Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4578-4585. doi:10.7150/jca.28040 This issue Cite
Research Paper
Knockout of MTF1 Inhibits the Epithelial to Mesenchymal Transition in Ovarian Cancer Cells
1. Department of Anatomy, College of Basic Medical Science, Harbin Medical University, Harbin, China.
2. Department of Pathology and Laboratory Medicine, the University of Tennessee Health Science Center, Memphis, TN, 38163, USA.
3. Center for Cancer Research, University of Tennessee Health Science Center, Memphis, TN, 38163, USA.
4. Department of Obstetrics and Gynecology, Hokkaido University School of Medicine, Hokkaido University, Sapporo, Japan.
Received 2018-6-24; Accepted 2018-8-8; Published 2018-11-11
Abstract
Due to peritoneal metastasis and frequent recurrence, ovarian cancer has the highest mortality among gynecological cancers. Epithelial to mesenchymal transition (EMT) contributes to ovarian tumor metastasis. In this study, we report for the first time that metal regulatory transcription factor 1 (MTF1) was upregulated in ovarian cancer, and its high expression was associated with poor patient survival and disease relapse. Knockout of MTF1 using lentiviral CRISPR/Cas9 nickase vector-mediated gene editing inhibited EMT by upregulating epithelial cell markers E-cadherin and cytokeratin 7, and downregulating mesenchymal markers Snai2 and β-catenin in ovarian cancer SKOV3 and OVCAR3 cells. Loss of MTF1 reduced cell proliferation, migration, and invasion in both SKOV3 and OVCAR3 cells. Knockout of MTF1 upregulated the expression of the KLF4 transcription factor, and attenuated two cellular survival pathways, ERK1/2 and AKT. Our studies demonstrated that MTF1 plays an oncogenic role and contributes to ovarian tumor metastasis by promoting EMT. MTF1 may be a novel biomarker for early diagnosis as well as a drug target for clinical therapy.
Keywords: MTF1, CRISPR/Cas9 nickase, lentiviral vector, ovarian cancer, epithelial to mesenchymal transition
Introduction
Ovarian cancer is a malignancy of women with high mortality. In the US alone, over 15,000 patients die and 20,000 new cases are diagnosed each year1,2. Ovarian cancer patients have no apparent symptoms at early stages, and the disease advances to the late stages before diagnosis. Ovarian tumors metastasize to multiple peritoneal organs from primary organ ovaries and frequently recur due to chemoresistance3-5. The epithelial to mesenchymal transition (EMT) is a biological process characterized by loss of epithelial cell polarity and cell-cell junctions, and the gain of migratory and invasive properties. Accumulating evidence indicates that EMT contributes to tumor metastasis and chemoresistance6-11. Therefore, identifying the genes or pathways involved in EMT will provide a novel approach for cancer therapy. EMT is regulated by multiple transcription factors including Snai1/2, ZEB1/2, and twist112, and signaling pathways including TGFβ, ERK1/2, AKT, Notch, and WNT/catenin13-16. For the first time, we have identified the metal responsive transcription factor 1 (MTF1) contributing to EMT in ovarian cancer cells.
MTF1 is a zinc finger transcription factor that promotes cell survival by activating downstream target genes, including the metal binding protein metallothionein (MT1), matrix metalloproteinases (MMPs), the zinc efflux protein ZnT-1, and the zinc influx regulator ZIP-117-20. MTF1 and its regulated gene MT1 can be activated by zinc and copper in the presence of p53 in breast cancer cells, but not in p53 inactivated cells21. MTF1 has not been well investigated in human cancers. MTF1 is upregulated in breast, lung and cervical cancers22. However, in ovarian cancer the role of MTF1 is largely unknown. MTF1 is regulated by zinc and copper, and the ratio of copper versus zinc and CA125 together have been used as biomarkers for ovarian cancer diagnosis 23,24. MTF1 is a biomarker for the prediction of the disease recurrence in advanced-stage head and neck carcinoma25.
In this study, we analyzed MTF1 expression in ovarian cancer patient samples and knocked out MTF1 in ovarian cancer SKOV3 and OVCAR3 cells using a lentiviral CRISPR/Cas9 nickase vector approach. We find that MTF1 is upregulated in ovarian cancer and knockout of MTF1 leads to inhibition of EMT in ovarian cancer cells, suggesting that MTF1 may contribute to ovarian tumor metastasis.
Materials and Methods
The paraffin-embedded (FFPE) blocks of fully de-identified ovarian serous carcinoma were obtained from the Tissue Services Core of the University of Tennessee Health Science Center (UTHSC). Hematoxylin and eosin (H&E) staining was performed by Histology Core of UTHSC and reviewed by a pathologist.
Cell culture
Ovarian cancer cell lines SKOV3 and OVCAR3 were obtained from ATCC and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (Hyclone; Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen; Carlsbad, CA). HEK293 FT cells were purchased from Invitrogen and cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% glutamine.
Generation of MTF1 KO ovarian cancer cells using lentiviral CRISP/Cas9 nickase vector
The lentiviral CRISPR/Cas9 nickase-mediated MTF1 gene editing vectors were constructed by synthesizing and annealing two gRNAs and subcloning them in the BsmII site of lentiviral Lentiguide-puro vector (#52963, Addgene). Two gRNA sequences, 5' GCCATTTGAGTGTGACGTGC and 5' CCTTCGTGTGCACTCGCACG, were designed to target upstream and downstream regions of exon 2 of MTF1. CRISPR/cas9 nickase was driven by EF1a promoter in LentiCas9-blast vector (#52962, Addgene). Lentivirus was produced by packaging in 293FT cells as published previously26. MTF1 stable KO cell lines were established by transducing the SKOV3 or OVCAR3 cells lentiCas9-blast Cas9 nickase vector and selected with 10 μg/ml blasticidin, and subsequently transduced with the lentiviral CRISPR/Cas9 nickase-mediated MTF1 gRNA vector and selected with 5 μg/ml puromycin. LentiCas9-blast was used as the control vector without gRNAs.
MTT assay
SKOV3 or OVCAR3 cells (8,000 cells/well), transduced with lentiviral CRISPR/Cas9 nickase for MTF1 gRNA and control vectors, were seeded into 96-well plates and cultured for 24, 48, and 72 h. MTT reagent (10 µl) was added to each well and incubated for ~4 h. Then reaction was stopped by adding 100 µl detergent reagent and the plates were incubated at room temperature in the dark for 2 h. Cell proliferation was assessed by measuring the absorbance at 570 nm wavelength.
Cell colony formation assay
MTF1 KO and control SKOV3 and OVCAR3 cells were plated into 6-well plates (400 cells/well) and cultured for 10 days. Cell colonies were stained with crystal violet and the number of colonies was counted.
Cell migration assay
The cell migration assay was performed using modified transwell chambers (BD Falcon™, San Jose, CA) inserted into 24-well cell culture plates. SKOV3 or OVCAR3 stable KO and control cells (3 × 104) were suspended in 300 µl serum-free DMEM and added into the upper chamber. DMEM containing 10% FBS (serving as the chemoattractant) was added into the lower chamber of each well and incubated for 24 h. The medium and non-migrated cells in the upper chamber were removed, while the migrated cells on the lower side of the membranes were fixed with methanol and stained with crystal violet. Pictures were taken at 10x magnification, and cells from at least three different fields were counted.
Cell invasion assay
SKOV3 and OVCAR3 MTF1 stable KO and control cells (5 × 105) were seeded in serum-free DMEM onto inserts precoated with Matrigel (BD BioCoatTM) using 24-well Tumor Invasion System (BD BioSciences, San Jose, CA). DMEM containing 10% FBS was added to the bottom chamber of the invasion system as the chemoattractant for 24h. The transwell inserts were fixed with methanol for 20 min, and stained for 5 min with H&E. Pictures were taken at 10x magnification and invaded cells were counted from at least three different fields.
Immunofluorescent staining
To detect MTF1 and EMT marker expression in human ovarian cancer specimens or cell lines, ovarian serous carcinoma sections were antigen-retrieved by heating sections in sodium citrate buffer(10mM sodium citrate,0.05%Tween, pH6.0) for 30 mins, whereas ovarian cancer cells were fixed in 4% paraformaldehyde for 30 mins. Sections or cells were incubated with blocking buffer (5% normal goat serum, 3% bovine serum albumin, and 0.1% Triton-X100 in PBS) for 1 h and then incubated overnight with primary antibodies to MTF1, PCNA, cytokeratin 7, and β-catenin (1:200 dilution, Cell Signaling, Danvers, MA). After rinsing three times for 5 min with PBST, Alexa 488- or 594-conjugated goat anti-rabbit (Invitrogen, Carlsbad, CA) antibodies were incubated for 1 h at room temperature. Cell nuclei were counterstained with DAPI (Vector Laboratories, Inc.; Burlingame, CA). Images were captured under a fluorescent microscope (Zeiss model LSM700, San Diego, CA). Imunofluoresent intensity was measured using Image J software on tumor and adjacent non-tumor area from six different sections of human ovarian serous carcinoma and mean intensity was used to compare the significance using Student's t-test.
Western blot
Ovarian cancer cells were collected in RIPA buffer (Thermo Scientific; Rockford, IL) containing 1% Halt Proteinase Inhibitor Cocktail (Thermo Scientific). Equal amounts of protein (40 µg/lane) were loaded onto 10% SDS-PAGE gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat milk for 1 h and incubated with primary antibodies against MTF1 (1:1000, Novus Biologicals), GAPDH (1:5000, Sigma; St. Louis, MO), β-catenin, KLF4, pERK1/2, ERK1/2, pAKT, AKT, E-cadherin, Snai2 (1:1000, Cell Signaling), and cytokeratin 7 (1:1000, Abcam).
Data query from The Human Protein Atlas
To evaluate the correlation of MTF1 mRNA expression with ovarian patient survival, we queried TCGA data based on RNA-seq using The Human Protein Atlas. The expression level of MTF1 was quantified using the FPKMs (number Fragments Per Kilobase of exon per Million reads) with a detection threshold of 1 FPKM. The median and maximally separated Kaplan-Meier plots are presented by comparing low and high expression groups in log-rank in the Human Protein Atlas27.
Analysis of relapse time using SurvExpress program
To analyze the correlation of MTF1 expression and relapse time, we queried TCGA ovarian cancer data using SurvExpress program28 and divided patients into low and high risk group based on MTF1 expression. The relapse time of ovarian cancer patients was plotted based on risk group.
Statistical analysis
Significant differences were determined from at least two independent experiments performed in triplicate by Student's t-test and data were presented as mean ± SD. P < 0.05 was considered significant.
Results
MTF1 was highly expressed in ovarian cancer and associated with poor patient survival and tumor relapse
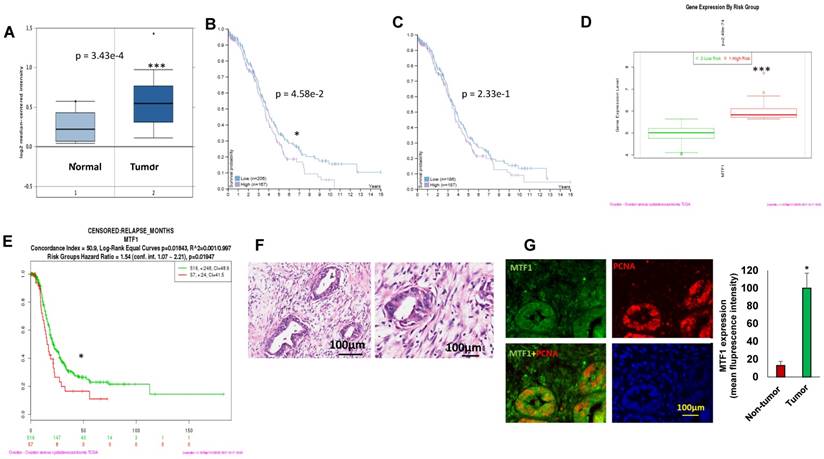
To assess MTF1 expression in ovarian cancer, we analyzed 185 ovarian serous carcinomas and 10 normal ovarian surface epithelia in the Oncomine database29. MTF1 mRNA expression was significantly higher (~1.2-fold increase) in tumors than that in normal ovarian surface epithelia (p = 3.43E-4) (Fig. 1A). Using the human protein atlas, we examined patient survival based on RNA sequencing data of MTF1 expression from a total of 373 patients. There were 167 patients with high and 206 with low MTF1 mRNA expression while a 2.9FPKM cut-off was used as the best separation whereas there were 187 patients with high and 186 with low MTF1 expression while a 2.7 FPKM cut-off was used as the median separation in TCGA database30. Patients with high MTF1 mRNA expression had significantly lower survival probability compared to patients with low MTF1 mRNA expression in the best separation (p=4.58e-2) (Fig. 1B) although not in the median separation (p=2.33e-1) (Fig.1C).
We also analyzed MTF1 gene expression based on risk group using SurvExpress program on TCGA data28, and found that MTF1 gene expression was higher in the high-risk group of 57 ovarian cancer patients as compared to the low-risk group of 516 (p=2.49e-74) (Fig. 1D). Moreover, ovarian serous carcinoma patients in the high-risk group had significantly shorter time to relapse compared to the low-risk group (p=0.01947) (Fig.1E). We first characterized as high-grade ovarian serous carcinoma by H&E staining (Fig.1F) and then verify the expression of MTF1 in ovarian cancer using immunofluorescent staining of tumor sections from six high-grade ovarian serous carcinoma. MTF1 was strongly stained in tumor cell cytoplasm and membranes from all of specimens, but not in the adjacent normal tissues (Fig. 1G). These data indicate that MTF1 is highly expressed in high-grade ovarian serous cancer, and high MTF1 expression is associated with poor patient survival and more frequent relapses in ovarian cancer.
Knockout of MTF1 expression using lentiviral CRISPR/Cas9 nickase vector-mediated editing resulted in the inhibition of EMT in ovarian cancer cells
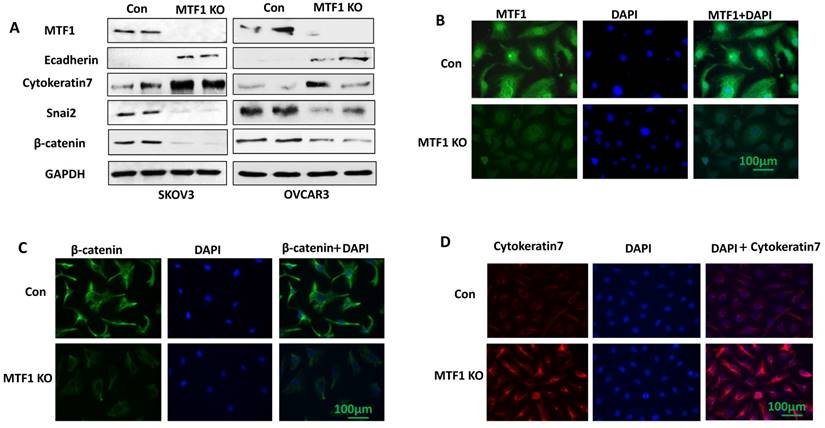
To investigate the function of MTF1 gene in ovarian cancer cells, we knocked out MTF1 gene by transducing SKOV3 and OVCAR3 cells using lentiviral CRISPR/Cas9 nickase, targeting a region of exon 2. Stable MTF1 KO and control cells were established by puromycin selection in both cell lines. MTF1 protein expression was depleted in both SKOV3 and OVCAR3 MTF1 KO cells compared to control, as shown by western blot (Fig. 2A). We also examined the expression of EMT-associated markers in both ovarian cancer cell lines. Knockout of MTF1 led to the upregulation of epithelial cell markers E-cadherin and cytokeratin 7, and downregulation of mesenchymal markers Snai2 and β-catenin in both MTF1 KO cell lines compared to control cells (Fig. 2A). We also performed immunostaining for MTF1 and EMT markers, cytokeratin 7 and β-catenin, in SKOV3 MTF1 KO and control cells. MTF1 was strongly stained in cell nuclei and cytoplasm in control cells, but not in SKOV3 MTF1 KO cells (Fig. 2B). β-catenin was strongly stained in cell cytoplasm and membrane of SKOV3 control cells but weak stained in SKOV3 MTF1 KO cells (Fig. 2C). Cytokeratin 7 was strongly stained in cell membrane of SKOV3 MTF1 KO cells, but weakly stained in control cells (Fig. 2D). As shown by these western blot and immunofluorescent staining experiments, loss of MTF1 inhibited EMT in ovarian cancer cells.
Knockout of MTF1 inhibited cell proliferation and survival in ovarian cancer cells
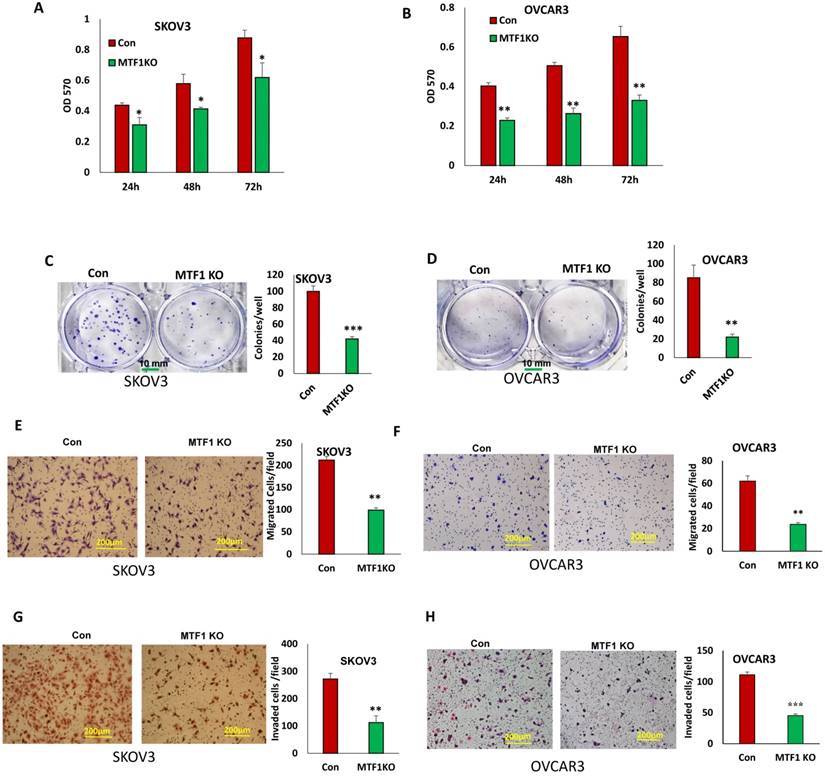
MTF1 function has not been well investigated and its role is completely unknown in ovarian cancer. To determine the function of MTF1 in ovarian cancer cells, we examined cell proliferation in MTF1 KO and control SKOV3 and OVCAR3 cells by performing MTT assays. Knockout of MTF1 significantly reduced cell proliferation in both SKOV3 and OVCAR3 cells compared to control cells at all three time points (24, 48, and 72 h) (Fig. 3A, B). We also examined cell survival using cell colony formation assays; knockout of MTF1 led to significant inhibition of colony formation in both SKOV3 (Fig. 3C) and OVCAR3 cells (Fig. 3D). Our data indicate that knockout of MTF1 in ovarian cancer cells led to inhibition of cell proliferation and colony formation.
MTF1 was highly expressed in ovarian serous carcinoma and associated with ovarian patient survival and relapses. A. MTF1 mRNA expression in ovarian serous carcinoma and normal ovary control in Oncomine database. 1 = normal ovary surface epithelium (n = 10); 2 = ovarian serous carcinoma (n = 185) (***p<0.001). B. C, High MTF1 mRNA expression is significantly associated with poor patient survival in best (B) separation group (*p<0.05), but not in the median(C) separation group based on MTF1 expression cut-off value. D. MTF1 mRNA expression in high risk and low risk ovarian serous carcinoma patients based on SurvExpress program (***p<0.001). E. Ovarian serous carcinoma patient relapse time in high and low risk group based on SurvExpress program (*p <0.05). F. H.E. staining of ovarian serous carcinoma specimens (left: low magnification; right: high magnification). G. Immunofluorescent staining of MTF1 and PCNA in ovarian serous carcinoma specimens and fluorescent intensity was compared between tumor and adjacent non-tumor tissues (*p<0.05, n = 6).
Knockout of MTF1 inhibited cell migration and invasion in ovarian cancer cells
EMT contributes to tumor cell invasion. We found that loss of MTF1 led to the inhibition of EMT. We tested whether loss of MTF1 affects cell motility and invasion. Using transwell plates, we examined cell migration in MTF1 KO and control ovarian cancer SKOV3 and OVCAR3 cells. Cell migration was significantly reduced in both MTF1 KO SKOV3 and OVCAR3 cells compared to control cells (Fig. 3E, F). Using Matrigel-coated transwell plates, we assessed cell invasion, and found that cell invasion was also significantly reduced in both SKOV3 and OVCAR3 MTF1 KO cells compared to the control cells (Fig. 3G, H). Our results show that MTF1 KO significantly reduced migration and invasion in both ovarian cancer cell lines.
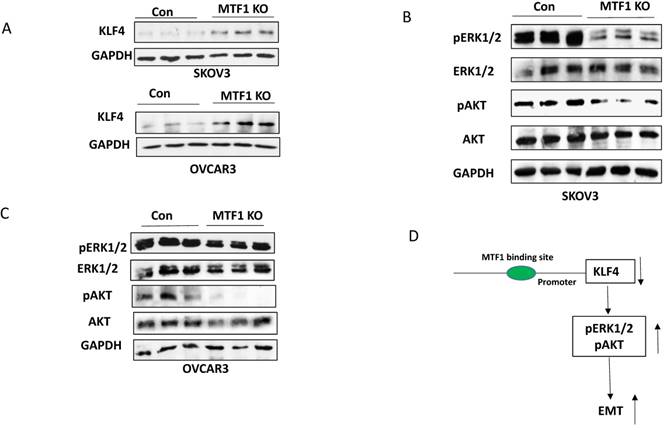
Knockout of MTF1 upregulated KLF4 expression and attenuated two cellular survival pathways in ovarian cancer cells
We showed previously that the transcription factor KLF4 inhibits EMT by attenuating the TGFβ pathway in ovarian cancer cells31. To understand the molecular mechanism underlying MTF1-mediated EMT, we examined KLF4 expression in both MTF1 KO and control SKOV3 and OVCAR3 cells by Western blot. We found that KLF4 expression was upregulated in MTF1 KO cells compared to control (Fig. 4A). We also examined two cellular survival pathways, including ERK1/2 and AKT in MTF1 KO and control SKOV3 and OVCAR3 cells. Loss of MTF1 led to attenuation of phospho-ERK1/2 and phospho-AKT in both cell lines, but total ERK1/2 and AKT were not significantly altered, as compared to control cells (Fig. 4B, C). Our results indicated a novel molecular mechanism underlying MTF1 mediated EMT that MTF1 transcriptionally repressed KLF4 expression and attenuated two cellular survival pathways ERK1/2 and AKT, thus inhibited EMT in ovarian cancer cells (Fig.4D).
Discussion
In this study, we demonstrated for the first time that MTF1 was highly expressed in ovarian cancer and its high expression was associated with poor patient survival and disease relapse, suggesting that MTF1 may contribute to ovarian cancer development and is a potential biomarker for ovarian cancer diagnosis and prognosis. MTF1 is highly expressed in head and neck carcinoma, and is associated with disease relapse25. MTF1 is also upregulated in colorectal cancer and is associated with copper homeostasis32.
Lentiviral CRISPR/Cas9 nickase vector-mediated MTF1 gene editing resulted in inhibition of EMT in ovarian cancer cells. A. MTF1 and EMT marker expression was examined in MTF1 KO and control SKOV3 and OVCAR3 cells by Western blot. B. Immunofluorescent staining of MTF1 expression in SKOV3 MTF1 KO and control cells. C. Mesenchymal marker β-catenin was stained in SKOV3 MTF1 KO and control cells. D. Epithelial marker cytokeratin 7 was stained in SKOV3 MTF1 KO and control cells.
Knockout of MTF1 led to reduced cell proliferation, migration, and invasion in ovarian cancer cells. A, B. Cell proliferation in MTF1 KO and control SKOV3 (A) and OVCAR3 (B) cells at different time points determined by MTT assay. C, D. Cell survival in MTF1 KO SKOV3 (C) and OVCAR3 (D) cells determined by colony formation assay. E, F. Cell migration in MTF1 KO and control SKOV3 (E) and OVCAR3 (F) cells determined using transwell plates. G, H. Cell invasion in MTF1 KO and control SKOV3 (G) and OVCAR3 (H) cells examined using Matrigel-coated plates. *p < 0.05; **p < 0.01;***p < 0.001
Knockout of MTF1 expression inhibited EMT in ovarian cancer cells. Knockout was accomplished using lentiviral CRISPR/Cas9 nickase vector-mediated gene editing. EMT acquisition contributes to aggressive cellular behavior, and is associated with tumor metastasis and chemoresistance. Our studies suggest that MTF1 may contribute to tumor metastasis and chemoresistance, thus leading to ovarian cancer relapse. Thus, MTF1 may be a promising drug target for ovarian cancer therapy. In particular, a small molecule inhibitor of MTF1 APTO-253 was recently approved for phase I clinical trials to treat several other cancers33. We examined the function of MTF1 in ovarian cancer SKOV3 and OVCAR3 KO and control cells by performing cell functional assays, and found that knockout of MTF1 resulted in reduced cell proliferation, colony formation, migration, and invasion in both SKOV3 and OVCAR3 cells.
MTF1, as a metal regulatory transcription factor, regulates gene expression by binding to the metal regulatory element (MRE) within the promoter of target genes, in response to excess zinc and other stresses 34,35. Although we do not yet know how MTF1 functions in response to zinc, we found that zinc can induce dose-dependent expression of MTF1 in ovarian cancer cells (data not shown). We are in the process of investigating the signaling of zinc/MTF1 axis in contributing to ovarian cancer metastasis and chemoresistance. However, we found that knockout of MTF1 resulted in upregulation of KLF4 expression in both ovarian cancer SKOV3 and OVCAR3 cells (Fig. 4A), indicating that MTF1 negatively regulates KLF4 expression. KLF4 functions as a tumor suppressor and inhibits EMT in ovarian cancer cells by targeting the TGFβ pathway31. Therefore, it is possible that MTF1 in its function as a transcription factor regulates KLF4 expression through transcriptional repression, which is a hypothesis requiring further analysis. MTF1 reportedly not only activates downstream target genes but also can repress the downstream target genes by interacting with co-repressors35. Interestingly, KLF4 was identified as one of target genes of MTF1 in human adenocarcinoma cell line HT-2936. Therefore, one of the molecular mechanisms underlying MTF1-mediated EMT in ovarian cancer cells may be through negatively regulating KLF4 expression. Previous studies showed that activation of ERK1/2 and AKT pathways contributes to EMT in numerous human cancers, including ovarian cancer16,37-40. Our studies indicated that knockout of MTF1 attenuated the ERK1/2 and PI3K/AKT pathways in both SKOV3 and OVCAR3 ovarian cancer cells. MTF1 can also regulate EMT by targeting these two pathways, although it is unknown whether and how MTF1 regulates these pathways via repressing KLF4 expression. Further studies are required to reveal the exact mechanism how MTF1/KLF4 axis regulates EMT in ovarian cancer cells.
In conclusion, we demonstrated that MTF1 is highly expressed in ovarian cancer, and high MTF1 expression is associated with poor ovarian cancer patient survival and disease relapse. We found that disrupting MTF1 expression inhibited EMT, proliferation, survival, migration, and invasion of ovarian cancer SKOV3 and OVCAR3 cells. Also, knockout of MTF1 upregulated KLF4 expression and attenuated both the ERK1/2 and AKT pathways, indicating that MTF1 may contribute to ovarian tumor metastasis by promoting EMT in ovarian cancer.
Acknowledgements
This study was supported by a grant (1R21CA216585-01A1) from National Institute of Health and grant from West Cancer Center to Dr. Yue. This study was partially supported by a grant from the National Natural Science Foundation of China (No. 81272503) to Dr. Zheng and the content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsoring organizations.
Knockout of MTF1 attenuated the cellular survival pathways in ovarian cancer cells. A. Western blot analysis of KLF4 expression in MTF1 KO and control SKOV3 and OVCAR3 cells. B, C. Western blot analysis of phospho- and total ERK1/2 and phospho- and total AKT in MTF1 KO and control SKOV3 (B) and OVCAR3 (C) cells, respectively. D. A schematic diagram of molecular mechanism underlying MTF1 mediated EMT: MTF1 transcriptionally represses KLF4 expression by binding KLF4 promoter, activates ERK1/2 and AKT pathways, and thus promotes EMT. Knockout of MTF1 upregulates KLF4 expression and attenuates both ERK1/2 and AKT pathways, thus inhibits EMT in ovarian cancer cells.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Su Z, Graybill WS, Zhu Y. Detection and monitoring of ovarian cancer. Clin Chim Acta. 2013;415:341-5
2. Lowe KA, Chia VM, Taylor A. et al. An international assessment of ovarian cancer incidence and mortality. Gynecol Oncol. 2013;130:107-14
3. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
4. Scalici JM, Arapovic S, Saks EJ. et al. Mesothelium expression of vascular cell adhesion molecule-1 (VCAM-1) is associated with an unfavorable prognosis in epithelial ovarian cancer (EOC). Cancer. 2017;123:977-84
5. Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256
6. Vergara D, Merlot B, Lucot JP. et al. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291:59-66
7. Amankwah EK, Lin HY, Tyrer JP. et al. Epithelial-Mesenchymal Transition (EMT) Gene Variants and Epithelial Ovarian Cancer (EOC) Risk. Genet Epidemiol. 2015;39:689-97
8. Ahmed N, Abubaker K, Findlay J. et al. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Curr Cancer Drug Targets. 2010;10:268-78
9. Helleman J, Smid M, Jansen MP. et al. Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol. 2010;117:170-6
10. Yoshida S, Furukawa N, Haruta S. et al. Expression profiles of genes involved in poor prognosis of epithelial ovarian carcinoma: a review. Int J Gynecol Cancer. 2009;19:992-7
11. Takai M, Terai Y, Kawaguchi H. et al. The EMT (epithelial-mesenchymal-transition)-related protein expression indicates the metastatic status and prognosis in patients with ovarian cancer. J Ovarian Res. 2014;7:76
12. Bogachek MV, Park JM, De Andrade JP. et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Reports. 2016;7:1140-51
13. Bocci F, Jolly MK, Tripathi SC. et al. Numb prevents a complete epithelial-mesenchymal transition by modulating Notch signalling. J R Soc Interface. 2017:14
14. Baldwin LA, Hoff JT, Lefringhouse J. et al. CD151-alpha3beta1 integrin complexes suppress ovarian tumor growth by repressing slug-mediated EMT and canonical Wnt signaling. Oncotarget. 2014;5:12203-17
15. Arend RC, Londono-Joshi AI, Straughn JM Jr. et al. The Wnt/beta-catenin pathway in ovarian cancer: a review. Gynecol Oncol. 2013;131:772-9
16. Choi JH, Hwang YP, Kim HG. et al. Saponins from the roots of Platycodon grandiflorum suppresses TGFbeta1-induced epithelial-mesenchymal transition via repression of PI3K/Akt, ERK1/2 and Smad2/3 pathway in human lung carcinoma A549 cells. Nutr Cancer. 2014;66:140-51
17. Gunther V, Lindert U, Schaffner W. The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823:1416-25
18. Zhao WJ, Song Q, Wang YH. et al. Zn-responsive proteome profiling and time-dependent expression of proteins regulated by MTF-1 in A549 cells. PLoS One. 2014;9:e105797
19. Hardyman JE, Tyson J, Jackson KA. et al. Zinc sensing by metal-responsive transcription factor 1 (MTF1) controls metallothionein and ZnT1 expression to buffer the sensitivity of the transcriptome response to zinc. Metallomics. 2016;8:337-43
20. Bi Y, Lin GX, Millecchia L. et al. Superinduction of metallothionein I by inhibition of protein synthesis: role of a labile repressor in MTF-1 mediated gene transcription. J Biochem Mol Toxicol. 2006;20:57-68
21. Ostrakhovitch EA, Olsson PE, von Hofsten J. et al. P53 mediated regulation of metallothionein transcription in breast cancer cells. J Cell Biochem. 2007;102:1571-83
22. Shi Y, Amin K, Sato BG. et al. The metal-responsive transcription factor-1 protein is elevated in human tumors. Cancer Biol Ther. 2010;9:469-76
23. Gal D, Lischinsky S, Friedman M. et al. Prediction of the presence of ovarian cancer at surgery by an immunochemical panel: CA 125 and copper-to-zinc ratio. Gynecol Oncol. 1989;35:246-50
24. Gao ZJ. [Diagnostic value of serum copper/zinc ratio in gynecologic tumors]. Zhonghua Zhong Liu Za Zhi. 1988;10:434-6
25. Pavon MA, Parreno M, Tellez-Gabriel M. et al. CKMT1 and NCOA1 expression as a predictor of clinical outcome in patients with advanced-stage head and neck squamous cell carcinoma. Head Neck. 2016;38(Suppl 1):E1392-403
26. Yue J, Sheng Y, Ren A. et al. A miR-21 hairpin structure-based gene knockdown vector. Biochem Biophys Res Commun. 2010;394:667-72
27. Uhlen M, Zhang C, Lee S. et al. A pathology atlas of the human cancer transcriptome. Science. 2017:357
28. Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E. et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250
29. Bonome T, Levine DA, Shih J. et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478-86
30. Kandoth C, McLellan MD, Vandin F. et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333-9
31. Chen Z, Wang Y, Liu W. et al. Doxycycline inducible Kruppel-like factor 4 lentiviral vector mediates mesenchymal to epithelial transition in ovarian cancer cells. PLoS One. 2014;9:e105331
32. Barresi V, Trovato-Salinaro A, Spampinato G. et al. Transcriptome analysis of copper homeostasis genes reveals coordinated upregulation of SLC31A1,SCO1, and COX11 in colorectal cancer. FEBS Open Bio. 2016;6:794-806
33. Cercek A, Wheler J, Murray PE. et al. Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors. Invest New Drugs. 2015;33:1086-92
34. Jackson KA, Valentine RA, Coneyworth LJ. et al. Mechanisms of mammalian zinc-regulated gene expression. Biochem Soc Trans. 2008;36:1262-6
35. Hogstrand C, Zheng D, Feeney G. et al. Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish. Biochem Soc Trans. 2008;36:1252-7
36. Kindermann B, Doring F, Budczies J. et al. Zinc-sensitive genes as potential new target genes of the metal transcription factor-1 (MTF-1). Biochem Cell Biol. 2005;83:221-9
37. Xie L, Law BK, Chytil AM. et al. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6:603-10
38. Strippoli R, Loureiro J, Moreno V. et al. Caveolin-1 deficiency induces a MEK-ERK1/2-Snail-1-dependent epithelial-mesenchymal transition and fibrosis during peritoneal dialysis. EMBO Mol Med. 2015;7:102-23
39. Al-Qasem A, Al-Howail HA, Al-Swailem M. et al. PAC exhibits potent anti-colon cancer properties through targeting cyclin D1 and suppressing epithelial-to-mesenchymal transition. Mol Carcinog. 2016;55:233-44
40. Li Y, Xie Y, Cui D. et al. Osteopontin Promotes Invasion, Migration and Epithelial-Mesenchymal Transition of Human Endometrial Carcinoma Cell HEC-1A Through AKT and ERK1/2 Signaling. Cell Physiol Biochem. 2015;37:1503-12
Author contact
![]() Corresponding authors: Jinhua Zheng, Department of Anatomy, College of Basic Medical Science, Harbin Medical University, 157 Baojian Road, Harbin, Heilongjiang, 150081, China, Tel: +86-451-86674508; Fax: +86-451-86672942; Email: jhzhenghrbmu.edu.cn OR Dr. Junming Yue, Department of Pathology, University of Tennessee Health Science Center, 19 S. Manassas St., Rm. 266, Memphis, TN 38163, Fax: 901-448-3910; Phone: 901-448-2091; Email: jyueedu
Corresponding authors: Jinhua Zheng, Department of Anatomy, College of Basic Medical Science, Harbin Medical University, 157 Baojian Road, Harbin, Heilongjiang, 150081, China, Tel: +86-451-86674508; Fax: +86-451-86672942; Email: jhzhenghrbmu.edu.cn OR Dr. Junming Yue, Department of Pathology, University of Tennessee Health Science Center, 19 S. Manassas St., Rm. 266, Memphis, TN 38163, Fax: 901-448-3910; Phone: 901-448-2091; Email: jyueedu

 Global reach, higher impact
Global reach, higher impact