Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(23):4341-4345. doi:10.7150/jca.27414 This issue Cite
Research Paper
Detection of Exon 12 and 14 Mutations in Janus Kinase 2 Gene Including a Novel Mutant in V617F Negative Polycythemia Vera Patients from Pakistan
1. Department of Zoology, Division of Science and technology, University of Education, Township, Lahore, Pakistan.
2. Department of biotechnology, Kinnaird College for Women, Lahore.
3. Department of Biotechnology, University of Sialkot, Sialkot, Pakistan.
4. School of biological sciences, University of The Punjab, Lahore, Pakistan.
5. Centre for Applied Molecular Biology (CAMB), University of the Punjab, Lahore, Pakistan.
6. Department of Hematology and Oncology, Institute of Nuclear Medicine and Oncology, lahore, Pakistan
7. Cancer and Medical Genetics, CAMS-A, King Saud Bin Abdulaziz University for Health Sciences & King Abdullah International Medical Research Centre (KAIMRC), King Abdulaziz Medical City, National Guard Health Affairs, Al Ahsa, Saudi Arabia.
8. KKUH, King Saud University, Riyadh, Saudi Arabia.
Received 2018-5-22; Accepted 2018-8-28; Published 2018-10-21
Abstract
The most frequently reported genetic aberration among polycythemia vera (PV) patients is a gain of function mutation V617F in exon 14 of Janus kinase 2 (JAK2) gene. However in many investigations, V617F negative PV patients have been reported to harbor mutations in JAK 2 exon 12. We investigated 24 patients with PV (diagnosed following 2016 WHO guidelines) to detect V617F mutation through allele specific PCR. The frequency of which was found to be 19/24 (79.2 %). Later on JAK2 exon 12 and 14 was amplified by conventional PCR in V617F negative patients and subjected to sequence analysis. A total of 03 mutated sites in exon 12 were detected in only two V617F-negative patients 2/5 (40%). All three substitutions were heterozygous i.e. F537F/I found in both patients and R528R/T, which is a novel mutation. In addition, one patient 1/5 (10%) manifested amino acid substitution V617A in JAK2 exon 14. Hematological parameters of individuals harboring mutations do not vary significantly than rest of the PV patients. Previous history and 2.3 years of follow-up studies reveal 15-year survival of V617F positive patients (n=19) to be 76%, while it is 94% for wild type V617 patients (n=05). Mean TLC of the patient cohort was 17.6± 9.1 x 109/L, mean platelet count was 552± 253 x 109/L, mean hemoglobin was 16.9± 3.2 g/dl, mean corpuscular volume (MCV) was 77.2± 13.0 fl and mean corpuscular hemoglobin (MCH) was 25.6± 3.9 pg. This is the very first attempt from Pakistan to screen JAK2-exon 12 mutations in PV patients. We further aim to investigate Jak2 exon 12 mutations in larger number of PV patients to assess their clinical relevance and role in disease onset, progression and transformation.
Keywords: Polycythemia vera, Janus kinase 2, V617F, JAK2 Exon 12 mutations, JAK2 exon 14 mutations
Introduction
Chronic myeloproliferative neoplasms (MPNs) are disorders of hematopoetic stem cells. According to the WHO classification, MPNs include Polycythemia vera (PV), Essential thrombocythemia (ET), primary myelofibrosis (PMF), Chronic Myeloid leukemia (CML), chronic eosinophilic leukemia (CEL), chronic neutrophilic leukemia (CNL), mast cell disease (MCD) and unclassified myeloproliferative neoplasms (MPN-u) [1]. Among them, PV is distinguished by uncontrolled proliferation of myeloid lineage cells and red blood cells in absence of erythropoietin. Molecular diagnosis of PV was made possible after discovery of V617F a gain of function mutation in janus kinase 2 gene [2] which is now an important criterion to identify Philadelphia negative myeloproliferative neoplasms (Ph -ve MPNs) [3]. This mutation is found in majority (95%) of polycythemia vera patients, leading to constitutive tyrosine kinase phosphorylation inducing cytokine hypersensitivity and erythrocytosis [4-7]. JAK2 is part of the janus kinase family having 7 JAK homology domains (JH1-JH7). The JH2 is a pseudokinase domain which actually suppresses the JAK2 kinase activity, where exon 14 of JAK2 encodes some of the JH2 autoinhibitory site. The V617F mutation in exon 14 of JAK2 kinase causes activation of tyrosine kinase by relieving the autoinhibition of pseudokinase domain [8]. Nucleotide substitutions other than V617F in JAK2 exon 14, like C616Y, C618R and D620E have been repeatedly reported in patients with MPNs [9]. Many mutations in exon 12 of JAK2 gene involved in the pathogenesis of PV have also been described since 2007[10, 11]. It is known that functionally similar JAK2 exon 12 mutations are involved in activation of erythropoietin signaling pathways. This exon is important to be studied as almost 3% of PV patients exhibit mutations in jak2 exon 12 [12, 13]. Patients exhibiting these mutations usually have erythroid hyperplasia with hypercellular bone marrow [14]. Patnaik and Tefferi[15] in their studies recommended screening for exon 12 mutations in Jak2 gene among PV patients who are negative for V617F and manifest erythropoiesis with decreased levels of erythropoietin.
Keeping in view the importance of JAK2 exon 12 and exon 14 mutations in diagnosis, and progression of polycythemia vera, we aimed to investigate their incidence in PV patients from Pakistan. In addition, the correlation of mutations with the patient characteristics and response outcomes is also the matter of interest in this study.
Methodology
Patients and DNA isolation
Peripheral blood samples of 24 polycythemia vera patients were collected, who were diagnosed following 2016 WHO guidelines. The diagnostic criteria includes elevated levels of blood hemoglobin (HB)/ hematocrit (16.5g/dl/ 49% in males and 16g/dl/ 48% in females) along with occurrence of JAK2 V617F mutation [16]. However, due to poor socioeconomic status most of the patients were unable to bear the cost for JAK2 mutation analysis. The samples were collected from Jinnah hospital, Lahore, and Institute of nuclear medicine and oncology (INMOL) Lahore, Pakistan after obtaining informed consent from the patients. The present study was approved by the ethical review committee, University of Education, Lahore. Available clinical history was noted since the disease diagnosis and follow up till 2.3 years was done to register patients' response towards therapy. There were 16 male and 08 female individuals with median age of 57 years. Blood samples collected in EDTA coated vacutainers were stored at -20 °C till further processing. Blood samples of healthy individuals (n=20) were also collected as control group (Table 1). DNA was isolated from collected blood samples by using GeneJET Genomic DNA Purification Kit (ThermoFisher Scientific CAT# K0721) following manufacturer's instructions. This was followed by the integrity check of isolated DNA through Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene amplification [17, 18].
Clinical characteristics of Polycythemia Vera patients included in the detection of JAK2 V617F and exon 12 mutations.
| Patient characteristics | Patients n=24(%) | |
|---|---|---|
| Gender | Males | 16 (66.7) |
| Females | 08 (33.3) | |
| Median age (years) | 57 | |
| Platelet count (x109/L) | 100-450 | 02 (8.3) |
| >450 | 22 (35.6) | |
| Mean Platelet count (x109/L) | 552± 253 | |
| Total leukocyte count (x109/L) | 4-11 | 05 (91.67) |
| >11 | 19 (79) | |
| Mean TLC (x109/L) | 17.6± 9.1 | |
| Mean Hematocrit (%) | 51.4±5.2 | |
| Hemoglobin level in peripheral blood | >16g/dl | 11 (45.8) |
| Mean corpuscular Volume (MCV) | <77fl | 18 (75) |
| Mean MCV | 77.2± 13.0 fl | |
| Mean corpuscular Hemoglobin (MCH) | <27pg | 20 (83.4) |
| Mean MCH | 25.6± 3.9 pg. | |
V617F Detection
Mutation screening of exon 12 and 14 of JAK2 gene was performed by direct sequencing technique. Prior to that, Allele Specific oligonucleotide (ASO) PCR was carried out for amplification of frequently associated V617F exon 14 mutation in PV patients to screen the V617F negative individuals. Primers used for this reaction were forward mutant (5'- AGCATTTGGTTTTAAATTAGGAGTATATT-3') yielding a 203 bp product, forward control i.e. wild type V617 (5'- ATCTATAGTCATGCTGAAAGTAGGAGAAAG-3') resulting in 364 bp product, with a common reverse primer (5'- CTGAATAGTCCACAGTGTTTTCAGTTTCA-3') [19]. The accuracy of the reaction was assured by running a positive and negative control reaction with each batch.
Amplification of JAK2 exon 12 and 14 and their bidirectional sequence analysis
Among V617F-negative PV patients, amplification of JAK2 exon 12 and 14 was performed. PCR reaction was carried out in 20 μL reaction volume with 5pMol each of forward (5'-CTCCTCTTTGGAGCAATTCA-3') and reverse (5'-GAGAACTTGGGAGTTGCGATA-3') primers for exon 12 yielding 496bp product. Whereas, forward (5'-TTCTTTGAAGCAGCAAGTATGATGA-3') and reverse (5'- CTGACACCTAGCTGTGATCC -3') for exon 14 giving an amplicon of 180bp. The amplified DNA fragments were purified and sequenced by Sanger method. Standard procedure for BigDye® Terminator v3.1 Cycle Sequencing Kit from Applied Biosystems, USA was followed as per manufacturer's instructions. Same set of primers were used for sequencing purposes. Sequence analysis software Geneious 11.1.3 was used to detect mutations in sequenced reads. The reference sequence to which we aligned our patient sequences was NCBI GeneBank accession number [NG_009904.1] (Homo sapiens Janus kinase 2 (JAK2). Similar procedure was followed for samples of healthy individuals to identify any single nucleotide polymorphisms (SNPs) in the population. To ensure our results, whole procedure was repeated for sequences in which heterozygous mutations were observed noisy data was found.
Statistical analysis
Overall survival of two groups i.e. V617F positive and V617F negative patients was estimated by Kaplan-Meier method. Association of mutations with patient characteristics was studied using Fisher's exact test (IBM SPSS version 19, Chicago, IL, USA.). A p-value of less than 0.05 was considered statistically significant.
Results and Discussion
In current investigation, the hallmark gain of function mutation JAK2-V617F for Polycythemia vera, was amplified successfully in 19/24 (79.2 %) patients, whereas five individuals were negative for that. More than 20 somatic mutations in JAK2 gene have been described since year 2005, with V617F being most important and frequent one. Frequency of this mutant is estimated at approximately 97% in PV patients, which makes it an important diagnostic criterion of disease [2, 20]. Many studies indicate that patients with non-CML MPNs having wild type 617 codon in JAK2 exon 14 manifest low levels of deletions in the same exon (JAK2 Δexon14). We screened whole exon 14 to detect possible deletions or mutations in our V617F negative patients (n=5). Only one (10%) of them was observed with V617A amino acid substitution instead of V617F. However no deletions were observed in our sequence reads. As these deletion bearing transcripts are very few as compared to the total JAK2 transcripts, sensitive techniques such as RT-PCR-based fluorescent fragment analysis may be required for detailed analysis rather than bi-directional sequencing[21]. In addition use of RNA instead of DNA provides edge to detect the additional abnormalities and transcript variants as well [21]. In less than 5% of PV cases who have unmutated JAK2 V617 codon, JAK2 exon 12 mutations can explain the disease genetics [22].
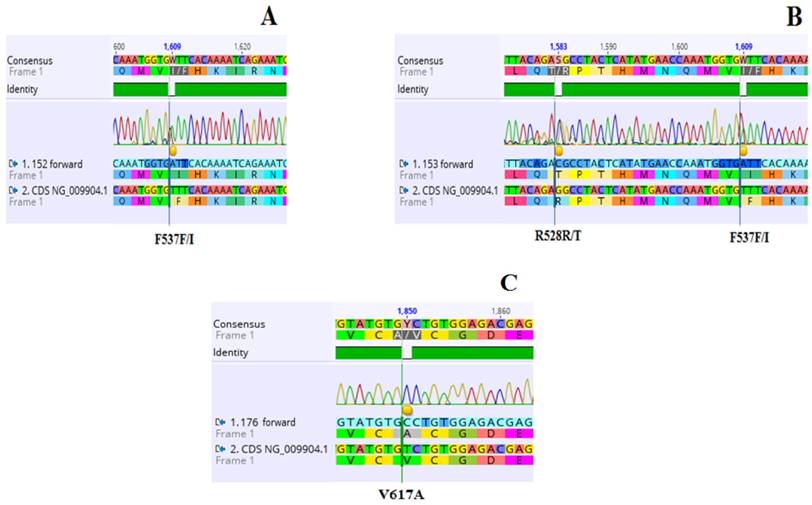
Previously almost 10 nucleotide alterations have been described in exon 12, most of which lay between codon 536-544. Among them some are replacements of 2 bp, three to six nucleotide deletions and some are duplications [12, 21, 23-25]. Many studies concluded that mutations in exon 12 are numerously found in only a small proportion of granulocytes in some patients [12, 25-28]. So owing to low allelic burden and heterogenic nature of mutations, the molecular diagnosis of exon 12 mutant individuals is complicated [29]. We examined nucleotide substitutions in exon 12 of JAK2 pseudo kinase domain. A total of 03 mutated sites in exon 12 were detected in only two V617F-negative patients 2/5 (40%) (Table 2, Figure 1). All three substitutions were heterozygous i.e. F537F/I found in both patients and R528R/T, which is a novel mutation. Screening of nucleotide alterations in JAK2 exon 12 in V617F negative PV patients with low erythropoietin levels and erythrocytosis was recommended in 2009 [15]. This practice is now part of the major criteria in diagnosis of polycythemia vera [1]. Keeping in view the wide variety of mutations in this domain, nucleotide sequencing is a facilitating tool to accomplish this [24].
Previous reports describe higher incidence upto 83% of exon 12 nucleotide alterations in unmutated V617 patients. This can be due to better analytical strategies or advanced sequencing techniques [12, 26, 30, 31]. In present study we have collected samples of PV cases from diverse health care centres and number of unmutated V617 patients is also very low (05) to declare a conclusive outcome. Due to this reason, clinical response of mutant exon 12 individuals is not found much different from that of unmutated ones. Similarly, hematological parameters of individuals harboring mutations do not vary significantly than rest of the PV patients. Fifteen year overall survival of wild type V617 PV patients (n=05) and those harboring V617F mutation (n=19) was estimated to be 94% and 76% respectively, which is not significantly different from one another. Concordantly, in reported literature the 10-15 year overall survival outcome of patients ranges from 65- 81%[32-34].
Conclusions
The gain of function mutation V617F is found in majority of our PV patients, which supports the previous suggestions. However, among patients negative for this hallmark mutation, sequence analysis of JAK2 exon 12 and 14 has revealed few underlying point mutations. This practice is a good tool for diagnosis of PV on genetic basis. This is the very first study of its kind from south asia, which will be of significant importance for physicians and clinicians of the region to treat Polycythemia vera. We aim to investigate these nucleotide substitutions in large group of PV patients to understand their role in disease biology, progression and clinical response outcomes.
Acknowledgements
The individuals diagnosed with Polycythemia vera who agreed to participate in this study are deeply acknowledged. We are also grateful to the School of biological sciences, University of The Punjab, Lahore for the lab facilities.
Clinical and biological characteristics of V617F-negative polycythemia vera cases screened for JAK2 exon 12 and 14 mutations.
| Pt. ID | Age (Yrs)/Sex | Time since dig. (Yrs) | Karyotype | Treatment | Nucleotide substitution | Exon 12 mutation detected | Exon 14 mutation detected | Previously reported by |
|---|---|---|---|---|---|---|---|---|
| PV147 | 44/M | 6.2 | 46/XY | Phlebotomy+ Hydroxyurea + Aspirin | - | - | ||
| PV 152 | 78/M | 3.3 | 46/XY | Hydroxyurea + Aspirin | 1609 T→A | F537F/I | - | [35] |
| PV153 | 6.5 | 46/XX | Phlebotomy+ Hydroxyurea + Aspirin | 1609 T→A 1583G→C | F537F/I R528R/T | - | [35] Novel | |
| PV176 | 52/M | 7.1 | 46/XY | Phlebotomy+ Hydroxyurea + Aspirin | 1850 T→C | V617A | [36] | |
| PV181 | 76/M | 4.5 | 46/XY | Hydroxyurea + Aspirin | - | - |
Yrs=Years, dig= Diagnosis, PV=Polycythemia Vera, M= Male, F= Female
Electropherogram showing three (03) heterozygous mutations in JAK2 exon 12 identified in two individuals (A and B), and exon 14 in one patient of PV (C). A: 1609 T→A substitution resulting in amino acid substitution from Phenylalanine to Isoleucine (F537F/I) along with the wild type transcripts in PV 152. B: Two mutated sites. Nucleotide change 1583G→C leading to amino acid substitution from Arginine to Threonine (R528R/T) along with wild type transcripts in PV 153. The mutant F537F/I is also identified in this patient. C: 1850 T→C substitution resulting in amino acid replacement from Valine to Alanine (V617A).NCBI gene accession number NG_009904.1 is the reference sequence.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Barbui T, Thiele J, Gisslinger H, Kvasnicka HM, Vannucchi AM, Guglielmelli P. et al. The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: document summary and in-depth discussion. Blood Cancer Journal. 2018;8:15
2. James C, Ugo V, Le Couédic J-P, Staerk J, Delhommeau F, Lacout C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144
3. Didone A, Nardinelli L, Marchiani M, Ruiz ARL, de Lima Costa AL, Lima IS. et al. Comparative study of different methodologies to detect the JAK2 V617F mutation in chronic BCR-ABL1 negative myeloproliferative neoplasms. Practical laboratory medicine. 2016;4:30-7
4. Zhao S, Zhang X, Xu Y, Feng Y, Sheng W, Cen J. et al. Impact of JAK2V617F mutation burden on disease phenotype in Chinese patients with JAK2V617F-positive polycythemia vera (PV) and essential thrombocythemia (ET). International journal of medical sciences. 2016;13:85
5. Passamonti F, Rumi E, Pietra D, Elena C, Boveri E, Arcaini L. et al. A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia. 2010;24:1574
6. Tefferi A, Rumi E, Finazzi G, Gisslinger H, Vannucchi A, Rodeghiero F. et al. Survival and prognosis among 1545 patients with contemporary polycythemia vera: an international study. Leukemia. 2013;27:1874
7. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM. et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391-405
8. Silvennoinen O, Hubbard SR. Molecular insights into regulation of JAK2 in myeloproliferative neoplasms. Blood. 2015;125:3388-92
9. Niranjan Y. Functional Characterization of the Kinase and Pseudokinase Domains in the Janus Tyrosine Kinase (JAK) 2. 2014.
10. Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118:1723-35
11. Schnittger S, Bacher U, Haferlach C, Geer T, Müller P, Mittermüller J. et al. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. haematologica. 2009;94:414-8
12. Pietra D, Li S, Brisci A, Passamonti F, Rumi E, Theocharides A. et al. Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)-negative myeloproliferative disorders. Blood. 2008;111:1686-9
13. Leszczynska A, Grzenkowicz-Wydra J, Chmielewska-Gorycka L, Bieniaszewska M, Hellmann A. Detection of JAK2 Exon 12 Mutations in JAK2 V617F-Negative Polycythemia Vera Patients by Cloning Technique. Acta haematologica. 2016;136:123-8
14. Lakey MA, Pardanani A, Hoyer JD, Nguyen PL, Lasho TL, Tefferi A. et al. Bone Marrow Morphologic Features in Polycythemia Vera With JAK2 Exon 12 Mutations. American Journal of Clinical Pathology. 2010;133:942-8
15. Patnaik M, Tefferi A. The complete evaluation of erythrocytosis: congenital and acquired. Leukemia. 2009;23:834
16. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM. et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016 blood-2016-03-643544
17. Akram AM, Iqbal Z, Akhtar T, Khalid AM, Sabar MF, Qazi MH. et al. Presence of novel compound BCR-ABL mutations in late chronic and advanced phase imatinib sensitive CML patients indicates their possible role in CML progression. Cancer biology & therapy. 2017;18:214-21
18. Asad S, Ijaz B, Ahmad W, Kausar H, Sarwar MT, Gull S. et al. Development of persistent HCV genotype 3a infection cell culture model in huh-7 cell. Virology journal. 2012;9:11
19. Bang S-M, Ahn JY, Park J, Yoo SJ, Park SH, Nam EM. et al. Diagnostic Usefulness of the Janus Kinase 2 Mutation in non BCR/ABL Myeloproliferative Disorders. The Korean Journal of Internal Medicine. 2006;21:219-24
20. Barbui T, Thiele J, Vannucchi AM, Tefferi A. Rationale for revision and proposed changes of the WHO diagnostic criteria for polycythemia vera, essential thrombocythemia and primary myelofibrosis. Blood Cancer J. 2015;5:e337
21. Ma W, Kantarjian H, Zhang X, Wang X, Zhang Z, Yeh C-H. et al. JAK2 Exon 14 Deletion in Patients with Chronic Myeloproliferative Neoplasms. PLoS ONE. 2010;5:e12165
22. Scott LM. The JAK2 exon 12 mutations: a comprehensive review. Am J Hematol. 2011;86:668-76
23. Kouroupi E, Zoi K, Parquet N, Zoi C, Kiladjian JJ, Grigoraki V. et al. Mutations in exon 12 of JAK2 are mainly found in JAK2 V617F-negative polycythaemia vera patients. British journal of haematology. 2008;142:676-9
24. Bernardi M, Ruggeri M, Albiero E, Madeo D, Rodeghiero F. Isolated erythrocytosis in V617F negative patients with JAK2 exon 12 mutations: report of a new mutation. American journal of hematology. 2009;84:258-60
25. Li S, Kralovics R, De Libero G, Theocharides A, Gisslinger H, Skoda RC. Clonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2-V617F mutations. Blood. 2008;111:3863-6
26. Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR. et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. New England Journal of Medicine. 2007;356:459-68
27. Percy MJ, Scott LM, Erber WN, Harrison CN, Reilly JT, Jones FG. et al. The frequency of JAK2 exon 12 mutations in idiopathic erythrocytosis patients with low serum erythropoietin levels. Haematologica. 2007;92:1607-14
28. Butcher C, Hahn U, To L, Gecz J, Wilkins EJ, Scott H. et al. Two novel JAK2 exon 12 mutations in JAK2V617F-negative polycythaemia vera patients. Leukemia. 2008;22:870
29. Laughlin TS, Moliterno AR, Stein BL, Rothberg PG. Detection of Exon 12 Mutations in the JAK2 Gene: Enhanced Analytical Sensitivity Using Clamped PCR and Nucleotide Sequencing. The Journal of molecular diagnostics: JMD. 2010;12:278-82
30. Pardanani A, Lasho T, Finke C, Hanson CA, Tefferi A. Prevalence and clinicopathologic correlates of JAK2 exon 12 mutations in JAK2V617F-negative polycythemia vera. Leukemia. 2007;21:1960
31. Passamonti F, Elena C, Schnittger S, Skoda RC, Green AR, Girodon F. et al. Molecular and clinical features of the myeloproliferative neoplasm associated with JAK2 exon 12 mutations. Blood. 2011;117:2813-6
32. Alvarez-Larrán A, Pereira A, Cervantes F, Arellano-Rodrigo E, Hernández-Boluda J-C, Ferrer-Marín F. et al. Assessment and prognostic value of the European LeukemiaNet criteria for clinicohematologic response, resistance, and intolerance to hydroxyurea in polycythemia vera. Blood. 2012;119:1363-9
33. Passamonti F, Rumi E, Pungolino E, Malabarba L, Bertazzoni P, Valentini M. et al. Life expectancy and prognostic factors for survival in patients with polycythemia vera and essential thrombocythemia. The American journal of medicine. 2004;117:755-61
34. Crisà E, Venturino E, Passera R, Prina M, Schinco P, Borchiellini A. et al. A retrospective study on 226 polycythemia vera patients: impact of median hematocrit value on clinical outcomes and survival improvement with anti-thrombotic prophylaxis and non-alkylating drugs. Annals of hematology. 2010;89:691-9
35. Sayyah J, Magis A, Ostrov DA, Allan RW, Braylan RC, Sayeski PP. Z3, a novel Jak2 tyrosine kinase small-molecule inhibitor that suppresses Jak2-mediated pathologic cell growth. Molecular cancer therapeutics. 2008;7:2308-18
36. Albitar M. Jak2 mutations. Google Patents. 2009
Author contact
![]() Corresponding author: afiamakramedu.pk, +9242-323 6009330
Corresponding author: afiamakramedu.pk, +9242-323 6009330

 Global reach, higher impact
Global reach, higher impact