3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4568-4577. doi:10.7150/jca.27655 This issue Cite
Research Paper
Systematic Analysis of Gene Expression Alteration and Co-Expression Network of Eukaryotic Initiation Factor 4A-3 in Cancer
1. Department of Medical Oncology, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, Guangxi 530021, People's Republic of China
2. Maternal and Child Health Hospital and Obstetrics and Gynecology Hospital of Guangxi Zhuang Autonomous Region, Guangxi 530003, People's Republic of China
3. The Fifth Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi 530022, People's Republic of China
4. Department of Biology, Pennsylvania State University, University Park, PA, 16802, USA
*These authors contributed equally to this work.
Received 2018-6-4; Accepted 2018-7-26; Published 2018-11-11
Abstract
Purpose: Eukaryotic initiation factor 4A-3 (EIF4A3) is an RNA-binding protein (RBP) that is a core component of the exon junction complex (EJC). It has been identified as an important player in post-transcriptional regulation processes. Recently, investigations have focused on EIF4A3 dysfunction in carcinogenesis. The present study aims to determine whether EIF4A3 can serve as a prognostic marker and potential regulatory mechanism in human cancers.
Materials and methods: EIF4A3 expression in various cancers was assessed using Oncomine. The Correlation between EIF4A3 expression and patient survival was evaluated using PrognoScan. EIF4A3 mutations in various cancers were investigated using cBioPortal. EIF4A3 co-expression networks in various cancers were established using Coexpedia. Finally, we analyzed potential functional roles of EIF4A3 using Gene Ontology and pathway enrichment analyses by FunRich V3.
Results: EIF4A3 was overexpressed in common malignancies at the transcription levels. High incidences of the breast, lung, and urinary cancers were closely related to the prognostic index for survival. The most prevalent mutation in EIF4A3 was E59K/Q. The tumor necrosis factor-α (TNF-α)/nuclear factor-κB (NF-κB) signaling pathway was affected by these mutations. Co-expression networks showed that EIF4A3 regulates apoptosis and cell cycle via several cancer-related signal pathways, and promotes tumor cell migration, invasion and drug resistance.
Conclusion: Our results suggest the potential role for EIF4A3 to serve as a diagnostic marker or therapeutic target for certain types of cancers.
Keywords: EIF4A3, cancer, prognosis, co-expression analysis
Introduction
According to the Estimated Cancer Incidence, Mortality and Prevalence Worldwide (GLOBOCAN2012) data, the burden of cancer will continue to rise because of the continuous increase of global population and epidemiological changes anticipated in the next few decades. Developing countries are expected more than 20 million new cases of cancer annually, as early as 2025[1]. Thus, cancer has become a worldwide problem and seeking new directions for cancer investigations are critical.
Gene expression is accomplished through the process of transcription and post-transcriptional modification; the metabolism of messenger RNA (mRNA) occupies a very important position in transcriptional regulation. In a eukaryotic cell, each mRNA is bound by a variety of proteins to form an mRNA-protein complex (mRNP). Together, these proteins impact nearly every step in the life cycle of an mRNA and are critical for the proper control of gene expression[2].
Eukaryotic initiation factor 4A-3 (EIF4A3) is an Asp-Glu-Ala-Asp (DEAD) box-family adenosine triphosphate (ATP)-dependent RNA helicase[3]. EIF4A3 functions as an RBP mainly localized in the nucleus that is a core component of the exon junction complex (EJC). As an ATP-dependent RNA clamp, EIF4A3 serves as an nucleation center to recruit other EJC components[4]. EIF4A3 plays essential roles in mRNA splicing, transport, translation, and surveillance [5, 6]. Among these functions, the most widely studied is Nonsense-Mediated Decay (NMD), which is a quality control system that degrades mRNAs containing premature termination codons, thereby preventing the accumulation of dysfunctional RNAs and proteins[7]. Knockdown of EIF4A3 leads to a defect in NMD[8, 9].
The function of EIF4A3 is complex and important, and its structural and functional anomalies directly lead to changes in downstream biological effects. Scientists begin to explore its association with malignancy: Michelle et al. reported that EIF4A3 controls several apoptotic regulators, such as Bcl-2 family proteins, which are alternatively spliced to produce isoforms with opposite functions[10]. Han et al. reported that EIF43A is highly expressed in colorectal cancer. After it binds to LncRNA H19, the mRNA of the cell cycle regulators cyclinD1, cyclin E1, and CK4 can no longer recruit EIF43A for correct post-transcriptional modification, leading to a loss of cell cycle progression and the acceleration of colon cancer cell growth[11]; in ovarian cancer, EIF43A binds to LncRNA CASC2 to form a negative feedback axis, and its high expression inhibits CASC2 and promotes the development of ovarian cancer[12]. Phosphoproteomic analysis of highly-metastatic hepatocellular carcinoma (HCC) showed that proteins involved in transcriptional regulation, mRNA processing, and RNA splicing were over-represented; EIF4A3 was identified as one of the crucial phosphoproteins in the metastasis of HCC[13]. Nevertheless, the exact role and underlying mechanism of EIF4A3 in the development and progression of cancer remains elusive.
Because of the rapid advances in high-throughput RNA sequencing (RNA-seq) technologies and free online database resources, we were able to perform a systematic and in-depth exploration of EIF4A3 in cancer. In this study, for the first time, we analyzed the expression, prognostic value, mutations, and co-expression network of EIF4A3 in multiple cancers to better understand the potential role and functions played by EIF4A3 in carcinogenesis.
Materials and Method
Oncomine analysis
The mRNA expression of EIF4A3 in various types of cancer in The Cancer Genome Atlas (TCGA) was analyzed using the Oncomine™ 4.5 database (https://www.oncomine.org/resource/main.html) [14]. The threshold was defined by the fold in mRNA expression in cancer tissue compared to normal tissue with the following parameters: p-value ˂0.05, fold change of 2, and gene ranking of 10%. Results of these analyses are summarized in Figure 1 and Supplementary Table 1.
PrognoScan analysis
The correlation between EIF4A3 expression in tumors and survival of patients with various types of cancer in TCGA was determined by Cox proportional hazards analysis using data from the PrognoScan database (http://gibk21.bse.kyutech.ac.jp/PrognoScan/index.html)[15]. The adjusted p-value threshold was <0.05. The results are summarized in Figure 2 and Supplementary Table 2.
c-BioPortal analysis
We conducted analysis of EIF4A3 mutations in various tumors in TCGA using the cBio Cancer Genomics Portal (http://cbioportal.org)[16]. The primary search parameters included alterations, such as amplification, deep deletion, and missense mutations, and copy number alterations from GISTIC and RNA-seq data with the default setting. Overall survival and disease-free survival were calculated according to cBioPortal's online instructions. The results are shown in Figures 3 and 4, and Supplementary Tables 3 and 4.
Co-expedia analysis
The co-expression networks of EIF4A3 in Gene Expression Omnibus (GEO) database were analyzed using the Coexpedia (http://www.coexpedia.org/) [17]. The visualization of the co-expression network, the enriched Gene Ontology (GO)-Disease Ontology (DO) term, and medical subject headings (MeSH) terms were ranked according to Coexpedia's online instructions. The enrichment analyses of GO - cellular component (CC), biological processes (BP), molecular function (MF) and biological pathway using FunRich V3[18]. The results are shown in Figures 5-7, and Supplementary Tables 5 and 6.
Results
EIF4A3 transcription expression in cancer
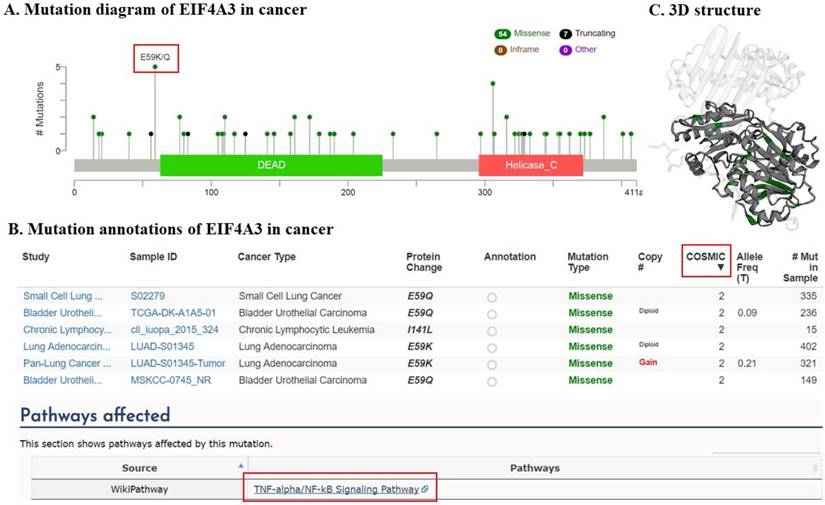
In order to explore its role in cancer, we queried the Oncomine database. EIF4A3 mRNA transcription level was compared between tumor and normal tissues. For OncomineTM 4.5 analysis of mRNA transcription, the threshold was set using the following parameters: p-value of 0.05, fold-change of 2, and gene ranking of 10% (Figure 1 and Supplementary Table 1); EIF4A3 was overexpressed in many common and high incidence solid tumors, such as cancers of the lung, breast, head and neck, brain and central nervous system, genitourinary tract (kidney, bladder, and prostate), and hematologic cancers such as lymphoma, but was low in leukemia and pancreatic cancer.
The transcription level of EIF4A3 in different cancer types (Oncomine). The data in the box chart shows a comparison of EIF4A3 mRNA expression between tumor tissues (right, dark blue) and normal tissues (left, light blue). The fold-change and p-value of EIF4A3 expression in these cancers was identified from our analyses shown in supplementary Table 1. The results show EIF4A3 expression in breast carcinoma relative to normal breast (A), in tongue squamous cell carcinoma relative to normal tongue (B), in squamous cell lung carcinoma relative to normal lung (C), in childhood acute B-cell lymphoblastic leukemia relative to normal hematogones (D), in superficial bladder cancer relative to normal bladder (E), in prostate intraepithelial neoplasia relative to normal prostate (F), in small cell lung carcinoma relative to normal lung (G), and in anaplastic large cell lymphoma, ALK-positive tissue relative to normal hematogones (H).
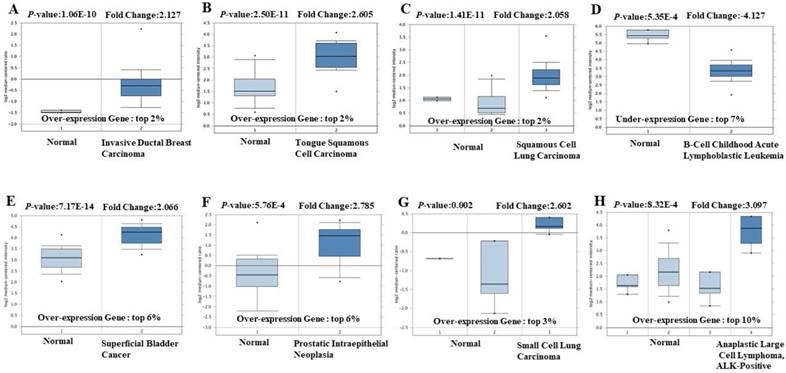
Prognostic value of EIF4A3 expression in various cancer types (PrognoScan). The survival curveswere plotted as the threshold of Cox p-value < 0.05. The survival curves comparing patients with EIF4A3 high (red) and low (blue) expression in bladder cancer (A, B), glioma (C), multiple myeloma (D), breast cancer (E, F) and lung cancer (G, H).
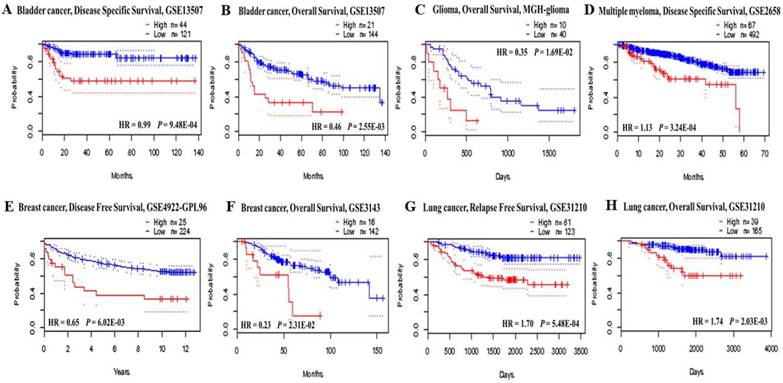
The frequency and type of EIF4A3 mutations in cancer (cBioPortal). The alteration frequency of EIF4A3 in various cancer studies; only cancer types containing >100 samples and an alteration frequency of >5% are shown (A). The alteration frequency of EIF4A3 in various cancer types; only cancer types containing >50 samples and an alteration frequency of >1% are shown(B). The alteration type included amplification (red), deep deletions (blue), multiple alterations (grey), and missense mutations (green).
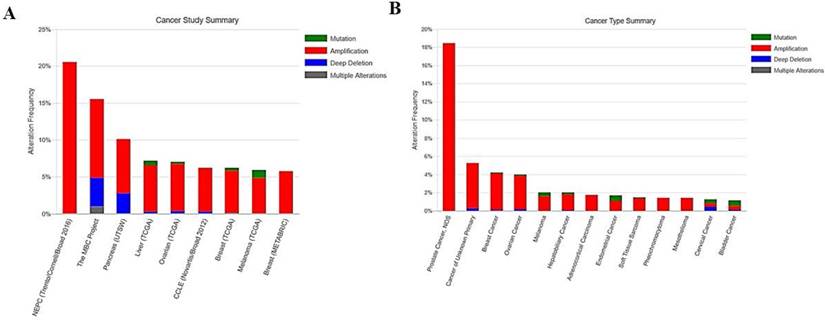
EIF4A3 mutations in cancer studies (cBioPortal). This graphical view shows the Pfam protein domains and the positions of 61 mutations; the hotspot (E59K/Q) represents the common founder mutation observed in the DEAD domain(A); the tabular view provides additional information about partial mutations. Hyperlinks to the COSMIC database show that mutations (E59K/Q) and (I141L) affect the TNF-α/NF-κB signaling pathway (B); 3-D protein structural analysis shows the locations of these mutation sites in EIF4A3 (green) (C).
Prognostic value of EIF4A3 transcription expression in cancer
The prognostic value of EIF4A3 mRNA transcription in cancer was analyzed using the PrognoScan database (Figure 2 and Supplementary Table 2). EIF4A3 overexpression was associated with poor prognosis in cancers of the breast, lung, brain, bladder, glioma, multiple myeloma, and liposarcoma. Especially in lung cancer and breast cancer, there were consistent results across three different cancer datasets: overexpression of EIF4A3 in GSE11121, GSE4922-GPL96, and GSE3143 in the breast cancer was correlated with shorter distant metastasis-free survival, disease-free survival, and overall survival, respectively. In the lung cancer, overexpression of EIF4A3 in GSE31210 was correlated with short relapse-free survival and overall survival, and jacob-00182-MSK and GSE4573 were simultaneously associated with decreased overall survival.
Analysis of EIF4A3 mutations in various cancers
The frequency and type of EIF4A3 mutation in cancer
We used the cBio Cancer Genomics Portal (cBioPortal) database to analyze the EIF4A3 mutation of 151 different cancer studies in TCGA database. The results showed nine different cancer studies that contained >5% alteration frequency and at least 100 samples in the dataset (Figure 3A and Supplementary Table 3). The first cancer study in the frequency of alteration was NEPC (Trento/Cornell/Broad 2016), which investigated whole exome and RNA-seq data of castration-resistant adenocarcinoma and castration- resistant neuroendocrine cancer; the database contained 107 samples and the total rate of alteration frequency in EIF4A3 was up to 20.56%. The second cancer study - The MBC Project - contained 103 samples of patients with metastatic breast cancer, and the EIF4A3 mutation rate was 15.53%. The third study, Pancreatic Cancer (UTSW), contained 109 samples of micro-dissected pancreatic ductal adenocarcinoma, and the EIF4A3 mutation rate was 10.09%. Subsequently, further reclassification of the above studies according to the pathological types of the tumors, the top three tumors with alteration frequency were prostate cancer (18.46%), cancer of unknown primary (5%), and breast cancer (4.01%); only cancer types containing >50 samples and an alteration frequency of >1% are shown (Figure 3B and Supplementary Table 4). At the same time, we also noticed that the most common alteration type of EIF4A3 was amplified, with a few deep deletion and mutations, which were consistent with either of the high-frequency alteration cancer studies or tumor types.
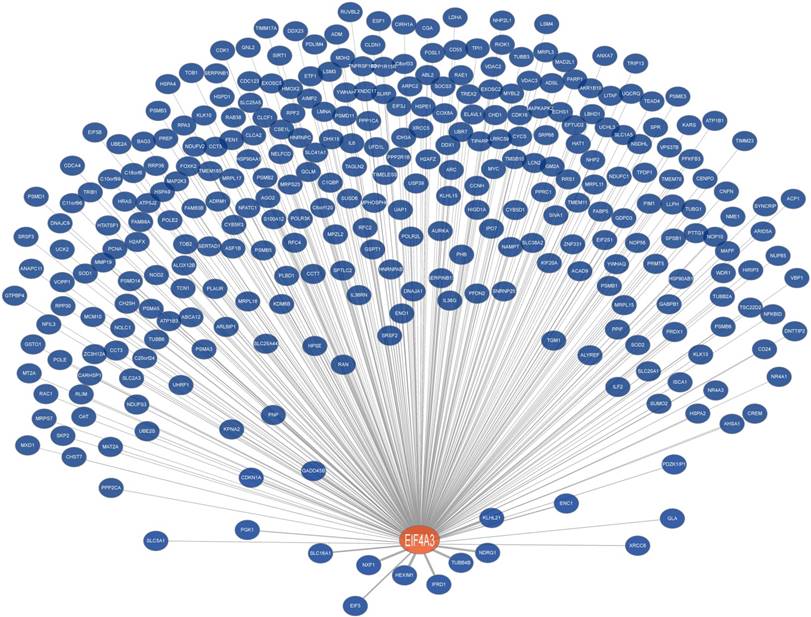
Co-expression network of EIF4A3 (Coexpedia). Co-expressed genes with EIF4A3 are ranked according to Coexpedia's online instructions.
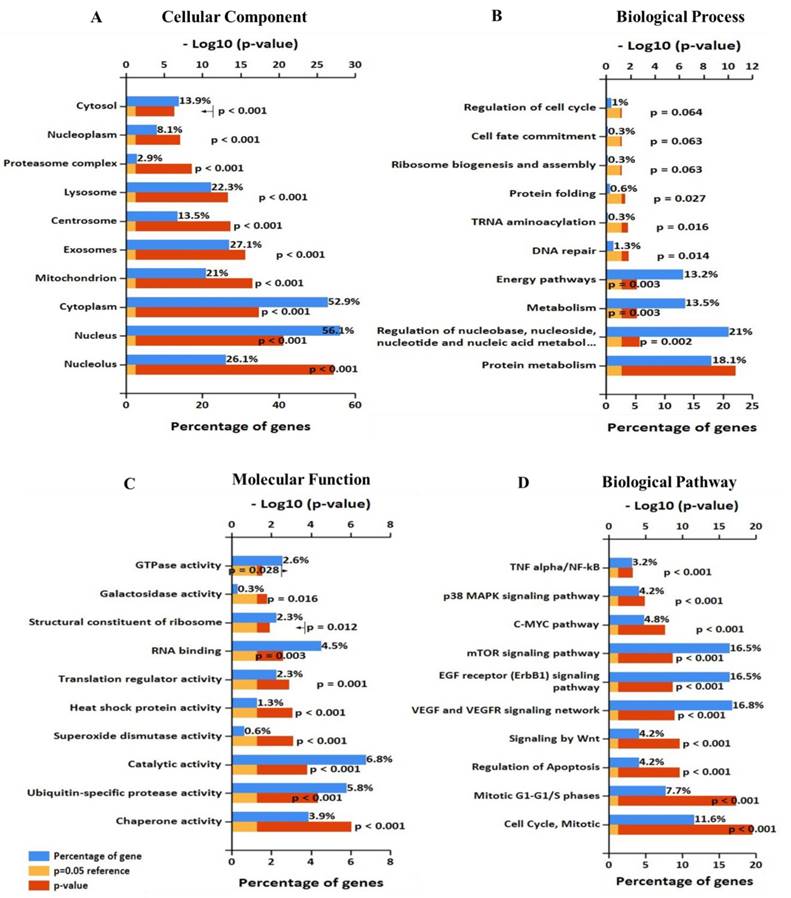
Significantly enriched Gene Ontology annotation of EIF4A3 including (A) cellular component, (B) biological process, and (C) molecular function. (D)biological pathway (FunRich V3).
EIF4A3 mutation annotations in cancer
Figure 4 summarizes the details of all mutations in 151 cancer studies: EIF4A3 had 54 missense mutations and 7 truncating mutations, of which 5 mutations occurred in a hotspot of E59K (substitution - missense, position 59, E➞K) and E59Q (substitution - missense, position 59, E➞Q), very close to the DEAD domain (Figure 4A). All five mutations reached mutation level 2 (the number of patients with the same mutation site) in the Catalogue Of Somatic Mutations In Cancer (COSMIC) database, suggesting that this is a critical region for its function. These mutations were from small cell lung cancer, lung adenocarcinoma, and bladder urothelial carcinoma samples. Interestingly, hyperlinks to the COSMIC database showed that mutations in this site affect the TNF-α/NF-κB signaling pathway (Figure 4B). Moreover, the mutation in the DEAD domain at amino acid I141L (substitution - missense, position 141, I➞L), which reached mutation level 2 in the COSMIC database, also was implicated in the TNF-α/NF-κB signaling pathway (Figure 4A, B). Consistently, the helicase C domain of EIF4A3 appeared to be frequently mutated.
EIF4A3 co-expression network in cancer
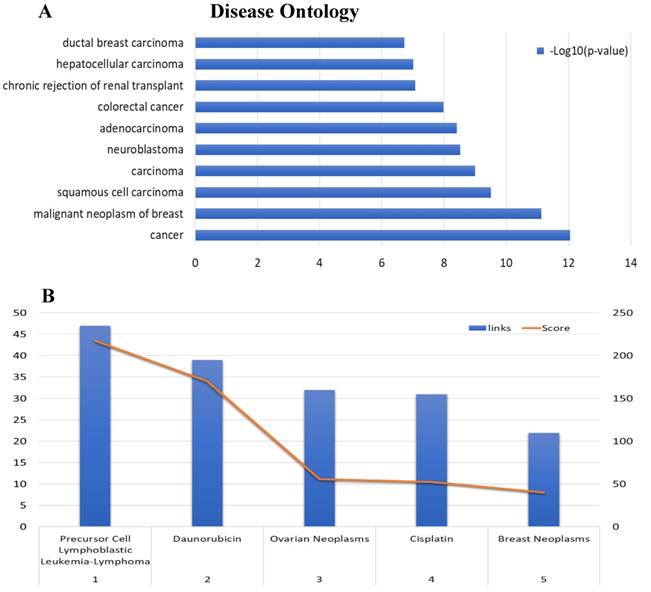
To further analyze the role and specific molecular network of EIF4A3 in disease, especially in cancer, the co-expression of genes functionally associated with EIF4A3 was predicted using the Coexpedia database; EIF4A3 was co-expressed with 312 genes (Figure 5 and Supplementary Table 5). Analysis of GO terms filtered for CC, BP, MF and biological pathway (Figure 6). Biological pathway showed cell cycle was highly enriched among EIF4A3 co-expressed partners, which were also closely related to regulation apoptosis, Wnt signaling, EGFR signaling, MAPK signaling and TNF/NF-κB signaling (Figure 6D). Next, GO-DO enrichment analysis returned 'cancer' and 'malignant neoplasm of the breast' terms as the diseases most frequently associated with EIF4A3, with squamous cell carcinoma, adenocarcinoma, colorectal carcinoma, and hepatocellular carcinoma within the top ten terms (Figure 7A).
The co-expression network associated with MeSH in the Coexpedia database, which is distinctive from other co-expression databases, provided biomedical information for disease and chemical relevance of EIF4A3 (Figure 7B and Supplementary Table 6). Interestingly, the neoplasm MeSH terms associated with EIF4A3 returned 'Precursor Cell Lymphoblastic Leukemia-Lymphoma' as the number one associated term; ovarian cancer and breast cancer were closely relevant malignancies in the top five terms. Furthermore, daunorubicin and cisplatin were suggested as highly associated with EIF4A3; these are commonly used in cancer chemotherapy. The analysis of daunorubicin came from the GEO Series of acute lymphoblastic leukemia cases, and the analysis of cisplatin came from the GEO Series of ovarian and gastric cancer cases associated with acquired resistance to chemotherapy.
Disease Ontology annotation and Enrichment after including 'neoplasm' as aMeSH term in the analysis of the EIF4A3 co-expression network (Coexpedia). The enriched GO-DO terms(A) and MeSH terms including 'neoplasm' (B)were ranked according to Coexpedia's online instructions.
Discussion
EIF4A3 is a core component of EJC that affects the fate of mRNAs [5, 6], which indicates that EIF4A3 may play an important role in cancer progression. However, little is known about whether it can serve as a marker for cancer diagnosis and prognosis. Our results from the Oncomine database showed that EIF4A3 was overexpressed in common malignancies at the mRNA transcription expression levels[14, 19]. At the same time, PrognoScan database analysis showed that high incidence cancers of the breast, lung and urinary system were closely related to the prognostic index for survival[15], suggesting that EIF4A3 might serve as a diagnostic marker for certain types of cancers. At the same time, we noticed that the relationship between EIF4A3 expression and prognosis was not identical in several different studies of the same cancer. For example, one of blood cancer study GSE4475 suggest that EIF4A3 high expression is associated with better prognosis (opposite direction from other studies), which may be closely related to specific subtypes. The study of blood cancer in PrognoScan includes four different pathological types: acute myeloid leukemia (AML) (GSE12417-GPL96, GSE12417-GPL570, GSE5122, GSE8970), B-cell lymphoma (GSE4475, E-TABM-346), follicular lymphoma (GSE16131-GPL96) and multiple myeloma (GSE2658). We also found that in another study of B-cell lymphoma, E-TABM-346, did not reach statistical significance, but the survival curve trend suggested that patients with high expression of EIF4A3 had better prognosis, which is consistent with GSE4475, suggesting that EIF4A3 is in B cell lymphoma may play a different role than other tumors. This result reminds us of the need to further combine clinical specimens and clinical pathology subtypes to study the potential value of EIF4A3 for cancer diagnosis, treatment and prognosis.
Genetic mutations usually cause phenotypic changes, which are closely related to DNA damage repair, carcinogenesis and aging[20, 21]. Therefore, analysis of EIF4A3 mutations has great significance in understanding the role of EIF4A3 in tumorigenesis and predicting treatment targets. Analysis of data from cBioPortal[16] showed that in cancers of the prostate, breast, pancreas, liver, and ovary, more than 5% had mutations in EIF4A3; gene amplification was the major type of mutation represented. It is noteworthy that EIF4A3 showed more than 20% variation in tumors from 107 patients with castration-resistant adenocarcinoma and castration-resistant neuroendocrine prostate cancer; this particular pathological subtype warrants further investigation.
Annotated mutations from cBioPortal showed that the most recurrent mutation in EIF4A3 was E59K/Q, which was detected in lung cancer and urothelial cancer samples. In addition, another noted mutation was I141L, which is located within the DEAD domain of EIF4A3, and TNF-α/NF-κB signaling pathway was affected by these two mutations. TNF-α is a pro-inflammatory cytokine that binds to cell membrane-expressed receptors of the TNF receptor family and activates the NF-κB pathway [22]. NF-kB comprises a family of transcription factors and aberrant or constitutive NF-κB activation has been detected in many human malignancies. Growing evidence supports a major role of NF-κB in oncogenesis as it regulates genes involved in the development and progression of cancer, such as proliferation, migration, apoptosis, drug resistance, and angiogenesis[23-27]. The TNF-α/NF-κB signaling pathway cross-talks with the MAPK pathway, Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway, and mitochondrial apoptosis pathway regulated by bcl-2 family proteins[28-30]. Thus, the TNF-α/NF-κB signaling pathway is a key signaling pathway that affects multiple phenotypes of cancer and is a potential target for cancer therapy. The close relationship between EIF4A3 mutations and this pathway suggests that EIF4A3 plays a complex and extensive role in the development of cancer. Additional experiments will be necessary to explore the extent to which the multifactorial modulation by EIF4A3 affects aberrant activation of the TNF-α/NF-κB signaling pathway.
The gene co-expression network constructed based on inter-gene expression data is an important way to study the molecular function of genes and the upstream and downstream signaling pathways. The difference between Coexpedia and other co-expression databases is that this database only contains co-expression through functional association, which is related to specific biological background, and it is the only co-expression database related to MeSH at present, which greatly facilitates the mining of biomedical co-expression information[17]. The disease that ranked first in the GO-DO analysis of EIF4A3 was cancer. The top 10 diseases were mainly subtyping of cancer. Further analysis using 'neoplasm' as a MeSH term in the top five showed that EIF4A3 correlated most closely with 'precursor cell lymphoblastic leukemia-lymphoma' and 'daunorubicin'. Co-expression analysis of high dimensional expression data provided direct evidence of the close relationship between EIF4A3 and a variety of common cancer types, especially those of the blood, even in the regulation of chemotherapy drug resistance. Statistical analysis of GO categories for EIF4A3 co-expression genes provides new directions for the study of EIF4A3 in carcinogenesis. The first is its role in cell cycle, particularly, in regulation of mitosis, which is consistent with previous studies: Fukumura et al. reported that EIF4A3 assembly on pre-mRNA supports fast and accurate splicing of a particular set of short introns in mitotic cell cycle-related genes[31]. The second is its function in the TNF-α/NF-κB signaling pathway, it is noteworthy that this pathway appears both in the pathway affected by the EIF4A3 mutation and in the biological pathway for the co-expression gene enrichment. The TNF-α/NF-κB signaling pathway is involved in many cellular processes associated with cancer[32, 33]. Tsumuraya et al. evaluated the effects of hippuristanol, an inhibitor of EIF4A, on adult T-cell leukemia[34]. Hippuristanol inhibited proliferation of cancer cells via induction of cell cycle arrest during G1 phase and induced apoptosis by inactivating NF-κB. These in vitro results validate our co-expression analysis that there is a close link between EIF4A3 and cell cycle, apoptosis and the TNF-α/NF-κB signaling pathways. The specific mechanism is not clear, by querying the starBase v2.0 database[35], we found that EIF4A3 is the RBP of many crucial genes in cancer related pathways, including NFKB1, EGFR, MAPK and mTOR. The possible hypothesis is that EIF4A3 leads to the abnormal expression of genes and the dysfunction of proteins through AS during carcinogenesis[4, 36].
In summary, as an important RBP, EIF4A3 is a core EJC component that plays important roles in multiple post-transcriptional regulation events. In cancer, EIF4A3 regulates cell cycle and cell apoptosis via the TNF-α/NF-ĸB signaling pathways and other important signaling pathways, promotes tumor cell migration and invasion, and development of drug resistance. In this study, we provide multi-level elucidation of the importance and role of EIF4A3 in carcinogenesis. The specific significance, role, and molecular mechanism of EJC in cancer deserve further exploration.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No: 81660498 ), the Guangxi Natural Science Foundation (Grant No: 2016GXNSFBA380090), the Guangxi Medical and Health Appropriate Technology Development and Application Project (Grant No: Grant No: S2017101 and S2016 34), the Basic Ability Enhancement Program for Young and Middle-aged Teachers of Guangxi (Grant No: 2017KY0120), the Key Research and Development project of Guangxi (Grant No: Guike AB16380215), the Project of Nanning Scientific Research and Technology Development Plan (Grant Number: 20183040-2), the China Scholarship Council (201608455001 and 201708455059) and a grant from Guangxi Medical University Training Program for Distinguished Young Scholars. Dr. Yingwei Mao is a receipt of NARSAD Young Investigator Award and American Heart Scientist Development Award.
Supplementary Material
Supplementary tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359-86
2. Rissland OS. The organization and regulation of mRNA-protein complexes. Wiley interdisciplinary reviews RNA. 2017:8
3. Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature reviews Molecular cell biology. 2011;12:505-16
4. Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonqueze O. et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124-31
5. Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536-48
6. Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature reviews Molecular cell biology. 2004;5:232-41
7. Ito M, Tanaka T, Cary DR, Iwatani-Yoshihara M, Kamada Y, Kawamoto T. et al. Discovery of Novel 1,4-Diacylpiperazines as Selective and Cell-Active eIF4A3 Inhibitors. Journal of medicinal chemistry. 2017;60:3335-51
8. Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753-7
9. Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346-51
10. Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M. et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Molecular and cellular biology. 2012;32:954-67
11. Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J. et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159-73
12. Zhang S, Leng T, Zhang Q, Zhao Q, Nie X, Yang L. Sanguinarine inhibits epithelial ovarian cancer development via regulating long non-coding RNA CASC2-EIF4A3 axis and/or inhibiting NF-kappaB signaling or PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2018;102:302-8
13. Tian M, Cheng H, Wang Z, Su N, Liu Z, Sun C. et al. Phosphoproteomic analysis of the highly-metastatic hepatocellular carcinoma cell line, MHCC97-H. International journal of molecular sciences. 2015;16:4209-25
14. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY). 2007;9:166-80
15. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC medical genomics. 2009;2:18
16. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1
17. Yang S, Kim CY, Hwang S, Kim E, Kim H, Shim H. et al. COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic acids research. 2017;45:D389-D96
18. Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA. et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-601
19. Ponten F, Schwenk JM, Asplund A, Edqvist PH. The Human Protein Atlas as a proteomic resource for biomarker discovery. Journal of internal medicine. 2011;270:428-46
20. Masuda M, Sawa M, Yamada T. Therapeutic targets in the Wnt signaling pathway: Feasibility of targeting TNIK in colorectal cancer. Pharmacol Ther. 2015;156:1-9
21. Saini N, Gordenin DA. Somatic mutation load and spectra: A record of DNA damage and repair in healthy human cells. Environ Mol Mutagen. 2018
22. Wu Z, Bruggeman LA. Assaying NF-kappaB activation and signaling from TNF receptors. Methods in molecular biology (Clifton, NJ). 2014;1155:1-14
23. De Simone V, Franze E, Ronchetti G, Colantoni A, Fantini MC, Di Fusco D. et al. Th17-type cytokines, IL-6 and TNF-alpha synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34:3493-503
24. Wang F, Ma J, Wang KS, Mi C, Lee JJ, Jin X. Blockade of TNF-alpha-induced NF-kappaB signaling pathway and anti-cancer therapeutic response of dihydrotanshinone I. International immunopharmacology. 2015;28:764-72
25. Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S. et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. The Journal of clinical investigation. 2014;124:528-42
26. You DJ, Park CR, Lee HB, Moon MJ, Kang JH, Lee C. et al. A splicing variant of NME1 negatively regulates NF-kappaB signaling and inhibits cancer metastasis by interacting with IKKbeta. The Journal of biological chemistry. 2014;289:17709-20
27. Liang S, Chen Z, Jiang G, Zhou Y, Liu Q, Su Q. et al. Activation of GPER suppresses migration and angiogenesis of triple negative breast cancer via inhibition of NF-kappaB/IL-6 signals. Cancer letters. 2017;386:12-23
28. Hu W, Shi L, Li MY, Zhou PH, Qiu B, Yin K. et al. Adrenomedullin protects Leydig cells against lipopolysaccharide-induced oxidative stress and inflammatory reaction via MAPK/NF-kappaB signalling pathways. Scientific reports. 2017;7:16479
29. Snyder M, Huang J, Huang XY, Zhang JJ. A signal transducer and activator of transcription 3.Nuclear Factor kappaB (Stat3.NFkappaB) complex is necessary for the expression of fascin in metastatic breast cancer cells in response to interleukin (IL)-6 and tumor necrosis factor (TNF)-alpha. The Journal of biological chemistry. 2014;289:30082-9
30. Giampazolias E, Zunino B, Dhayade S, Bock F, Cloix C, Cao K. et al. Mitochondrial permeabilization engages NF-kappaB-dependent anti-tumour activity under caspase deficiency. Nature cell biology. 2017;19:1116-29
31. Fukumura K, Wakabayashi S, Kataoka N, Sakamoto H, Suzuki Y, Nakai K. et al. The Exon Junction Complex Controls the Efficient and Faithful Splicing of a Subset of Transcripts Involved in Mitotic Cell-Cycle Progression. International journal of molecular sciences. 2016:17
32. Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985-99
33. Mitchell JP, Carmody RJ. NF-kappaB and the Transcriptional Control of Inflammation. International review of cell and molecular biology. 2018;335:41-84
34. Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J. et al. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochemical pharmacology. 2011;81:713-22
35. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014;42:D92-7
36. Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume AM. et al. Exon Junction Complexes Show a Distributional Bias toward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins. Cell reports. 2016;16:1588-603
Author contact
![]() Corresponding authors: Donghua Zou, The Fifth Affiliated Hospital of Guangxi Medical University, No 89 Qixing Road, Nanning, Guangxi 530022, People's Republic of China. Tel +86 771 2636189; Fax +86 771 2617892; Email: danvor0922com OR Yingwei Mao, Pennsylvania State University, 214 Life Sciences Building, University Park, PA, 16802, USA. Tel: 1-814 -867-4739; Fax: 1-814-863-1357; Email: yzm1edu
Corresponding authors: Donghua Zou, The Fifth Affiliated Hospital of Guangxi Medical University, No 89 Qixing Road, Nanning, Guangxi 530022, People's Republic of China. Tel +86 771 2636189; Fax +86 771 2617892; Email: danvor0922com OR Yingwei Mao, Pennsylvania State University, 214 Life Sciences Building, University Park, PA, 16802, USA. Tel: 1-814 -867-4739; Fax: 1-814-863-1357; Email: yzm1edu

 Global reach, higher impact
Global reach, higher impact