Cell Signaling Technology, Beverly, MA, USA) , AKT (#4691, Cell Signaling Technology) and GAPDH (sc-47724, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (NXA931-1ML and NA934-1ML, GE Healthcare Life Sciences, Beijing, China) at room temperature for 2 h. The blots were visualized by Chemiluminescence Reagents (Sigma, St-Louis, MO, USA) and pictures were captured by Amersham™ Imager 600 machine (GE Healthcare Life Sciences).
Tumor Xenograft model
BALB/c male nude mice (aged 4-5 weeks, 18-20g) were bought from Shanghai SLAC Laboratory Animal Co. Ltd (Shanghai, China). Hep3B cells (1×106 per injection) that were transfected with control inhibitors and miR-376c-3p inhibitors were implanted into the flank of the mice via subcutaneous injection (n=5 each group). Tumor volumes were recorded every 3 days after being apparently observed and calculated with the following formula: Volume = (length × width2)/2. After 3 weeks, all mice were sacrificed under anesthesia. Tumor tissues were harvested and detected by qRT-PCR and immunoblotting for miR-376c-3p and ARID2 expression. For in vivo metastasis assay, Hep3B cells with miR-376c-3p knockdown and control cells were injected into nude mice via tail vein (2×106 cells per injection, n=5 per group). The mice were sacrificed 30 days after implantation. The lung tissues that were harvested and fixed by 10% neutral phosphate buffered formalin followed by H&E staining. The number of metastatic tumor corpuscles counted. The animal protocols were reviewed approved by the Animal Care and Use Committee of Xi'an Jiaotong University.
Luciferase reporter assay
pEZX-MT06 luciferase reporter vector containing 3'-UTR of ARID2 was purchased from GeneCopoeia, Inc (HmiT004640-MT06, Guangzhou, China). The predicted binding sequences for miR-376c-3p were mutated using a Quick-change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The luciferase reporter vector containing wild type or mutant 3'-UTR of ARID2 and miR-376c-3p or control mimics were co-transfected into SMMC-7721 and Hep3B cells. The relative luciferase activities were measured using a Luc-Pair™ Luciferase Assay Kit (GeneCopoeia, Inc).
Statistical analysis
The quantitative data were shown as Mean ± SD and analyzed using GraphPad Prism 6.0 Software (GraphPad Inc., San Diego, CA, USA). Student's t-test (2 groups) and one-way ANOVA (multiple groups) were conducted to analyze the difference. Overall survival curves were plotted using the Kaplan-Meier method and estimated by the log-rank test. The correlation between miR-376c-3p and ARID2 mRNA expression in HCC was analyzed using Pearson correlation analysis. Differences were defined as statistically significant if p<0.05.
Results
Upregulation of miR-376c-3p expression in HCC
To determine the expression pattern of miR-376c-3p, 60 pairs of HCC and corresponding tumor-adjacent liver tissues were detected by qRT-PCR. miR-376c-3p expression level was marked elevated in HCC compared to non-tumor tissues (P=0.0005, Figure 1A). Otherwise, we examined the level of miR-376c-3p in four HCC cell lines, including Hep3B, HepG2, Bel-7402 and SMMC-7721 as well as LO2. As shown in Figure 1B, the levels of miR-376c-3p in all these HCC cell lines were aberrantly higher than that in LO2 cells (P<0.05). Next, our cohort of HCC were divided in to two subgroups (low/high miR-376c-3p level) with the median level of miR-376c-3p as a cut-off value. The higher expression of miR-376c-3p was correlated with large tumor size (P=0.037, Table 1) and venous infiltration (P=0.039, Table 1). Survival analysis was performed using our follow-up data of HCC patients. These results demonstrated that HCC patients with high miR-376c-3p expression showed a marked poorer overall survival as compared with miR-376c-3p low-expressing patients (P=0.0387, Figure 1C). All together, these data suggest a close relationship between miR-376c-3p upregulation and reduced survival of HCC patients.
miR-376c-3p contributes to proliferation, migration and invasion of HCC cells
The clinical significance of miR-376c-3p promoted us to disclose its role in the malignant behaviors of HCC cells. Loss-of-function experiments were performed in HCC cells. The level of miR-376c-3p was prominently knocked down in both SMMC-7721 and Hep3B cells after transfecting miR-376c-3p inhibitors (P<0.05, Figure 2A). The results of CCK-8 and colony formation assay demonstrated that cell proliferation was dramatically repressed by miR-376c-3p knockdown (P<0.05, Figure 2B and 2C). Moreover, miR-376c-3p knockdown led to G1 arrest in HCC cells (P<0.05, Figure 2D). Meanwhile, we observed that HCC cells with miR-376c-3p knockdown showed a weaker migratory and invasive capacities compared to control cells (P<0.05, Figure 3). Moreover, miR-376c-3p overexpression significantly promoted cell proliferation and cell cycle progression of Bel-7402 cells in vitro (P<0.05, Supplementary Figure 1A-1D). miR-376c-3p-overexpressing Bel-7402 cells were treated with mitomycin C (10 μg/ml) for 2h to rule out the effect of cell proliferation in Transwell assay. The results showed that miR-376c-3p truly facilitated cell migration and invasion (P<0.05, Supplementary Figure 1E). Thus, miR-376c-3p exhibits a tumor promoting role in HCC.
Clinicopathological correlation analysis of miR-376c-3p expression in hepatocellular carcinoma (n=60)
| Clinicopathologic features | n | miR-376c-3p expression | P | ||
|---|---|---|---|---|---|
| Low level (n=30) | High level (n=30) | ||||
| Age (year) | ≤50 | 24 | 14 | 10 | 0.292 |
| >50 | 36 | 16 | 20 | ||
| Sex | Male | 48 | 26 | 22 | 0.197 |
| Female | 12 | 4 | 8 | ||
| HBV infection | No | 12 | 7 | 5 | 0.519 |
| Yes | 48 | 23 | 25 | ||
| Serum AFP level (ng/mL) | ≤20 | 16 | 10 | 6 | 0.243 |
| >20 | 44 | 20 | 24 | ||
| Tumor size (cm) | ≤5 | 15 | 11 | 4 | 0.037* |
| >5 | 45 | 19 | 26 | ||
| No. of tumor nodules | 1 | 51 | 27 | 24 | 0.472 |
| ≥2 | 9 | 3 | 6 | ||
| Cirrhosis | Absent | 21 | 9 | 12 | 0.417 |
| Present | 39 | 21 | 18 | ||
| Venous infiltration | Absent | 30 | 19 | 11 | 0.039* |
| Present | 30 | 11 | 19 | ||
| Edmondson-Steiner grading | I+II | 45 | 25 | 20 | 0.136 |
| III+IV | 15 | 5 | 10 | ||
| TNM tumor stage | I+II | 48 | 27 | 21 | 0.053 |
| III+IV | 12 | 3 | 9 | ||
HBV, hepatitis B virus; AFP, alpha-fetoprotein; TNM, tumor-node-metastasis. *Statistically significant
ARID2 is a direct target of miR-376c-3p
Large amount of studies have confirmed that miRNAs directly bind to 3'-UTR of target mRNAs and subsequently repressing their expression, and then exhibit corresponding function[24]. Next, we used three miRNA databases (TargetScan, miRDB and miRSearch) to predict candidate targets for miR-376c-3p. Forty-four candidate targets, which had the binding sites of miR-376c-3p in 3'-UTR, were shown in all three databases (Table 3). Among these, only ARID2 is recognized as a well-characterized tumor suppressor in HCC. The luciferase reporter system revealed that miR-376c-3p directly bound to the seed sequence region in the 3'-UTR of ARID2 and inhibited its luciferase activity in SMMC-7721 and Hep3B cells (P<0.05, Figure 4A and 4B). miR-376c-3p silencing prominently up-regulated ARID2 expression in SMMC-7721 and Hep3B cells (P<0.05, Figure 4C). Moreover, the expression of ARID2 mRNA was elevated in HCC as compared with tumor-adjacent tissues and showed an inverse correlation with miR-376c-3p expression (P<0.05, Figure 4D and 4E). The expression ARID2 protein in miR-376c-3p low expressed HCC tissues was obviously higher compared to cases with high miR-376c-3p level (P<0.05, Figure 4F). Previous study indicates that ARID2 may mediate AKT phosphorylation in HCC cells[25]. In our study, we also found that miR-376c-3p knockdown reduced p-AKT expression in both SMMC-7721 and Hep3B cells (Supplementary Figure 2). Collectively, these data demonstrate that miR-376c-3p represses the expression of ARID2, which depends on the binding to 3'-UTR of ARID2.
miR-376c-3p knockdown restrains HCC growth and metastasis in mice
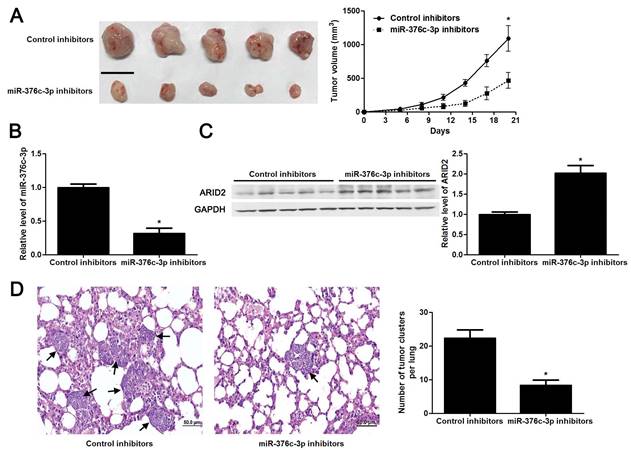
The tumors of xenograft mice models were presented in Figure 5A. Consistent with in vitro data, the tumor growth curves verified that miR-376c-3p knockdown led to HCC growth restriction in mice (P<0.05, Figure 5A). Tumor tissues harvested from miR-376c-3p knockdown group had an obvious lower miR-376c-3p expression and higher expression of ARID2 protein compared to control group (P<0.05. Figure 5B and 5C). Moreover, the lung metastases of HCC in miR-376c-3p knockdown group was prominently less than those in control group (P<0.05, Figure 5D). Our results suggest that miR-376c-3p contributes to HCC tumorigenesis and metastasis in vivo.
ARID2 mediates the oncogenic role of miR-376c-3p in HCC cells
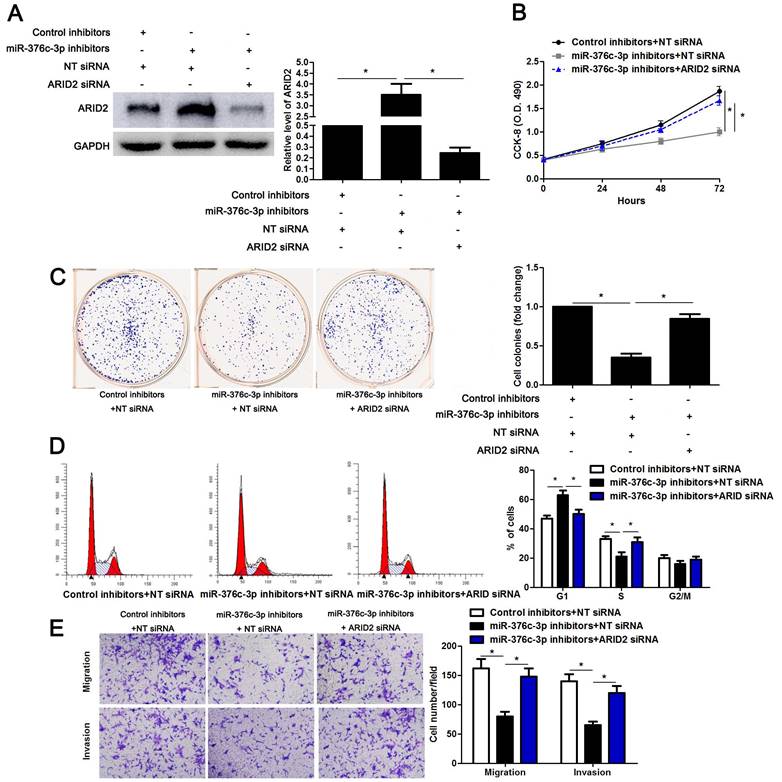
To detect whether ARID2 participates in miR-376c-3p induced HCC cell proliferation and mobility, ARID2 was overexpressed in Hep3B cells (P<0.05, Supplementary Figure 3A). ARID2 restoration significantly reduced cell cycle progression, proliferation, migration and invasion of Hep3B cells (P<0.05, Supplementary Figure 3B-3E), which was consistent with the effect of miR-376c-3p knockdown. Then, miR-376c-3p inhibitors and ARID2 siRNA were co-transfected into Hep3B cells (P<0.05, Figure 6A). Interestingly, the effects of miR-376c-3p knockdown on cell proliferation and mobility were attenuated by co-transfection with ARID2 siRNA (P<0.05, Figure 6B-6E). These data show that ARID2 is essential for miR-376c-3p-induced proliferation and mobility of HCC cells.
Discussion
Accumulating data have verified that miRNAs play an essential role in hepatocarcinogenesis and metastasis[24, 26]. For instance, miR-519a, miR-1468 and miR-367 promote cell growth and inhibit apoptosis of HCC cells[7, 27, 28]. On the other hand, miR-1296 and miR-542-3p repress epithelial-mesenchymal transition, and resulted in decreased metastatic ability of HCC cells[8, 9]. Recent studies have focused on the clinical value and biological role of miR-376c-3p in the tumorigenesis and metastasis of human cancer[12-14]. Yet, the expression pattern and biological function of miR-376c-3p remain unclear in HCC. Here, miR-376c-3p was found to be over-expressed in HCC tissues and predicted poor prognosis of patients. Additionally, functional assays revealed that miR-376c-3p knockdown inhibited cell cycle progression, proliferation, migration and invasion of HCC cells in vitro and restrained HCC growth and metastasis in nude mice. Meanwhile, miR-376c-3p overexpression facilitated the growth and mobility of HCC cells. These data reveal that miR-376c-3p plays a tumor promoting role in HCC.
Expression of miR-376c-3p in HCC. (A) The levels of miR-376c-3p were assessed in HCC tissues (n=80) and adjacent normal liver tissues (n=80). P=0.0005 by t test. (B) The levels of miR-376c-3p were increased in HCC cells lines (Bel-7402, HepG2, Hep3B and SMMC-7721) compared to LO2 cells. n=three independent repeats, *P<0.05 by ANOVA. (C) The cohort of HCC were divided in to two subgroups (low/high miR-376c-3p level) with the median level of miR-376c-3p as a cut-off value. HCC patients with high miR-376c-3p level (n=30) showed an obvious poorer over survival compared with cases with low miR-376c-3p level (n=30). P<0.05 by Log-rank test.
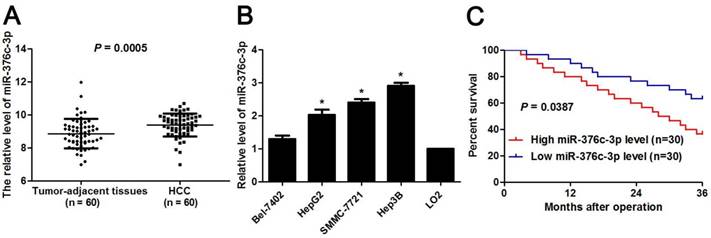
miR-376c-3p knockdown suppresses proliferation and cell cycle progression of HCC cells. (A) SMMC-7721 and Hep3B cells that were transfected with miR-376c-3p inhibitors or control inhibitors were subjected to qRT-PCR for miR-376c-3p expression. n=three independent repeats, *P<0.05 by t test. (B) miR-376c-3p knockdown inhibited proliferation of HCC cells as detected by CCK-8 assay. n=three independent repeats, *P<0.05 by ANOVA. (C) The colony formation capacity was weakened by miR-376c-3p knockdown in SMMC-7721 and Hep3B cells. n=three independent repeats, *P<0.05 by t test. (D) miR-376c-3p knockdown resulted in G1 arrest in HCC cells. n=three independent repeats, *P<0.05 by t test.
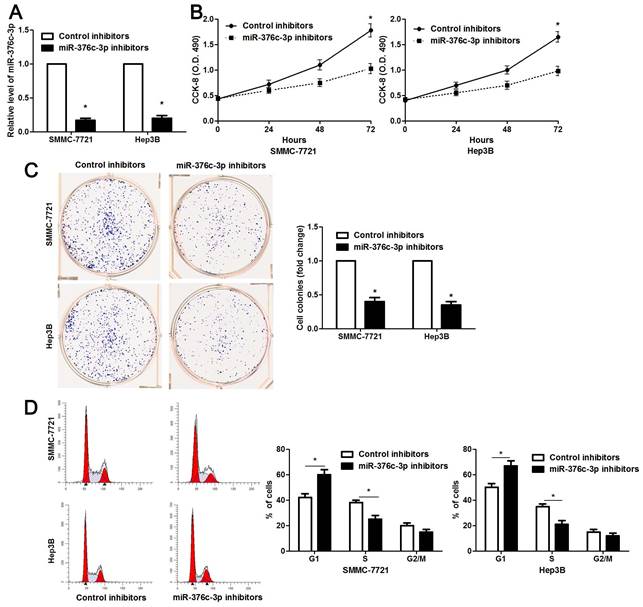
miR-376c-3p knockdown suppresses migration and invasion of HCC cells. SMMC-7721 and Hep3B cells were transfected with miR-376c-3p inhibitors or control inhibitors. Transwell assay indicated that miR-376c-3p knockdown significantly decreased the number of migrated and invaded HCC cells. n= five fields of three independent repeats, *P<0.05 by t test.
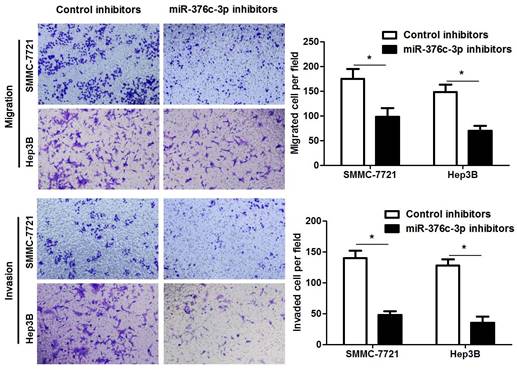
miR-376c-3p directly binds to the 3'-UTR of ARID2 in HCC cells. (A) The complementary pairing sequences between miR-376c-3p and 3'-UTR of ARID2. The mutant sites in the 3'-UTR of ARID2 were underlined. (B) SMMC-7721 and Hep3B cells co-transfected with wild type (wt) or mutant (mt) 3'-UTR of ARID2 and miR-376c-3p mimics. Overexpression of miR-376c-3p reduced the fluorescence intensity of vectors containing wt 3'-UTR of ARID2, but did not impact vectors containing mt 3'-UTR of ARID2 in HCC cells. n=three independent repeats, *P<0.05 by t test. (C) SMMC-7721 and Hep3B cells were transfected with miR-376c-3p inhibitors or control inhibitors. Knockdown of miR-376c-3p increased ARID2 expression in HCC cells. n=three independent repeats, *P<0.05 by t test. (D) 60 pairs of HCC tissues and matched noncancerous tissues were detected by qRT-PCR for ARID2 mRNA expression. P=0.0444 by t test. (E) A prominent inverse correlation between miR-376c-3p and ARID2 mRNA expression was found in HCC tissues. n=60, P<0.0001 by Pearson correlation analysis. (F) Six cases of HCC tissues with low or high miR-376c-3p level were detected by immunoblotting for ARID2 protein. ARID2 protein expression in miR-376c-3p low expressing HCCs was significantly higher than that in cases with high miR-376c-3p expression. *P<0.05 by t test.
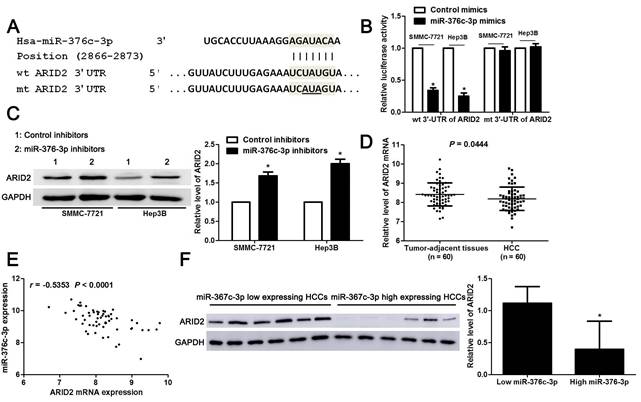
Knockdown of miR-376c-3p inhibits HCC growth and metastasis in vivo. (A) Hep3B cells that were transfected with control inhibitors or miR-376c-3p inhibitors were implanted into the right flank of the mice via subcutaneous injection (n=5 per group). the results indicated that miR-376c-3p knockdown led to HCC growth restriction in mice. *P<0.05 by ANOVA. Scale bar: 1 cm. (B) qRT-PCR analysis of miR-376c-3p expression in tumor tissues harvested from mice. n=5, *P<0.05 by t test. (C) Immunoblotting analysis of the expression of ARID2 protein in xenograft tissues arising from mice. n=5, *P<0.05 by t test. (D) Hep3B cells that were transfected with control inhibitors or miR-376c-3p inhibitors were implanted into nude mice via tail vein injection (n=5 per group). miR-376c-3p reduced the lung metastases of HCC in mice. *P<0.05 by t test. Black arrows indicated lung metastases. Scale bar: 50μm.
ARID2 silencing abolishes the effect of miR-376c-3p knockdown in HCC cells. (A) miR-376c-3p inhibitors and ARID2 siRNA were co-transfected into Hep3B cells. The protein level of ARID2 was determined using immunoblotting. n=three independent repeats, *P<0.05 by ANOVA. (B) miR-376c-3p knockdown suppressed proliferation of Hep3B cells, and ARID2 silencing subsequently promoted cell proliferation. n=three independent repeats, *P<0.05 by ANOVA. (C) ARID2 silencing increased the number of cell colonies in miR-376c-3p silenced Hep3B cells. n=three independent repeats, *P<0.05 by ANOVA. (D) miR-376c-3p knockdown led to G1 arrest, and ARID2 silencing subsequently promoted cell cycle progression in Hep3B cells. n=three independent repeats, *P<0.05 by ANOVA. (E) The migration and invasion capacities were repressed by miR-376c-3p knockdown, and accordingly reversed by ARID2 silencing in Hep3B cells. n= five filed of three independent repeats, *P<0.05 by ANOVA.
ARID2 is recognized as a novel tumor suppressor in HCC[29]. ARID2 loss impairs DNA repair process and subsequently leads to hepatocarcinogenesis[30]. ARID2 restoration restrains HCC cell proliferation and invasion via interferon induced transmembrane protein 1 (IFITM1)[31]. Moreover, ARID2 blocks G1/S transition and represses cell proliferation by suppressing cyclin D1 and cyclin E1 expression[32]. Gene mutation, transcriptional and posttranscriptional regulation are responsible for downregulation of ARID2 in HCC[31, 33-35]. Previous study reports that inactivation mutations of ARID2 is detected in 1 of 50 (2.0%) HBV-related HCC and 6 of 43 (14.0%) hepatitis C virus (HCV)-related HCC, respectively[33]. Meanwhile, another study shows an observation of ARID2 mutations in 7 of 120 (5.8%) HCC[35]. Hbx induces ARID2 transcriptional repression via inhibiting its promoter region, which contains atonal bHLH transcription factor 1 (ATOH1) binding sites in HCC[34]. Interestingly, two studies have revealed that ARID2 is posttranscriptionally modulated by miR-208-3p and miR-155 in HCC[25, 31]. Here, we demonstrated that miR-376c-3p supperssed ARID2 expression via directly pairing with the 3'-UTR of ARID2. Furthermore, the expression of miR-376c-3p was negatively associated with ARID2 mRNA level in HCC cases. Though several targets including HOXB7[14], ALK7[10], RUNX2[12] and ARID4A[20] are identified in other human cancers, this is the first study to report miR-376c-3p regulation of ARID2 in HCC. Notably, ARID2 restoration showed a similar effect to miR-376c-3p knockdown, which inhibited proliferation, cell cycle progression, migration and invasion of HCC cells. Notably, interference of ARID2 could reverse the effect of miR-376c-3p knockdown on HCC cell cycle progression, proliferation, migration and invasion of HCC cells. Our data may provide a new insight into miR-376c-3p/ARID2 axis in HCC.
To conclude, our study verified that miR-376c-3p expression was up-regulated in HCC and indicated a poor prognosis of patients. miR-376c-3p facilitated cell growth and metastasis through posttranscriptionally repressing ARID2 in HCC. Our data suggest that miR-376c-3p may be a potential prognostic marker and therapeutic target for HCC.
The candidate targets for hsa-miR-376c-3p
| Gene name | Gene name | Gene name |
|---|---|---|
| ARFGEF1 | HHIP | RAVER2 |
| ARFGEF2 | KIAA0247 | RFX6 |
| ARID2 | KIAA1239 | RPS6KA5 |
| B3GNT9 | KIAA2022 | SASH1 |
| BAG5 | LIMCH1 | SYF2 |
| C10orf118 | LIX1L | TMEM38B |
| CACNA1G | NCOA1 | VEZF1 |
| CCDC85C | NEUROD6 | VLDLR |
| DUSP11 | OPN3 | WDR44 |
| EDN1 | PAPSS2 | WNT3 |
| EN2 | PCBP2 | YTHDF1 |
| ERO1L | PHLDA2 | ZMYM2 |
| FRY | PPARGC1B | ZNF146 |
| GULP1 | PSPC1 | ZNF746 |
| HBEGF | PTBP2 |
Abbreviations
HCC, hepatocellular carcinoma; miRNAs, microRNAs; 3'-UTR, 3'-untranslated region; HNSCC, head and neck squamous cell carcinoma; CRC, colorectal cancer; OSCC, oral squamous cell carcinoma; ALK7, activin receptor-like kinase 7; lncRNA, long noncoding RNA; RUNX2, runt related transcription factor 2; HBV, hepatitis B virus; ARID2, AT-rich interaction domain 2; CCK-8, cell counting kit-8; qRT-PCR, quantitative real-time PCR; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF, polyvinylidene difluoride; HRP, horseradish peroxidase; HCV, hepatitis C virus; ATOH1, atonal bHLH transcription factor 1.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by the grant from the Social Development Science and Technology Research Project of Shaanxi Province (2013k12-05-07).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Diaz-Gonzalez A, Reig M, Bruix J. Treatment of Hepatocellular Carcinoma. Dig Dis. 2016;34:597-602
2. Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH. et al. A decade's studies on metastasis of hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2004;130:187-96
3. Desai JR, Ochoa S, Prins PA, He AR. Systemic therapy for advanced hepatocellular carcinoma: an update. Journal of gastrointestinal oncology. 2017;8:243-55
4. Finn RS, Zhu AX, Farah W, Almasri J, Zaiem F, Prokop LJ. et al. Therapies for Advanced Stage Hepatocellular Carcinoma with Macrovascular invasion or Metastatic Disease: a Systematic Review and Meta-analysis. Hepatology. 2017
5. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
6. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590-610
7. Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L. et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49
8. Tao J, Liu Z, Wang Y, Wang L, Yao B, Li Q. et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother. 2017;93:420-8
9. Xu Q, Liu X, Liu Z, Zhou Z, Wang Y, Tu J. et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16:103
10. Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y. et al. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci. 2011;124:359-68
11. Jin Y, Peng D, Shen Y, Xu M, Liang Y, Xiao B. et al. MicroRNA-376c inhibits cell proliferation and invasion in osteosarcoma by targeting to transforming growth factor-alpha. DNA and cell biology. 2013;32:302-9
12. Chang WM, Lin YF, Su CY, Peng HY, Chang YC, Lai TC. et al. Dysregulation of RUNX2/Activin-A Axis upon miR-376c Downregulation Promotes Lymph Node Metastasis in Head and Neck Squamous Cell Carcinoma. Cancer Res. 2016;76:7140-50
13. Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8:5286-97
14. Wang K, Jin J, Ma T, Zhai H. MiR-376c-3p regulates the proliferation, invasion, migration, cell cycle and apoptosis of human oral squamous cancer cells by suppressing HOXB7. Biomed Pharmacother. 2017;91:517-25
15. Huang Q, Wang C, Hou Z, Wang G, Lv J, Wang H. et al. Serum microRNA-376 family as diagnostic and prognostic markers in human gliomas. Cancer biomarkers: section A of Disease markers. 2017;19:137-44
16. Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P. et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941-50
17. Huo D, Clayton WM, Yoshimatsu TF, Chen J, Olopade OI. Identification of a circulating microRNA signature to distinguish recurrence in breast cancer patients. Oncotarget. 2016;7:55231-48
18. Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A. et al. Circulating microRNAs in plasma as early detection markers for breast cancer. International journal of cancer. 2013;132:1602-12
19. Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J. et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PloS one. 2013;8:e76729
20. Hung PS, Chen CY, Chen WT, Kuo CY, Fang WL, Huang KH. et al. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12:e0177346
21. Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY. et al. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PloS one. 2012;7:e33608
22. Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y. et al. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Molecular cancer. 2012;11:44
23. Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D. et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer. 2017;17:805
24. Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137-51
25. Zhang L, Wang W, Li X, He S, Yao J, Wang X. et al. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol. 2016;48:2425-34
26. Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603-18
27. Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C. et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123
28. Tu K, Liu Z, Yao B, Han S, Yang W. MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int J Oncol. 2016;48:965-74
29. Zhao H, Wang J, Han Y, Huang Z, Ying J, Bi X. et al. ARID2: a new tumor suppressor gene in hepatocellular carcinoma. Oncotarget. 2011;2:886-91
30. Oba A, Shimada S, Akiyama Y, Nishikawaji T, Mogushi K, Ito H. et al. ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. Journal of hepatology. 2017;66:942-51
31. Yu P, Wu D, You Y, Sun J, Lu L, Tan J. et al. miR-208-3p promotes hepatocellular carcinoma cell proliferation and invasion through regulating ARID2 expression. Experimental cell research. 2015;336:232-41
32. Duan Y, Tian L, Gao Q, Liang L, Zhang W, Yang Y. et al. Chromatin remodeling gene ARID2 targets cyclin D1 and cyclin E1 to suppress hepatoma cell progression. Oncotarget. 2016;7:45863-75
33. Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nature genetics. 2011;43:828-9
34. Gao Q, Wang K, Chen K, Liang L, Zheng Y, Zhang Y. et al. HBx protein-mediated ATOH1 downregulation suppresses ARID2 expression and promotes hepatocellular carcinoma. Cancer science. 2017;108:1328-37
35. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44:760-4
Author contact
![]() Corresponding author: Prof. Shuangsuo Dang, Department of Infectious Diseases, the Second Affiliated Hospital of Xi'an Jiaotong University, 157 Xiwu Road, Xi'an 710004, China. Email: dangshuangsuocom
Corresponding author: Prof. Shuangsuo Dang, Department of Infectious Diseases, the Second Affiliated Hospital of Xi'an Jiaotong University, 157 Xiwu Road, Xi'an 710004, China. Email: dangshuangsuocom

 Global reach, higher impact
Global reach, higher impact