Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(22):4234-4241. doi:10.7150/jca.26050 This issue Cite
Research Paper
P53 Codon 72 Polymorphism and Risk for Squamous Cell Carcinoma of the Penis: A Caucasian Case-Control Study
1. Institute of Pathology, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany;
2. Department of Otorhinolaryngology - Head and Neck Surgery, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany;
3. Institute of Pathology, RWTH Aachen University, Aachen, Germany;
4. Institute of Pathology, Hameln, Germany;
5. Institute of Pathology, Ansbach, Germany;
6. Department of Urology, Caritas St. Josef Medical Center, University of Regensburg, Regensburg, Germany;
7. Department of Urology, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany
Received 2018-3-14; Accepted 2018-7-21; Published 2018-10-20
Abstract
Squamous cell carcinoma of the penis is a rare but often aggressive disease. A large proportion of penile cancers are associated with HPV infection, mainly with HPV high-risk subtypes 16 and 18. From other HPV-related malignancies a link between a functional SNP in the p53 gene (rs1042522, p.Arg72Pro) and a higher disease risk in the presence of HPV is documented. The p53 p.Arg72 variant was described as a risk factor for developing a malignancy in combination with the presence of HPV as the p.72Arg variant is more prone to HPV E6 protein-mediated degradation than the p.72Pro variant. For penile carcinoma there are only sparse data available on this topic. We therefore analyzed the distribution of this p53 codon 72 SNP in a cohort of 107 penile cancer patients and a healthy control group (n=194) using Restriction Fragment Length Polymorphism (RFLP) analysis. After DNA isolation a PCR amplicon including the variant nucleotide was generated. Based on the variant nucleotide this amplicon can be cleaved into two parts or remain unaffected by a restriction enzyme. Subsequent electrophoresis allowed the discrimination of SNP alleles in the investigated sample. Comparison of the allelic variants revealed no significant differences in the distribution of this SNP between cases and controls (p=0,622). There was also no difference in SNP distribution between cases with/without HPV infection (p=0,558) or histologic variants (p=0.339). In order to strengthen the impact of our data we performed a combined analysis of all published data on this topic with our results. This ended up in SNP distribution data from 177 cases and 1149 controls. Overall, there were also no significant differences in the allelic distribution of the p53 codon 72 SNP between either cases and controls (p=0,914) or HPV-positive and HPV-negative cases (p=0,486). From this most comprehensive data available to date we conclude that there is no influence of the p53 codon 72 SNP on the risk of development of penile carcinoma in Caucasians even in the presence of HPV.
Keywords: penile cancer, HPV, squamous cell carcinoma, p53, polymorphism, RFLP
Introduction
Penile cancer is a rare but biologically aggressive disease especially in the metastatic situation. The mean incidence is less than 1 case per 100.000 males in Europe and North America [1]. Nevertheless, cancer of the penis adds up to 17% of all malignant diseases in men in developing countries [2]. The predominating histological subtype is the squamous cell carcinoma (SCC) and accounts for more than 95% of all malignant lesions of the penis [2]. Although approximately 80% of the cases can be cured if there is only limited involvement of the lymph nodes [3], more advanced disease might demand partial or complete amputation with dramatic effects on patient's quality of life [4].
Epidemiological studies revealed several risk factors for penile carcinoma like low standards of hygiene, phimosis, high number of sexual partners (early age at first sexual intercourse), presence of Human Papilloma Virus (HPV) infection, circumcision practice, recurrent balanitis, strong tobacco consumption, genital ultraviolet radiation and penile trauma. Interestingly, the actual data suggests that the critical exposure period for certain etiologic factors is already before puberty [1, 5] .
Penile cancer is a very heterogeneous disease. The molecular background of penile tumorigenesis is still not fully understood but recent comprehensive molecular profiling analyses allowed a deeper insight into prominent alterations in penile cancer [6, 7]. Although various other molecular mechanisms causing penile carcinoma independently of HPV probably represent the most common events, a large proportion of penile carcinomas can be attributed to HPV infection [8]. In 30%-40% of the analyzed penile cancer cases HPV was detectable worldwide [9]. It is well known that infection with HPV is a common risk factor for various tumor types. Especially presence of HPV high risk subtypes (e.g. HPV 16 and HPV 18) correlates closely with the development of cervical, vaginal, oropharynx or anal cancers to name only a few [9]. The oncogenic properties of HPV 16 and 18 are based on the expression of the viral E6 and E7 oncoproteins. While E7 protein binds and inactivates the tumor suppressor Rb [10], the E6 protein binds to the cellular p53 protein followed by leading in degradation by the ubiquitin proteasome pathway [11, 12]. This E6 protein guided degradation is also influenced by a functional single nucleotide polymorphism (SNP) in exon 4 of the p53 gene. At position 215 of the p53 cDNA either a guanine (5'-CGC-3', encoding for the amino acid arginine) or a cytosine (5'-CCC-3', encoding for the amino acid proline) base is located resulting in a polymorphic amino acid site at codon 72 of P53 (p.Arg72Pro). The p.Arg72 variant was shown to be more prone to the E6 protein guided degradation than the p.72Pro variant. In contrast, cells harboring the p.Pro72 variant were more resistant to apoptotic stimuli due to a slower traffic of this variant to the mitochondria [13-15]. The identification of this functional p53 SNP led to a multitude of case-control studies investigating the role of p53 codon 72 SNP as a risk factor in various tumor entities [16]. Indeed, in several HPV-related cancers (e.g. oral cancer, cervical cancer, vulvar and vaginal carcinoma) disease risk was shown to be influenced by the p53 p.Arg72Pro SNP [17-19] suggesting the p.72Arg variant as a risk factor for cancer development in high risk HPV positive individuals. But also in non HPV-related tumor types p53 codon 72 SNP was shown to influence disease risk [20-22]. These data gave a rationale for testing a hypothetical role of the p53 codon 72 SNP as a risk factor for penile carcinoma, too. Only sparse data from analyses of only small cohorts are available to date [23, 24]. The aim of our study was the combined investigation of HPV status and distribution of the p53 codon 72 SNP in a large Caucasian case-control study of penile carcinoma patients.
Methods
Patient material
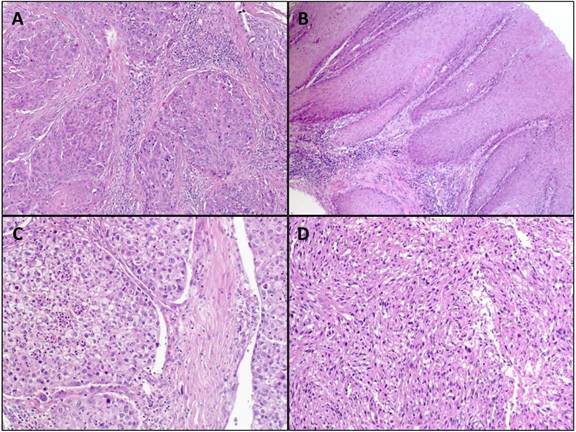
Archival formalin-fixed paraffin-embedded (FFPE) non-tumorous tissue from 107 cases of penile squamous cell carcinoma was used for the study. The tumors of the patients were classified and staged according to the WHO classification of penile tumors and the current AJCC/TNM-classification system [25]. Representative images of penile cancers are shown in figure 1. As a control group 194 blood samples from a male cohort of patients without any malignancy were investigated. The genotyping results of this control group were already described recently and also used for this study [26].
Clinicopathological characteristics of the cases are shown on Table 1. Prior IRB approval (University Hospital Erlangen, Germany) was obtained for the scientific use of archival material.
Microdissection of tissue and DNA isolation
Microdissection and isolation of genomic DNA was carried out from FFPE tissue as described previously [27]. In brief, 5 µm thick serial sections of the tissue were dewaxed and stained with 0.1% methylene blue for 15 seconds. Using an inverted microscope non-tumorous tissue (identified through matching with a marked H&E-stained section reviewed by an experienced surgical pathologist) was scraped off with a sterile needle. Isolation of genomic DNA from the microdissected tissue was performed using the Blood DNA Preparation Kit (Maxwell® 16 System, Promega, Mannheim, Germany) according to the manufacturer's instructions.
Representative images of H&E stained penile squamous cell carcinoma. A: usual type (magnification: x100); B: verrucous carcinoma (magnification: x100); C: clear cell squamous cell carcinoma (magnification: x200); D: sarcomatoid squamous cell carcinoma (magnification: x200).
Clinico-pathological characteristics of the analyzed cohorts
| Cohorts / Characteristics | Cases (n=107) | Controls (n=194) |
|---|---|---|
| Age, years | ||
| Median age | 67 | 69 |
| Mean age | 67,3 ± 11,8 | 67,3 ± 10,6 |
| Range | 39 - 93 | 34 - 88 |
| Tumor Stage | (n=) | not applicable |
| pTis | 8 | |
| pTa | 1 | |
| pT1 | 50 | |
| pT2 | 31 | |
| pT3 | 13 | |
| pT4 | 1 | |
| unknown | 3 | |
| Tumor Grade | (n=) | not applicable |
| 1 | 23 | |
| 2 | 36 | |
| 3 | 40 | |
| CIS | 8 | |
| HPV Status | (n=) | not determined |
| positive | 41 | |
| negative | 64 | |
| unknown | 2 | |
| Histopathology of penile SCC | ||
| Histological subtype | HPV negative (n) | HPV positive (n) |
| usual type | 34 | 7 |
| verrucous | 13 | 0 |
| basaloid | 3 | 14 |
| warthy-basaloid | 3 | 8 |
| pseudohyperplastic | 6 | 1 |
| warthy | 1 | 2 |
| lymphoepithelioma-like | 0 | 2 |
| clear cell | 1 | 1 |
| sarcomatoid | 1 | 0 |
| carcinoma cuniculatum | 1 | 0 |
| unknown: n=1 |
p53 codon 72 polymorphism analysis
SNP analysis was performed using restriction fragment length polymorphism analysis (RFLP) of the polymorphic region in exon 4 of the p53 gene which contains a Bst UI recognition site (5' -CG ↓ CG-3') in the presence of the G-allele (p.72Arg) as described previously [28]. Presence of the p.72Arg allele resulted in digestion of the PCR product (196bp = 113 + 83bp), whereas PCR products containing the C-allele (p.72Pro) remained unaffected.
Amplification of variable region and RFLP analysis
The SNP region was amplified by PCR in a total volume of 25 µl containing 100 ng DNA. Detailed information on primer sequences and exact PCR conditions were described previously [26]. PCR products were incubated for 6 h with 5 U Bst UI (New England Biolabs, Frankfurt/Main, Germany) at 60 ° C in a total volume of 30 µl to ensure complete digestion. Restriction fragments were separated by capillary electrophoresis using ABI Prism 3500 Genetic Analyzer and the fragment analysis was performed using the GeneMapper Software Version 4.1 (Applied Biosystems, Foster City, Calif., USA).
HPV PCR analysis of penile tumors
Tumor cells were microdissected and DNA was extracted as described above. Detection of HPV DNA was performed using GP5+/6+ primers as described previously [29]. Subclassification of HPV species was done using type-specific primers as described elsewhere [30, 31].
Statistics
To test if the genotype distribution followed Hardy-Weinberg equilibrium, the public software at http://ihg.gsf.de/cgi-bin/hw/hwa1.pl was used. Х2 statistics (2-sided exact significance) were used to evaluate case-control differences in the distribution of genotypes and to analyze associations between genotypes and HPV status. To determine the distribution of the risk allele versus the non-risk allele, Fisher's exact test (2-sided exact significance) was used. Statistical analysis was carried out using SPSS version 13.0 (SPSS, Chicago, IL, USA). P values less than 0.05 were interpreted as statistically significant.
Results
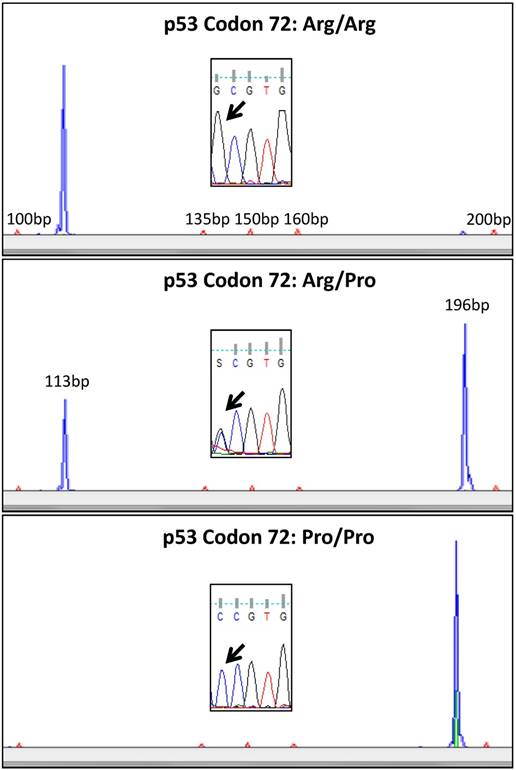
A comprehensive validation of the used method was already described in a prior study [26]. We therefore used only 5 cases for verification of the RFLP results by direct sequencing in the actual analysis. RFLP analysis was confirmed by sequencing in all cases. Representative examples for genotyping are shown in Figure 2.
Representative examples of p53 codon 72 analysis using RFLP. In the upper lane a case homozygous for the p.72Arg variant is shown (blue RFLP signal only at 113bp). In the middle lane a heterozygous case is shown with blue RFLP signals at 113bp and 196bp. The lower lane shows a case homozygous for the p.72 Pro variant (blue RFLP signal at 196bp only). Sanger sequencing confirmed the allelic status (black arrow) detected with RFLP.
Distribution of the p53 codon 72 SNP in control groups from the literature compared to results of the control group from this work.
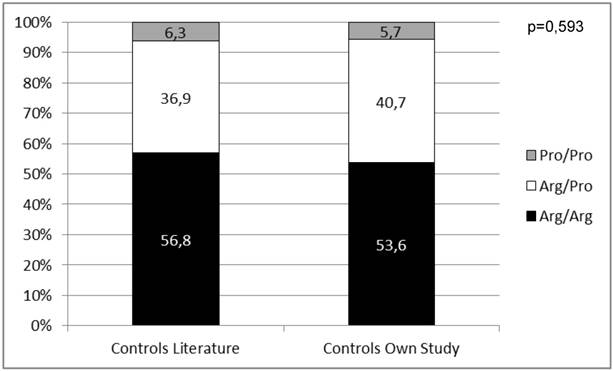
We first wanted to know if our male control cohort is representative for Caucasians regarding the distribution of the p53 codon 72 SNP. Therefore, we compared our results to already published data (n=955) on genotyping of the p53 codon 72 SNP in healthy Caucasian males [23, 24, 32, 33]. There was no significant difference in genotype distribution (p=0,593) strengthening the validity of our controls as a representative Caucasian male control group (Figure 3).
Determination of HPV status was successful in 105/107 cases. In 2/107 cases internal DNA control for HPV testing was not successful. In these cases HPV status could be determined. Positive HPV status was found in 39% (n=41/105) of the cases with interpretable results (Table 1). HPV status of the healthy male control group was not determined.
Overall, 85/107 samples from penile carcinoma cases gave an interpretable result in the RFLP analysis. In 22 cases the SNP region could not be amplified due to insufficient DNA quality (degradation of DNA). The genotype distribution in both penile carcinoma cohort (p=0,579) and control group (p=0,575) followed the Hardy-Weinberg equilibrium.
The distribution of the p53 p.Arg72Pro SNP did not differ significantly between cases and controls (p=0,622; Table 2). There was also no correlation between genotype distribution and HPV status of penile carcinoma cases (p=0,558; Table 3).
In addition, the new WHO classification of penile carcinoma defined several histologic variants according to the HPV status [25]. We also analyzed the distribution of the p53 p.Arg72Pro SNP in the histologic variants but found no significant differences (p=0,339; data not shown), too.
Genotype distribution of the p53 codon 72 SNP in cases (showing interpretable results) and controls.
| p53 p.72Arg/Arg | p53 p.72Arg/Pro | p53 p.72Pro/Pro | p-value | |
|---|---|---|---|---|
| Controls (n=194) | 53,6% (n=104) | 40,7% (n=79) | 5,7% (n=11) | |
| Cases (n=85) | 55,3% (n=47) | 36,5% (n=31) | 8,2% (n=7) | 0,622 |
Genotype distribution of the p53 codon 72 SNP in HPV positive and negative cases (cases with interpretable results: n=85).
| p53 p.72Arg/Arg | p53 p.72Arg/Pro | p53 p.72Pro/Pro | p-value | |
|---|---|---|---|---|
| HPV positive (n=34) | 50% (n=17) | 38,2% (n=13) | 11,8% (n=4) | |
| HPV negative (n=51) | 58,8% (n=30) | 35,3% (n=18) | 5,9% (n=3) | 0,558 |
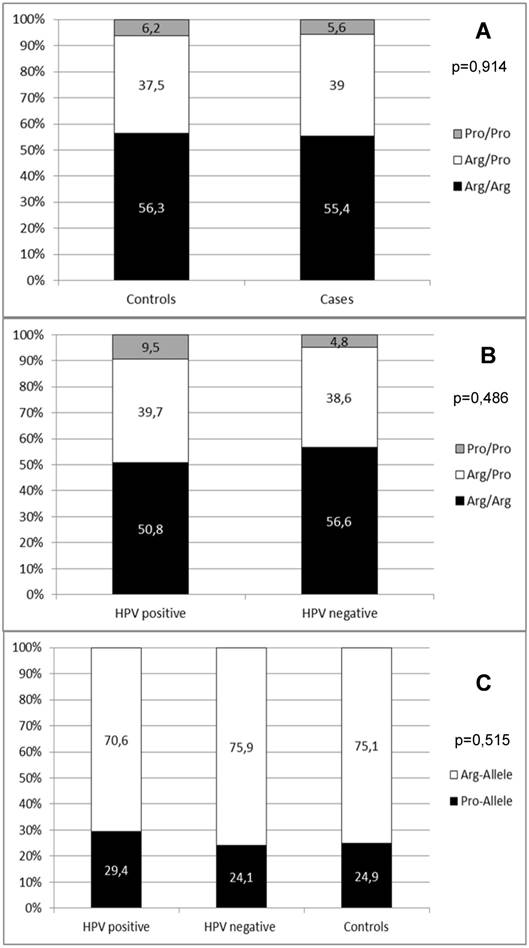
In a second step, we wanted to summarize the available data on the role of the p53 codon 72 SNP in penile carcinoma in Caucasians. We therefore combined all published data [23, 24, 32, 33] with the results of our study. In summary, data from 1149 controls and 177 penile carcinoma cases were available. With this combined data we repeated the statistical analyses as investigation of a larger number of cases and controls should increase the impact of the SNP distribution analyses. The genotype distribution of these combined cohorts did not differ from the Hardy-Weinberg equilibrium in cases (p=0,841) and controls (p=1,000). This combined analysis also showed no significant difference in p53 codon 72 SNP distribution between cases and controls (p=0,914; Figure 4A). Also, the genotype distribution between HPV positive and HPV negative penile carcinoma cases did not show a significant difference (p=0,486; Figure 4B). Regarding the distribution of the specific risk alleles (Arg allele vs. Pro allele) there was also no significant difference between controls, HPV positive and HPV negative cases (p=0,515; Figure 4C).
(A) Distribution of p53 codon 72 SNP in combined data from published studies and this work. (B) Distribution of p53 codon 72 SNP in combined data from published studies according to HPV positive and HPV negative cases. (C) Distribution of p53 codon 72 alleles in combined data from published studies according to HPV positive and HPV and controls.
Discussion
In the presented work, we analyzed the distribution of the functional p53 codon 72 SNP in a case/control study to further clarify the influence of this SNP on penile carcinoma risk in general and in association with the presence of HPV. By combining data from our cohorts with already published results on smaller Caucasian cohorts we were able to present the most comprehensive data on this topic yet. Here we could show that the p53 codon 72 SNP has no influence on penile carcinoma risk even in HPV-positive cases. Therefore, the presence of the variable SNP alleles seemed not to be a risk factor for this rare disease as described in other HPV-related tumors. To date, little is known about the role of molecular predisposing factors in penile carcinoma. A PubMed search for “penile cancer“and “SNP” or “penile cancer” and “polymorphism” revealed no other available publications than the already mentioned studies of the p53 codon 72 SNP. This underlines the lack of current knowledge of the role of patient's molecular background as a risk factor for this partly fatal disease. The reason for this might be the low incidence of penile carcinoma especially in developed countries urging on multi-institutional and international cooperative efforts.
Although the most prominent p53 variant plays no important role in penile carcinogenesis p53 alterations are a major feature of penile carcinomas. Consisting data from several studies showed that p53 protein overexpression in the tumors analyzed by immunohistochemistry is associated with disease progression, higher risk for lymph node metastasis and worse prognosis [34]. However, inconsistent data can be found on the frequency of p53 mutations. Previous studies reported only a lower frequency (17% - 25%) of p53 mutations, whereas recent genomic profiling studies reported p53 alterations in up to 76% of the cases analyzed [6, 7, 35-37]. These discrepancies might be explained with the methodological background of the studies (e.g. different sensitivity levels for detection of mutations for SSCP, Sanger and next-generation sequencing), ethnical differences (Asian and Caucasian cohorts), HPV status and number of analyzed samples or histopathological characteristics of the tumors. Moreover, the p53 signaling pathway could be identified as the most significantly altered pathway in penile cancer recently [38]. But despite inconsistent findings p53 dysregulation obviously plays a crucial role in the development of penile carcinoma whose elucidation would be important for a better understanding of this disease.
A critical point of our study might be the combined analysis of our data with already published studies. While Humbey et al. and Tornesello et al. used DNA from penile carcinoma tissue for genotyping, we used DNA from non-tumor tissue for the analysis [23, 24]. We therefore cannot rule out the possibility that loss of heterozygosity (LOH) at the p53 gene locus (chromosome 17p.13.1) in the tumors of our cases might have led to differing results. Especially, in heterozygous individuals LOH could cause an enrichment of one allele in the tumors that biased the frequencies of allelic distribution between cases and controls. But as we wanted to analyze the general risk of the p53 codon 72 SNP for development of a penile carcinoma it was reasonable to analyze the germline situation of the patients. Nevertheless, several points strengthen our approach. All three studies gave very similar results either analyzing tumor or non-tumor tissue without any enrichment of one variant. Humbey et al. performed a LOH analysis on a subset of their cases and did not detect any allelic losses [24]. Rocha et al. performed FISH analyses for the p53 gene locus in 297 penile carcinomas and did not find any case with a deletion [35]. Alves and colleagues studied genetic imbalances in penile squamous cell carcinoma using a comparative genomic hybridization approach. A loss of the chromosomal region 17p.13.1 or a complete loss of chromosome 17p was detected in only 7,7% of the cases analyzed [39]. Two additional studies reported cytogenetic analyses of single cases (n=3) of penile squamous cell carcinoma and they also reported rarely abnormalities of chromosome 17 [40, 41]. Interestingly, the generally detected high frequency of p53 expression in penile tumors often associated with HPV negativity was not reflected in a concordant high frequency of p53 mutations yet. Therefore the high expression of wild-type p53 in penile carcinoma cells induced by DNA damage or other stress factors (e.g. inflammation after laser-assisted evaporation of pre-neoplastic lesions, inactivation of p53 degeneration pathways) might be taken in consideration as it was already discussed for penile carcinoma and in other cancer entities [27, 42]. All these data argue against a frequent loss of the p53 gene locus in penile carcinoma and underline the validity of our practice.
Summarizing all these data our comprehensive analysis of the role of the p53 codon 72 SNP in penile carcinoma revealed no influence of these functional variants on disease risk even in cases with a HPV involvement in Caucasians. This is contrary to data from other HPV-related cancers and might suggest other interactions between HPV-encoded proteins, p53 and additional cellular components in penile carcinogenesis.
Acknowledgements
The authors thank Verena Popp, Stefanie Herlein, Daniela Renner, Petra Badorf and Claudia Schmied for excellent technical assistance. Parts of this study were presented at the 101st annual meeting of the German Association of Pathology in Erlangen, Germany from June 22 to 24, 2017. We acknowledge support by Deutsche Forschungsgemeinschaft and Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU) within the funding programme Open Access Publishing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hakenberg OW, Comperat EM, Minhas S, Necchi A, Protzel C, Watkin N. et al. EAU guidelines on penile cancer: 2014 update. European urology. 2015;67:142-50
2. Kroon BK, Horenblas S, Nieweg OE. Contemporary management of penile squamous cell carcinoma. Journal of surgical oncology. 2005;89:43-50
3. Srinivas V, Morse MJ, Herr HW, Sogani PC, Whitmore WF Jr. Penile cancer: relation of extent of nodal metastasis to survival. The Journal of urology. 1987;137:880-2
4. Sosnowski R, Kulpa M, Kosowicz M, Wolski JK, Kuczkiewicz O, Moskal K. et al. Quality of life in penile carcinoma patients - post-total penectomy. Central European journal of urology. 2016;69:204-11
5. Barski D, Georgas E, Gerullis H, Ecke T. Metastatic penile carcinoma - an update on the current diagnosis and treatment options. Central European journal of urology. 2014;67:126-32
6. Ali SM, Pal SK, Wang K, Palma NA, Sanford E, Bailey M. et al. Comprehensive Genomic Profiling of Advanced Penile Carcinoma Suggests a High Frequency of Clinically Relevant Genomic Alterations. The oncologist. 2016;21:33-9
7. McDaniel AS, Hovelson DH, Cani AK, Liu CJ, Zhai Y, Zhang Y. et al. Genomic Profiling of Penile Squamous Cell Carcinoma Reveals New Opportunities for Targeted Therapy. Cancer research. 2015;75:5219-27
8. Calmon MF, Tasso Mota M, Vassallo J, Rahal P. Penile carcinoma: risk factors and molecular alterations. TheScientificWorldJournal. 2011;11:269-82
9. Alemany L, Cubilla A, Halec G, Kasamatsu E, Quiros B, Masferrer E. et al. Role of Human Papillomavirus in Penile Carcinomas Worldwide. European urology. 2016;69:953-61
10. Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934-7
11. Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76-9
12. Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer science. 2007;98:1505-11
13. Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F. et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229-34
14. Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Molecular and cellular biology. 1999;19:1092-100
15. Murphy ME, Leu JI, George DL. p53 moves to mitochondria: a turn on the path to apoptosis. Cell cycle. 2004;3:836-9
16. Khan MH, Khalil A, Rashid H. Evaluation of the p53 Arg72Pro polymorphism and its association with cancer risk: a HuGE review and meta-analysis. Genetics research. 2015;97:e7
17. Hou J, Gu Y, Hou W, Wu S, Lou Y, Yang W. et al. P53 codon 72 polymorphism, human papillomavirus infection, and their interaction to oral carcinoma susceptibility. BMC genetics. 2015;16:72
18. Makni H, Franco EL, Kaiano J, Villa LL, Labrecque S, Dudley R. et al. P53 polymorphism in codon 72 and risk of human papillomavirus-induced cervical cancer: effect of inter-laboratory variation. International journal of cancer Journal international du cancer. 2000;87:528-33
19. Qvick A, Sorbe B, Helenius G, Karlsson MG, Lillsunde Larsson G. Does p53 codon 72 polymorphism have a prognostic value in carcinoma of the vulva and vagina? Medical oncology. 2017;34:36
20. Qiao Q, Hu W. The association between TP53 Arg72Pro polymorphism and lung cancer susceptibility: evidence from 30,038 subjects. Lung. 2013;191:369-77
21. Lu Y, Liu Y, Zeng J, He Y, Peng Q, Deng Y. et al. Association of p53 codon 72 polymorphism with prostate cancer: an update meta-analysis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:3997-4005
22. Hori Y, Miyabe K, Yoshida M, Nakazawa T, Hayashi K, Naitoh I. et al. Impact of TP53 codon 72 and MDM2 SNP 309 polymorphisms in pancreatic ductal adenocarcinoma. PloS one. 2015;10:e0118829
23. Tornesello ML, Duraturo ML, Guida V, Losito S, Botti G, Pilotti S. et al. Analysis of TP53 codon 72 polymorphism in HPV-positive and HPV-negative penile carcinoma. Cancer letters. 2008;269:159-64
24. Humbey O, Cairey-Remonnay S, Guerrini JS, Algros MP, Mougin C, Bittard H. et al. Detection of the human papillomavirus and analysis of the TP53 polymorphism of exon 4 at codon 72 in penile squamous cell carcinomas. European journal of cancer. 2003;39:684-90
25. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. European urology. 2016;70:93-105
26. Rogler A, Rogenhofer M, Borchardt A, Lunz JC, Knoell A, Hofstaedter F. et al. P53 codon 72 (Arg72Pro) polymorphism and prostate cancer risk: association between disease onset and proline genotype. Pathobiology: journal of immunopathology, molecular and cellular biology. 2011;78:193-200
27. Weyerer V, Schneckenpointner R, Filbeck T, Burger M, Hofstaedter F, Wild PJ. et al. Immunohistochemical and molecular characterizations in urothelial carcinoma of bladder in patients less than 45 years. Journal of Cancer. 2017;8:323-31
28. Faur N, Araud L, Laroche-Clary A, Kanno J, Toutain J, Yamori T. et al. The association between the T309G polymorphism of the MDM2 gene and sensitivity to anticancer drug is dependent on the p53 mutational status in cellular models. British journal of cancer. 2009;101:350-6
29. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. The Journal of general virology. 1995;76( Pt 4):1057-62
30. Lin CY, Chao A, Yang YC, Chou HH, Ho CM, Lin RW. et al. Human papillomavirus typing with a polymerase chain reaction-based genotyping array compared with type-specific PCR. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2008;42:361-7
31. Sahiner F, Kubar A, Yapar M, Sener K, Dede M, Gumral R. Detection of major HPVs by a new multiplex real-time PCR assay using type-specific primers. Journal of microbiological methods. 2014;97:44-50
32. Henner WD, Evans AJ, Hough KM, Harris EL, Lowe BA, Beer TM. Association of codon 72 polymorphism of p53 with lower prostate cancer risk. The Prostate. 2001;49:263-6
33. Koushik A, Tranah GJ, Ma J, Stampfer MJ, Sesso HD, Fuchs CS. et al. p53 Arg72Pro polymorphism and risk of colorectal adenoma and cancer. International journal of cancer Journal international du cancer. 2006;119:1863-8
34. Zargar-Shoshtari K, Sharma P, Spiess PE. Insight into novel biomarkers in penile cancer: Redefining the present and future treatment paradigm?. Urologic oncology. 2017
35. Rocha RM, Ignacio JA, Jordan J, Carraro DM, Lisboa B, Lopes A. et al. A clinical, pathologic, and molecular study of p53 and murine double minute 2 in penile carcinogenesis and its relation to prognosis. Human pathology. 2012;43:481-8
36. Yanagawa N, Osakabe M, Hayashi M, Tamura G, Motoyama T. Detection of HPV-DNA, p53 alterations, and methylation in penile squamous cell carcinoma in Japanese men. Pathology international. 2008;58:477-82
37. Kashofer K, Winter E, Halbwedl I, Thueringer A, Kreiner M, Sauer S. et al. HPV-negative penile squamous cell carcinoma: disruptive mutations in the TP53 gene are common. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2017;30:1013-20
38. Feber A, Worth DC, Chakravarthy A, de Winter P, Shah K, Arya M. et al. CSN1 Somatic Mutations in Penile Squamous Cell Carcinoma. Cancer research. 2016;76:4720-7
39. Alves G, Heller A, Fiedler W, Campos MM, Claussen U, Ornellas AA. et al. Genetic imbalances in 26 cases of penile squamous cell carcinoma. Genes, chromosomes & cancer. 2001;31:48-53
40. Xiao S, Feng XL, Shi YH, Liu QZ, Li P. Cytogenetic abnormalities in a squamous cell carcinoma of the penis. Cancer genetics and cytogenetics. 1992;64:139-41
41. Ornellas AA, Ornellas MH, Otero L, Simoes F, Campos MM, Harab RC. et al. Karyotypic findings in two cases of moderately differentiated squamous cell carcinomas of the penis. Cancer genetics and cytogenetics. 1999;115:77-9
42. Castren K, Vahakangas K, Heikkinen E, Ranki A. Absence of p53 mutations in benign and pre-malignant male genital lesions with over-expressed p53 protein. International journal of cancer Journal international du cancer. 1998;77:674-8
Author contact
![]() Corresponding author: Prof. Dr. Robert Stoehr, Institute of Pathology, University Hospital, Friedrich-Alexander University Erlangen-Nuremberg, Krankenhausstr. 8-10, D-91054 Erlangen, Germany. Phone: +49 9131 85 43610, e-mail: robert.stoehrde
Corresponding author: Prof. Dr. Robert Stoehr, Institute of Pathology, University Hospital, Friedrich-Alexander University Erlangen-Nuremberg, Krankenhausstr. 8-10, D-91054 Erlangen, Germany. Phone: +49 9131 85 43610, e-mail: robert.stoehrde

 Global reach, higher impact
Global reach, higher impact