3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(23):4355-4362. doi:10.7150/jca.26902 This issue Cite
Research Paper
Prognostic Values of LAPTM4B-35 in Human Cancer: A Meta-analysis
1. Department of Breast Surgery, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China;
2. Department of Oncology, The Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, China;
3. School of Life Science and Technology, Xi'an Jiaotong University, Xi'an 710049, China;
4. Department of Biochemistry and Molecular Medicine, The George Washington University Medical School, Washington, DC, USA.
*LH Z, C D and T T contributed equally to this work.
Received 2018-4-26; Accepted 2018-8-15; Published 2018-10-21
Abstract
Background: Lysosome-associated protein transmembrane-4β-35(LAPTM4B-35) has been observed overexpressed in multiple malignant tumors. However, the prognostic value of LAPTM4B-35 remains controversial. Therefore, we conducted a meta-analysis to evaluate the prognostic value of LAPTM4B-35 in human cancers.
Methods: The relevant publications were obtained by systematically searching the PubMed, Web of Science, Embase, Wanfang, and China National Knowledge Infrastructure (CNKI) databases. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated for the prognosis value of LAPTM4B-35 for cancer patient.
Results: Our result suggest that LAPTM4B-35 overexpression is significantly associated with poor overall survival (OS) (HR = 2.49, 95% CI = 1.87-3.32, p < 0.001), disease-free survival (DFS) (HR = 2.43, 95% CI = 1.35-4.35, p = 0.003), and progression-free survival (PFS) (HR = 4.12, 95% CI = 2.30-7.37, p < 0.001). Moreover, subgroup analysis revealed significant association with poor OS in lung (HR = 2.05, 95% CI = 1.37-3.06, p < 0.001), gastric carcinoma (HR = 1.88, 95% CI = 1.01-3.50, p < 0.047) and ovarian cancer (HR = 4.94, 95% CI = 1.44-16.94, p = 0.011).
Conclusion: LAPTM4B-35 may be a novel predictive biomarker and a potential target for treatment.
Keywords: LAPTM4B-35, cancer, prognosis, meta-analysis
Introduction
Cancer is one of the leading causes of death in the world. According to the most recent global cancer statistics, in 2017, there were 1,688,780 newly diagnosed cancer cases and 600,920 cancer-related deaths in the United States[1]. It is generally believed that highly specific biomarkers could be helpful to cancer diagnosis. However, many biomarkers are not specific enough for cancers. Therefore, it is important to find new biomarkers for cancer diagnosis.
As a member of the mammalian 4-tetra-transmembrane spanning protein superfamily, lysosome-associated protein transmembrane-4β-35 (LAPTM4B-35) was firstly found in hepatocellular carcinoma[2]. The LAPTM4B-35 is rich in proline at the N- and C-termini while LAPTM4B-35consisting of seven exons and six introns which locate on chromosome 8g22[3]. Previous studies have shown that LAPTM4B-35 is upregulated in many types of cancer, including breast [4, 5], lung [6, 7], gastric [8], pancreatic [9], colorectal [10], ovarian [11,12], and hepatocellular cancer[13]. LAPTM4B-35 is a novel oncoprotein that decreases apoptosis, promotes progression and metastasis, and induces tumor angiogenesis [14]. Moreover, it can induce multidrug resistance by activating PI3K/AKT signaling pathway [15, 16].
Multiple studies have shown that LAPTM4B-35 expression is associated with cancer prognosis [4, 6, 17]. However, most studies just focused on the association between LAPTM4B-35 and single cancer. In this article, we conducted a meta-analysis to assess the association between LAPTM4B-35 and multiple cancer.
Materials and Methods
Identification of eligible studies
Eligible studies which were published before March 2018, were selected from the PubMed, Web of Science, Embase, Wanfang, and China National Knowledge Infrastructure (CNKI) databases. The search phrases “cancer or carcinoma or tumor or neoplasm”, “LAPTM4B or Lysosome-associated protein transmembrane-4β”, “LAPTM4B-35 or Lysosome-associated protein transmembrane-4β-35”, and “prognosis or survival” were used. All references were screened to ensure relevant studies were included.
Inclusion and exclusion criteria
The included studies followed the criteria: (1) the source and size of the study population were clearly described, (2) a clear pathological diagnosis of cancer was mentioned, (3) the association with overall survival (OS), disease-free survival (DFS) or progression-free survival (PFS) in cancer patient were declared, (4) hazard ratios (HRs) with 95% confidence intervals (CIs) or survival curves were contained in the study. The excluded studies were classified into categories including: (1) duplicate studies, (2) case reports, reviews, letters, or meta-analyses, (3) no available data, or (4) the experiment was done with organisms other than humans.
Data extraction
Two investigators (Linghui Zhou and Cong Dai) independent performed data extraction. Discussions were held with senior investigators until a consensus was reached to eliminate differences. For each study, the following information was extracted: first author, year of publication, the source of patients, study size, types of specimens, method of detection, types of cancer, LAPTM4B-35 expression, median follow-up, prognostic outcome, analysis method, and HRs with 95% CIs.
Quality assessment
We adopt the Newcastle-Ottawa Scale (NOS) scoring criteria to evaluate the quality of each study[18]. A study with a score of 0-5 was considered a low-quality study and that with a score of 6-9 was considered a high-quality study. Two investigators (Linghui Zhou and Cong Dai) performed independent quality assessments.
Statistical methods
The association between LAPTM4B-35 and patient prognosis was assessed based on pooled HRs and the corresponding 95% CIs. The HR and corresponding 95% CIs were acquired directly from the study or calculated based on Kaplan-Meier survival curves (by Engauge Digitizer version 4.1) [19]. The chi-square-based Q statistic was conducted to evaluate heterogeneity among the studies. In cases where p < 0.10 or I² > 50, the random effects model was used to assess the pooled HRs[20].Otherwise, the fixed effects model was used. Subgroup analyses were conducted based on the cancer type and analysis method[21]. We used Begg's test, Egger's test, and Begg's funnel plot to evaluate publication bias[22, 23]. If p >0.05 and the funnel plots were symmetrical, it means no publication bias. Additionally, sensitivity analysis was calculated by omitting each study in turn. All the data analysis was performed by Stata 12.0 (Stata Corporation, College Station, TX, USA).
Results
Study Characteristics
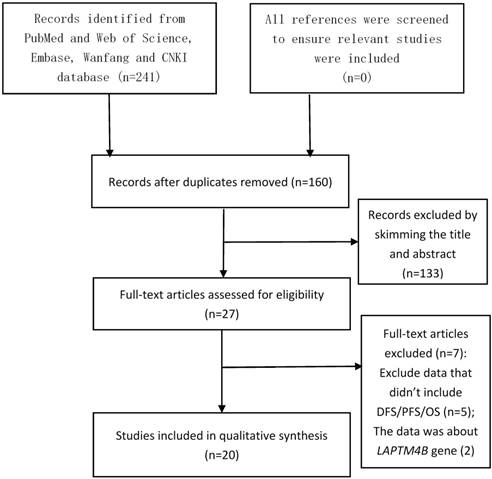
After searching the PubMed, Web of Science, Embase, Wanfang, and China National Knowledge Infrastructure (CNKI) databases, we obtained 160 different articles without duplication. As shown in Figure 1, after reviewing the title and abstract, 133 studies were excluded. The full text of the remaining 27 articles was analyzed, which excluded another 7 articles. After these analyses, there were 20 eligible studies remained[4-14, 17, 24-31].
The main characteristics of the 20 studies are shown in Table 1. All the studies evaluated the association between LAPTM4B-35 and OS in cancer patients. In addition, 7 studies assessed the relationship between LAPTM4B-35 and PFS, and 8 studies assessed the link between LAPTM4B-35 and DFS. Together, the studies assessed 3,274 cancer patients from China, USA, and Japan. The number of patients per study ranged from 39 to 652. The abundance of LAPTM4B-35 was quantified by immunohistochemistry (IHC) staining. The 20 studies analyzed many types of cancer, including pancreatic, ovarian, lung, hepatocellular, glioblastoma, gastric, endometrial, colorectal, cervical, breast, and bladder cancer.
Association between LAPTM4B-35 and OS in cancer patients
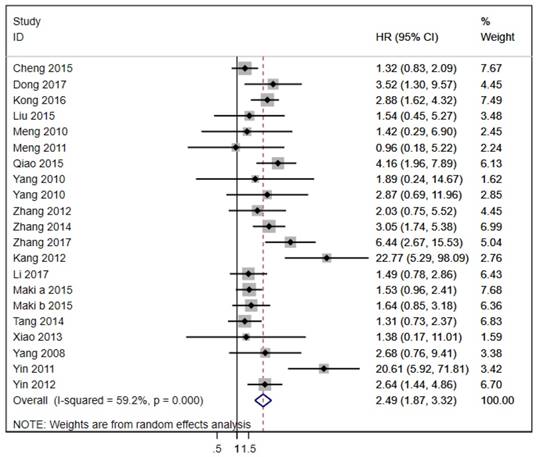
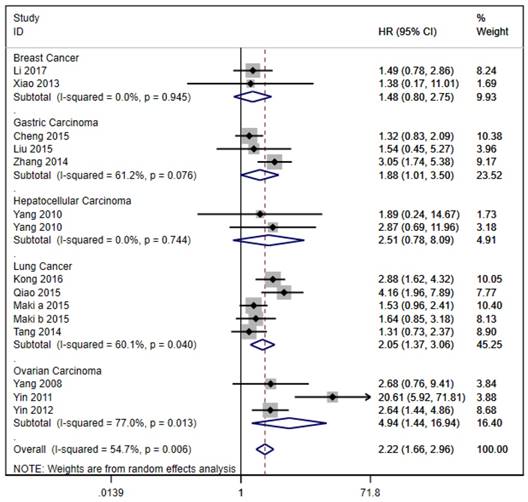
The 20 studies assessed the association between LAPTM4B-35 and OS in cancer patients. Due to significant heterogeneity (p < 0.001, I² = 59.2%), we used the random effects model to pool the HRs (Table 2). As shown in Figure 2, LAPTM4B-35 overexpression was significantly associated with poorer OS (HR = 2.49, 95% CI = 1.87-3.32, p < 0.001). Moreover, subgroup analyses by type of cancer revealed a significant association with poorer OS in patients with lung (HR = 2.05, 95% CI = 1.37-3.06, p < 0.001), gastric carcinoma (HR = 1.88, 95% CI = 1.01-3.50, p < 0.047) and ovarian cancer (HR = 4.94, 95% CI = 1.44-16.94, p = 0.011) (Figure 3).
The flow diagram of the meta-analysis. CNKI: China National Knowledge Infrastructure.
Association between LAPTM4B-35 and DFS PFS in cancer patients
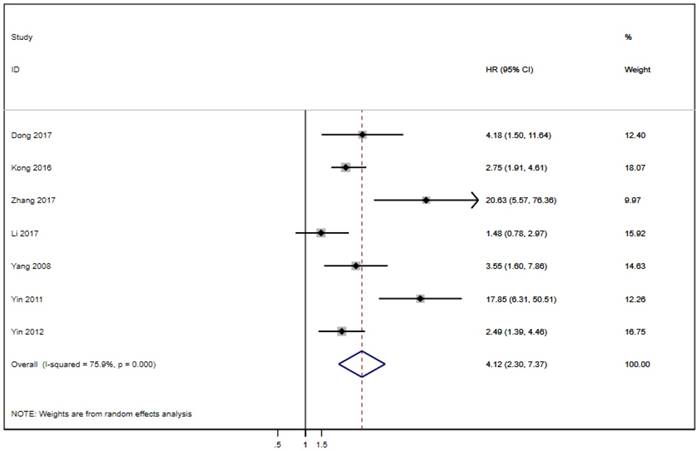
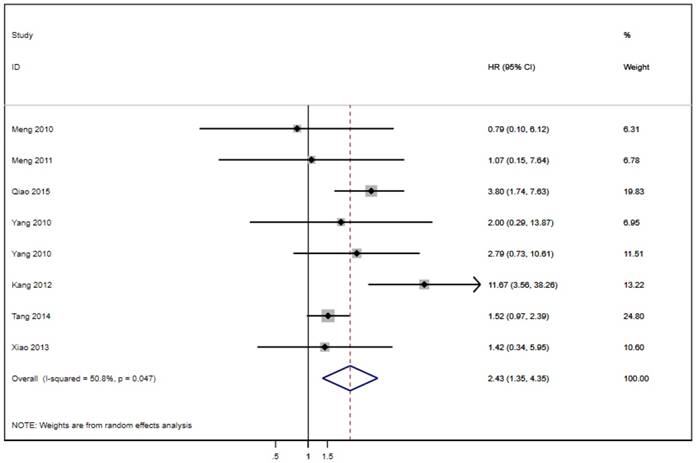
Seven of the twenty studies assessed the link between LAPTM4B-35 and PFS in cancer patients. Because of significant heterogeneity (p < 0.001, I² = 75.9%), the random effects model was employed to pool the HRs (Table 2). The results suggest that LAPTM4B-35 overexpression was significantly associated with poor PFS (HR = 4.12, 95% CI = 2.30-7.37, p < 0.001) (Figure 4). In addition, eight studies evaluated the association between LAPTM4B-35 and DFS in cancer patients. Heterogeneity analysis also showed significant heterogeneity (p = 0.047, I² = 50.8%) (Table 2), so the random-effects model was used to pool the HRs. The pooled data indicated that LAPTM4B-35 overexpression was significantly related to poor DFS (HR = 2.43, 95% CI = 1.35-4.35, p = 0.003) (Figure 5).
Sensitivity analyses
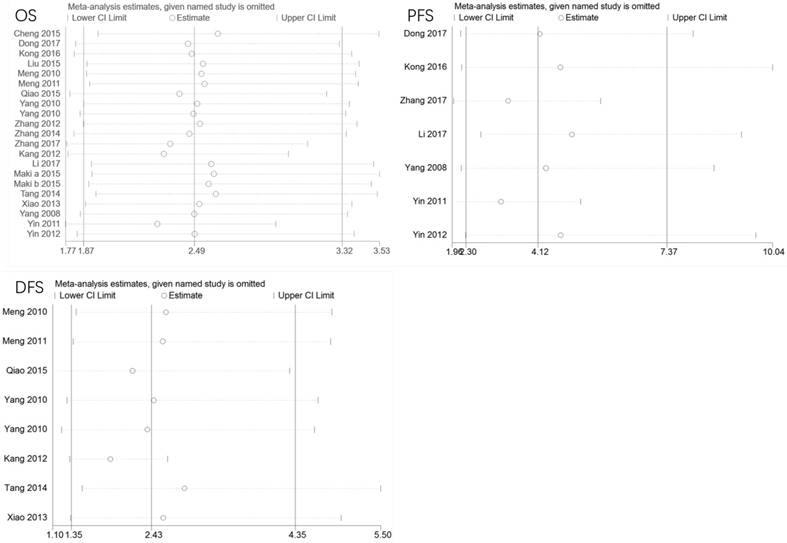
To evaluate the stability of the meta-analysis, we conducted sensitivity analyses by removing each study in turn. As shown in Figure 6, the meta-analysis is reliable and no individual study skewed the pooled HRs values of OS, PFS, or DFS.
Publication bias
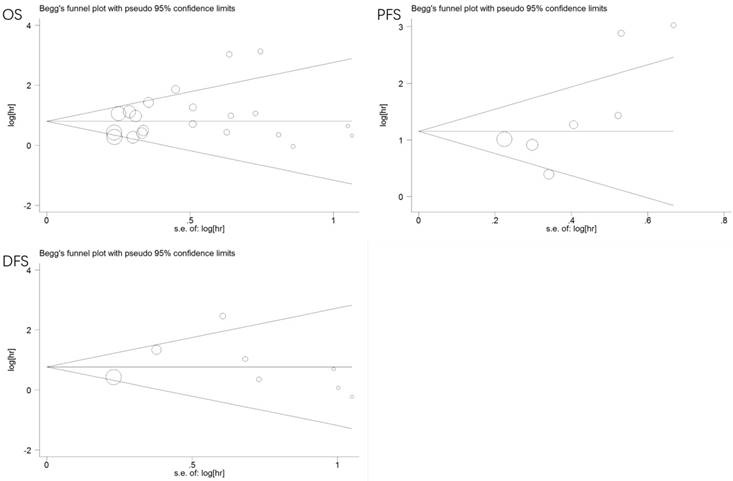
We used Begg's and Egger's tests to evaluate publication bias of the eligible studies. No publication bias was observed for OS (p = 0.381, 0.159), PFS (p = 0.072, 0.069), or DFS (p = 0.266, 0.370). Additionally, the funnel plots were symmetrical (Figure 7).
Discussion
Many studies have shown that the levels of LAPTM4B-35 are significantly upregulated in multiple cancer. Moreover, a study conducted by Li et al. suggested that the overexpression of LAPTM4B-35 could induce multidrug resistance, which could be eliminated by knockdown of LAPTM4B-35 expression [16]. These findings indicate that LAPTM4B-35 is a potential predictive biomarker for many types of cancers, as well as a promising target for knockdown by iRNA to decrease drug resistance.
Our meta-analysis, with 20 studies and 3,274 cancer patients, suggests that LAPTM4B-35 overexpression is significantly associated with poor OS, DFS, and PFS. Moreover, subgroup analysis showed a significant association with poor OS in lung, gastric and ovarian cancer. There were an insufficient number of studies in the meta-analysis to determine if there is a significant association between LAPTM4B-35 and other types of cancer, so the results should be interpreted carefully. Subgroup analysis suggests that the analysis method may lead to significant heterogeneity in the meta-analysis. The results of the sensitivity analysis prove that the meta-analysis is reliable. No publication bias was detected by either the Begg's or the Egger's test.
Characteristics of the studies included for the meta-analysis.
| First author | Year | Patient source | Number of patient | Method | Specimen | Tumor types | expression (%) | Median(range) Follow-up(month) | Outcome | M/ U | HR(95%CI) | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang | 2017 | China | 102 | IHC | tissue | BDC | 45.1 | — | OS | M | 6.439(2.67-15.531) | 6 |
| Zhang | 2017 | China | 102 | IHC | tissue | BDC | 45.1 | — | PFS | M | 20.631(5.574-76.36) | 6 |
| Zhang | 2014 | China | 652 | IHC | tissue | GC | 72.23 | — | OS | M | 3.055(1.739-5.382) | 6 |
| Zhang | 2012 | China | 98 | IHC | tissue | PC | 78.6 | 15 ( 3 - 65 ) | OS | U | 2.03(0.75-5352) | 6 |
| Yin | 2011 | China | 113 | IHC | tissue | OC | 70.8 | — | OS | M | 20.611(5.916-71.808) | 8 |
| Yin | 2012 | China | 102 | IHC | tissue | OC | 75.49 | — | OS | M | 2.644(1.439-4.858) | 8 |
| Yin | 2011 | China | 113 | IHC | tissue | OC | 70.8 | — | PFS | M | 17.852(6.31-50.52) | 8 |
| Yin | 2012 | China | 102 | IHC | tissue | OC | 75.49 | — | PFS | M | 2.489(1.388-4.461) | 8 |
| Yang | 2010 | China | 35 | IHC | tissue | HC | 57.14 | 16 ( 4 - 36 ) | DFS | M | 2(0.29-13.87) | 6 |
| Yang | 2010 | China | 71 | IHC | tissue | HC | 71.83 | 36 ( 3 - 93 ) | DFS | U | 2.79(0.73-10.61) | 6 |
| Yang | 2010 | China | 35 | IHC | tissue | HC | 57.14 | 16 ( 4 - 36 ) | OS | M | 1.89(0.24-14.67) | 6 |
| Yang | 2010 | China | 71 | IHC | tissue | HC | 71.83 | 36 ( 3 - 93 ) | OS | U | 2.87(0.69-11.96) | 6 |
| Yang | 2008 | China | 85 | IHC | tissue | OC | 63.53 | — | OS | U | 2.68(0.76-9.41) | 6 |
| Yang | 2008 | China | 85 | IHC | tissue | OC | 63.53 | — | PFS | U | 3.55(1.6-7.86) | 6 |
| Xiao | 2013 | China | 194 | IHC | tissue | BC | 74.74 | 62 (9 - 74 ) | DFS | M | 1.42(0.34-5.95) | 8 |
| Xiao | 2013 | China | 194 | IHC | tissue | BC | 74.74 | 61 (9 - 74 ) | OS | M | 1.38(0.17-11.01) | 8 |
| Tang | 2014 | China | 186 | IHC | tissue | LC | 69.35 | — | DFS | M | 1.52(0.97-2.39) | 8 |
| Tang | 2014 | China | 186 | IHC | tissue | LC | 69.35 | — | OS | M | 1.31(0.73-2.37) | 8 |
| Qiao | 2015 | China | 88 | IHC | tissue | LC | 50 | 66 | DFS | U | 3.802(1.737-7.632) | 8 |
| Qiao | 2015 | China | 88 | IHC | tissue | LC | 50 | 66 | OS | U | 4.16(1.964-7.893) | 8 |
| Meng | 2011 | China | 113 | IHC | tissue | CC | 72.57 | 64 ( 14-78) | DFS | M | 1.07(0.15-7.64) | 6 |
| Meng | 2011 | China | 113 | IHC | tissue | CC | 72.57 | 64 ( 14-78) | OS | M | 0.96(0.18-5.22) | 6 |
| Meng | 2010 | China | 165 | IHC | tissue | EC | 70.91 | 70 ( 9-78) | DFS | M | 0.79(0.1-6.12) | 9 |
| Meng | 2010 | China | 165 | IHC | tissue | EC | 70.91 | 70 ( 9-78) | OS | M | 1.42(0.29-6.9) | 9 |
| Maki b | 2015 | America | 123 | IHC | tissue | LC | 37.4 | — | OS | M | 1.64(0.85-3.18) | 6 |
| Maki a | 2015 | America | 245 | IHC | tissue | LC | 47.4 | — | OS | M | 1.53(0.96-2.41) | 6 |
| Liu | 2015 | China | 148 | IHC | tissue | GC | 89.19 | — | OS | M | 1.54(0.45-5.27) | 6 |
| Li | 2017 | China | 110 | IHC | tissue | BC | 56.36 | 49 (10-67) | OS | M | 1.49(0.78-2.86) | 9 |
| Li | 2017 | China | 110 | IHC | tissue | BC | 56.36 | 50 (10-67) | PFS | M | 1.48(0.78-2.97) | 9 |
| Kong | 2016 | China | 107 | IHC | tissue | LC | 50.47 | 64.5 | OS | M | 2.879(1.621-4.318) | 7 |
| Kong | 2016 | China | 107 | IHC | tissue | LC | 50.47 | 64.5 | PFS | M | 2.75(1.911-4.607) | 7 |
| Kang | 2012 | China | 136 | IHC | tissue | CRC | 21.15 | — | DFS | M | 11.674(3.562-38.263) | 6 |
| Kang | 2012 | China | 136 | IHC | tissue | CRC | 37.38 | — | OS | M | 22.774(5.287-98.091) | 6 |
| Dong | 2017 | Japan | 39 | IHC | tissue | G | 56.41 | 12.5 | OS | U | 3.52(1.3-9.57) | 8 |
| Dong | 2017 | Japan | 39 | IHC | tissue | G | 56.41 | 5.13 | PFS | U | 4.18(1.5-11.64) | 8 |
| Cheng | 2015 | China | 240 | IHC | tissue | GC | 71.52 | 26.2 (2.4-119.0) | OS | U | 1.32(0.83-2.09) | 8 |
Abbreviations: CI :confidence interval; BDC: Bladder Cancer; BC: Breast Cancer; CC: Cervical Carcinoma; CRC: Colorectal Carcinoma; EC: Endometrial Carcinoma; GC: Gastric Carcinoma; G: Glioblastoma; HC: Hepatocellular Carcinoma; LC: Lung Cancer; OC: Ovarian Carcinoma; PC: Pancreatic Carcinoma; NOS: Newcastle-Ottawa Scale; M: multivariate; U: univariate; OS: overall survival, DFS: disease-free survival; PFS: progression-free-survival; IHC: immunohistochemistry. a, b: different studies in the same article. expression (%): Percentage of patients with high expression LAPTM4B-35 in total patients. -: data is not available.
Main meta-analysis results
| Analysis | No of studies | Model | HR (95% CI) | P-value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I² (%) | P-value | |||||
| OS | 20 | Random | 2.49(1.87-3.32) | <0.001 | 59.20% | <0.001 |
| Tumor types | ||||||
| LC | 5 | Random | 2.05(1.37-3.06) | <0.001 | 60.10% | 0.04 |
| OC | 3 | Random | 4.94(1.44-16.94) | 0.011 | 77.00% | 0.013 |
| GC | 3 | Random | 1.88(1.01-3.50) | 0.047 | 61.2% | 0.076 |
| HR estimate | ||||||
| M | 14 | Random | 2.53(1.75-3.65) | <0.001 | 64.90% | <0.001 |
| U | 6 | Fixed | 2.11(1.53-2.89) | <0.001 | 44.20% | 0.111 |
| DFS | 8 | Random | 2.43(1.35-4.35) | 0.003 | 50.80% | 0.047 |
| PFS | 7 | Random | 4.12(2.30-7.37) | <0.001 | 75.90% | <0.001 |
Abbreviations: CI, confidence interval; LC: Lung Cancer; OC: Ovarian Carcinoma; HR, hazard ratio; M: multivariate; U: univariate; OS, overall survival; DFS: disease-free survival; PFS: progression-free-survival. GC: Gastric Carcinoma
A meta-analysis of the association between LAPTM4B-35 expression and cancer prognosis has not been previously reported other than two meta-analyses of the LAPTM4B polymorphism [32, 33]. Therefore, we conducted this meta-analysis to assess the prognostic and predictive values of LAPTM4B-35 in many types of human cancer.
Although our meta-analysis provided strong evidence that LAPTM4B-35 overexpression was significantly associated with poor prognosis, there were some limitations. First, some of the studies used did not include the survival data. Thus, we had to obtain the HRs indirectly using the Kaplan-Meier survival curves, which could influence the accuracy of data. In addition, significant heterogeneity was observed in our study and subgroup analysis suggests that the analysis method may contribute to the significant heterogeneity in the meta-analysis. Due to lack of data about age, sex, diet, living environment, smoking, family inheritance, we could not find the exact source of heterogeneity. In addition, we could not verify the conclusions about cancer types other than lung and ovarian cancer due to limited numbers of studies. Therefore, more studies on the prognosis of patients with other cancers are indispensable to confirm our conclusions.
Forest plot of HR for the association of LAPTM4B-35 overexpression and OS. CI: confidence interval; HR: hazard ratio; OS: overall survival.
Stratified analysis based on cancer types for the association between LAPTM4B-35 overexpression and OS. CI: confidence interval; HR: hazard ratio; OS: overall survival.
Forest plot of HR for the association of LAPTM4B-35 overexpression and PFS. CI: confidence interval; HR: hazard ratio; OS: overall survival.
Forest plot of HR for the association of LAPTM4B-35 overexpression and DFS. CI: confidence interval; HR: hazard ratio; OS: overall survival.
Conclusion
Overall, our study showed that the overexpression of LAPTM4B-35 is significantly associated with poor prognosis. As such, LAPTM4B-35 may be a new prognostic marker for many types of cancer. Due to the limitations, our results should be interpreted with caution. And further clinical studies on the prognosis of patients with many types of cancers are necessary to verify our conclusion.
Sensitivity analysis of pooled HRs on the association between LAPTM4B-35 expression and OS/PFS/DFS. CI: confidence interval; HR: hazard ratio; OS: overall survival. DFS: disease-free survival; PFS: progression-free survival.
Funnel plots of publication bias for all the included studies reported with OS/PFS/DFS. CI: confidence interval; HR: hazard ratio; OS: overall survival. DFS: disease-free survival; PFS: progression-free survival.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 81471670); the International Cooperative Project of Shaanxi province, China (No. 2016KW-008) and the Key research and development plan, Shaanxi Province, China (2017ZDXM-SF-066).
Author Contributions
L.-H. Z., C.D. and T.T. are contributed equally to this work. Z.-J.D. designed the study. L.-H.Z., C.D. and T.T. wrote the main manuscript text, M.W., S.L and Y.-J.D. performed figures and tables, P.X., Q.H., Y.W., T.-L.Y. and W.-G.Z. reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
2. Shao GZ, Zhou RL, Zhang QY, Zhang Y, Liu JJ, Rui JA. et al. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene. 2003;22:5060-9
3. Liu X, Zhou R, Zhang Q, Zhang Y, Shao G, Jin Y. et al. [Identification and characterization of LAPTM4B encoded by a human hepatocellular carcinoma-associated novel gene]. Beijing da xue xue bao Yi xue ban = Journal of Peking University Health sciences. 2003;35:340-7
4. Li S, Wang L, Meng Y, Chang Y, Xu J, Zhang Q. Increased levels of LAPTM4B, VEGF and survivin are correlated with tumor progression and poor prognosis in breast cancer patients. Oncotarget. 2017;8:41282-93
5. Xiao M, Jia S, Wang H, Wang J, Huang Y, Li Z. Overexpression of LAPTM4B: an independent prognostic marker in breast cancer. Journal of cancer research and clinical oncology. 2013;139:661-7
6. Kong F, Gao F, Chen J, Sun Y, Zhang Y, Liu H. et al. Overexpressed LAPTM4B-35 is a risk factor for cancer recurrence and poor prognosis in non-small-cell lung cancer. Oncotarget. 2016;7:56193-9
7. Maki Y, Fujimoto J, Lang W, Xu L, Behrens C, Wistuba II. et al. LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells. Scientific reports. 2015;5:13846
8. Liu L, Xu X, Jing L, Zhou G, Cao Z, Han Y. et al. Lysosomal-associated protein transmembrane 4 Beta-35 overexpression is a novel independent prognostic marker for gastric carcinoma. PloS one. 2015;10:e0118026
9. Zhang G, Liang Y, Huang Y, Chen Y, Zhou R. Elevated lysosome-associated protein transmembrane-4beta-35 is an independent prognostic marker in pancreatic carcinoma. The Journal of international medical research. 2012;40:1275-83
10. Kang Y, Yin M, Jiang W, Zhang H, Xia B, Xue Y. et al. Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. American journal of surgery. 2012;204:677-83
11. Yin M, Lou C, Zhang W, Meng F, Zhang H, Ning X. et al. LAPTM4B overexpression is a novel independent prognostic marker for metastatic ovarian tumors. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2012;22:54-62
12. Yin M, Li C, Li X, Lou G, Miao B, Liu X. et al. Over-expression of LAPTM4B is associated with poor prognosis and chemotherapy resistance in stages III and IV epithelial ovarian cancer. Journal of surgical oncology. 2011;104:29-36
13. Yang H, Lin M, Xiong F, Yang Y, Nie X, McNutt MA. et al. Combined lysosomal protein transmembrane 4 beta-35 and argininosuccinate synthetase expression predicts clinical outcome in hepatocellular carcinoma patients. Surgery today. 2011;41:810-7
14. Tang H, Tian H, Yue W, Li L, Li S, Gao C. et al. Overexpression of LAPTM4B is correlated with tumor angiogenesis and poor prognosis in non-small cell lung cancer. Medical oncology (Northwood, London, England). 2014;31:974
15. Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y. et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nature medicine. 2010;16:214-8
16. Li L, Wei XH, Pan YP, Li HC, Yang H, He QH. et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene. 2010;29:5785-95
17. Dong X, Tamura K, Kobayashi D, Ando N, Sumita K, Maehara T. LAPTM4B-35 is a novel prognostic factor for glioblastoma. Journal of neuro-oncology. 2017;132:295-303
18. Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F. et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. Journal of evidence-based medicine. 2015;8:2-10
19. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in medicine. 1998;17:2815-34
20. Dai C, Wang M, Lu J, Dai Z, Lin S, Yang P. et al. Prognostic and predictive values of PD-L1 expression in patients with digestive system cancer: a meta-analysis. OncoTargets and therapy. 2017;10:3625-34
21. Deng Y, Ge P, Tian T, Dai C, Wang M, Lin S. et al. Prognostic value of flotillins (flotillin-1 and flotillin-2) in human cancers: A meta-analysis. Clinica chimica acta; international journal of clinical chemistry. 2018;481:90-8
22. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315:629-34
23. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
24. Lei Z. Study on LAPTM4B expression, clinical pathology, and prognostic significance in bladder cancer. Capital medical university: master's thesis. 2017
25. Qiao M, Hu G. Lysosome-associated protein transmembrane-4beta-35 is a novel independent prognostic factor in small cell lung cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:7493-9
26. Cheng X, Zheng Z, Bu Z, Wu X, Zhang L, Xing X. et al. LAPTM4B-35, a cancer-related gene, is associated with poor prognosis in TNM stages I-III gastric cancer patients. PloS one. 2015;10:e0121559
27. Zhang H, Tian B, Yu H, Yao H, Gao Z. LAPTM4B-35 protein as a potential therapeutic target in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:12737-42
28. Yang H, Xiong FX, Lin M, Yang Y, Nie X, Zhou RL. LAPTM4B-35 overexpression is a risk factor for tumor recurrence and poor prognosis in hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2010;136:275-81
29. Yang H, Xiong F, Qi R, Liu Z, Lin M, Rui J. et al. LAPTM4B-35 is a novel prognostic factor of hepatocellular carcinoma. Journal of surgical oncology. 2010;101:363-9
30. Meng FL, Yin MZ, Song HT, Yang H, Lou G, Zhou RL. LAPTM4B-35 overexpression is an independent prognostic marker in endometrial carcinoma. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2010;20:745-50
31. Meng F, Luo C, Hu Y, Yin M, Lin M, Lou G. et al. Overexpression of LAPTM4B-35 in cervical carcinoma: a clinicopathologic study. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2010;29:587-93
32. Xia LZ, Yin ZH, Ren YW, Shen L, Wu W, Li XL. et al. The relationship between LAPTM4B polymorphisms and cancer risk in Chinese Han population: a meta-analysis. SpringerPlus. 2015;4:179
33. Mo C, Lu Y, Deng Y, Wang J, Xie L, Li T. et al. LAPTM4B polymorphism increases susceptibility to multiple cancers in Chinese populations: a meta-analysis. BMC genetics. 2014;15:48
Author contact
![]() Corresponding authors: Zhijun Dai, Department of Breast Surgery, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China (E-Mail: dzj0911com), or Wenge Zhu, Department of Biochemistry and Molecular Medicine, The George Washington University Medical School, Washington, DC, USA (E-Mail: wz6812edu).
Corresponding authors: Zhijun Dai, Department of Breast Surgery, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China (E-Mail: dzj0911com), or Wenge Zhu, Department of Biochemistry and Molecular Medicine, The George Washington University Medical School, Washington, DC, USA (E-Mail: wz6812edu).

 Global reach, higher impact
Global reach, higher impact