Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4642-4649. doi:10.7150/jca.27686 This issue Cite
Research Paper
Hedgehog Interacting Protein 1 is a Prognostic Marker and Suppresses Cell Metastasis in Gastric Cancer
1. Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, 200032, China
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 200032, China
3. Institute of Pathology, Fudan University, Shanghai 200032, China
* These authors contributed equally to this work.
Received 2018-6-6; Accepted 2018-9-14; Published 2018-11-24
Abstract
Background: The gene Hedgehog interacting protein (HHIP) is a pivotal morphogen for multiple developmental processes. However, the expression and clinical correlation of HHIP in gastric cancer (GC) has not been fully investigated. Here, we aimed to explore the expression of HHIP in gastric cancer (GC) and evaluate its clinicopathological and functional correlations.
Methods: The expression of HHIP mRNA was first determined in the Human Protein Atlas (HPA) and The Cancer Genome Atlas (TCGA) GC database and then validated by RT-qPCR (n = 41) and immunohistochemistry (IHC, n = 95) in a cohort of in-house GC patients and in 29 cases of gastric intraepithelial neoplasia (GIN). The clinicopathological and functional relationship of HHIP with GC were also analyzed.
Results: We found that HHIP mRNA were significantly downregulated in GC in the TCGA and HPA databases, as well as in our in-house cohort (P < 0.05). HHIP mRNA is mainly located in the cell nucleus, while HHIP protein is mainly located in the cell cytoplasm. Moreover, the HHIP protein level in the GIN tissues was significantly higher than that in the GC tissues (P < 0.001) and significantly lower than that in adjacent normal controls (P < 0.001). In addition, low HHIP expression was correlated with lymphatic metastasis (P = 0.041), pTNM stage (P = 0.007) and nervous system invasion (P = 0.001). Furthermore, we observed strong positive correlations between HHIP protein expression and overall survival (P < 0.001) and disease-free survival (P = 0.027) in GC patients. HHIP protein expression was an independent prognostic factor for overall survival (P < 0.001). Functional experimental results showed that overexpression of HHIP attenuated the migration and invasion ability of GC cells (P < 0.01).
Conclusion: HHIP may be a promising tumor metastatic-suppressor and prognostic biomarker for gastric cancer.
Keywords: gastric cancer, HHIP, expression, metastasis, survival
Introduction
Although the incidence of gastric cancer (GC) is decreasing worldwide, it is still relatively high in eastern Asia, especially in China [1]. In China, as the dominant types of cancer in the age group 60 to 74 years, GC is the leading cause of death and a major digestive system public health problem, with 0.50 million deaths and 0.68 million new cases in 2015 [2]. Although prevention efforts are critical to reduce the long-term burden of cancer, any effects of these efforts will not be seen in the near future. For this purpose, improving the access and availability of optimal prognostic and therapeutic biomarkers may hold the greatest potential to provide effective molecules for clinical management of GC patients and to facilitate early intervention and individualized/precise treatment.
The Hedgehog interacting protein (HHIP) gene is located on chromosome 4q31.21, and it encodes a member of the hedgehog-interacting protein (HHIP) family. As evolutionarily conserved proteins, the hedgehog (HH) proteins are key mediators of many fundamental processes in embryonic development. HH signals can be transduced and/or regulated by multiple cell-surface receptors [3]. Abnormal activation of the hedgehog signaling pathway is one pivotal cause of the oncogenesis and development of human malignancies. By binding to all three hedgehog proteins, Sonic Hedgehog (SHH), Indian hedgehog (IHH) and Desert hedgehog (DHH) HHIP proteins function as negative regulators in the hedgehog pathway and exhibit significant roles in human malignancies [4-7]. Cumulative evidence has illustrated the role of HHIP in cancer. For example, downregulation of HHIP in stromal cells increased the proliferation of leukemic cells [4]; HHIP-overexpression attenuated the activation of HGF/ MET and HH pathways and made lung adenocarcinoma cells more susceptible to stress conditions [5]. It has been shown that HHIP-overexpression inhibited GC cell proliferation, migration and invasion [7]. However, the relationship between HHIP expression and GC has never been illuminated.
In the present study, we first evaluated the dysregulation of HHIP mRNA in the HPA and TCGA GC databases. Then, we validated the results in our cohort and analyzed the correlation of HHIP protein with clinicopathological characteristics and prognosis of GC patients. Additionally, we assessed the role of HHIP on GC cell migration and invasion in vitro.
Materials and Methods
Patients
The HPA database (http://www.proteinatlas.org/), which includes 354 GC and 193 unpaired normal gastric mucosa samples, and the TCGA gene expression database (http://cancergenome.nih.gov/), which includes 360 GC and 37 unpaired normal gastric mucosa samples, were used for primary study. The Reads per kilobase of exon per million reads mapped (RPKM) of HHIP were extracted for analysis.
A total of 165 cases were collected in our Fudan University Shanghai Cancer Center (FUSCC) cohort. Forty-one paired, frozen, fresh GC and normal gastric tissue samples, 95 cases of formalin fixed paraffin embedded (FFPE) GC and normal gastric tissue samples from patients who underwent resection of primary GC at FUSCC and 29 cases of FFPE tissue samples from gastric intraepithelial neoplasia (GIN) patients who underwent endoscopic biopsy at FUSCC were studied. All samples were obtained from the tissue bank of FUSCC. This study was approved by The Research Ethics Committee of FUSCC, and all patients had signed informed consent. The GC diagnosis was histopathologically confirmed by two pathologists. None of the patients underwent preoperative treatment. The data collected from all subjects included gender, age, and GC features (e.g., histologic stage, tumor size, tumor depth, status of lymphatic metastasis, vascular invasion, nervous invasion, and peritoneal metastasis). The GC clinical stage was evaluated using the tumor-node-metastasis (TNM) classification system [20]. The follow-up of GC patients who donated FFPE samples (n = 95) were performed every three months during the first postoperative year and one year thereafter until March 30, 2018. Overall survival (OS) was calculated from the date of surgery to the date of death. Disease-free survival (DFS) was defined as the length of time between the surgery and tumor progression or death from the cancer. During the follow-up, 24 patients died of GC.
Cell lines and culture conditions
The human GC cell lines AGS, MGC-803, MKN-45, SGC-7901 and HGC-27, and the human normal gastric mucosa cell line GES-1 were cultured in 90% RMPI-1640 (Gibco) and 90% DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 50 U/ml penicillin and 50 μg/ml streptomycin (Gibco) in a humidified atmosphere at 37°C and 5% CO2, as appropriate.
Transient transfection
The full-length HHIP sequence was amplified by PCR from the cDNA and then subcloned into a pENTER vector (Transheep, Shanghai, China). SGC-7901 and MGC-803 cells were transfected with plasmid vectors using NamipoTM (Transheep, Shanghai, China) according to the manufacturer's protocol.
Cell migration and invasion assays
Cell motility and invasion were detected using a wound-healing assay and Transwell assay. The experiments were carried out and assessed as described previously [21].
RNA extraction, reverse transcription, and qRT-PCR analysis
Total RNA was isolated from 41 gastric cancer samples using TRIzol® reagent (Invitrogen). Real-time RT-PCR was performed as previously described [21]. β-actin was used as an internal control. The primers for RT-PCR analysis were synthesized by Transheep (Shanghai, China). Primers were as follows: HHIP-F, 5'- TCTCAAAGCCTGTTCCACTCA -3', HHIP-R, 5'-GCCTCGGCAAGTGTAAAAGAA-3'; U6-F, 5'- TGCTCGCTTCGGCAGCACAT-3', U6-R, 5'-CTTGCGCAGGGGCCATGCTA-3'; MT RNR1-F, 5'- CCTCCCCAATAAAGCTAAAA -3', MT RNR1-R, 5'- GCTATTGTGTGTTCAGATAT-3'; β-actin-F, 5'- AGTCATTCCAAATATGAGATGCGTT-3', β-actin-R, 5'- TGCTATCACCTCCCCTGTGT-3'.
Western blotting
Antibodies against HHIP (OAGA00579) were purchased from Aviva Systems Biology (San Diego, CA, USA). GAPDH (#2118) was purchased from Cell Signaling Technology (Cambridge, MA, USA). Cells were lysed in RIPA buffer (Sigma-Aldrich) supplemented with a protease inhibitor (Roche, Basel, Switzerland) and a phosphatase inhibitor (Roche). Protein concentrations were measured using a BCA protein assay kit (Thermo Scientific, MA, USA) as described previously [19]. Protein expression levels were normalized to that of GAPDH.
Immunohistochemistry (IHC) staining
FFPE samples were selected from the archives of the Pathology Department of FUSCC. The 10×12 tissue microarrays (TMA) were made by FUSCC Tissue Bank. IHC was performed on 5-μm-thick TMA sections using the HHIP antibody (HPA012616, Sigma, 1:200 dilution). Each case had one repeat core to preclude loss of samples. IHC staining was performed as described previously [21]. Sections were evaluated and scored by two pathologists independently. The Immunoreactive Score (IRS) was the multiplicity of the staining intensity and positive cancer percentage and was calculated by the mean value of two cores. Finally, the assessment of HHIP protein expression was defined as low (≤1.5+) and high (>1.5+ to ≤3+).
Localization
Nuclear and cytoplasmic RNA were separately extracted from the AGS and MGC-803 cell lines using the Protein and RNA Isolation System (PARIS) Kit (Life Technologies, USA), and nuclear and cytoplasmic RNA were converted to cDNA and analyzed by qRT-PCR as described previously [22]. Of the control genes, the mitochondrial gene MT RNR1 was expressed in the cytoplasm, whereas the nuclear transcript U6 was expressed in the nucleus. β-actin was used for normalization.
Statistical Analysis
The data were calculated using SPSS 22.0 or GraphPad Prism and reported as the mean ± SD. All experiments were performed at least in triplicate. Comparisons between groups were determined by a paired t test, Mann-Whitney test, two-tailed χ2 test or one-way ANOVA followed by Tukey's multiple comparison tests, as appropriate. The survival curves were estimated by Kaplan-Meier curves with a log rank test. Univariate and multivariate Cox proportional backwards hazards regressions were applied to estimate the hazard ratio (HR) for the survival, and the hazard ratio (HR) and 95% confidence interval (CI) were measured. All P values were two-sided, and P < 0.05 was considered statistically significant.
Results
HHIP expression in GC tissues
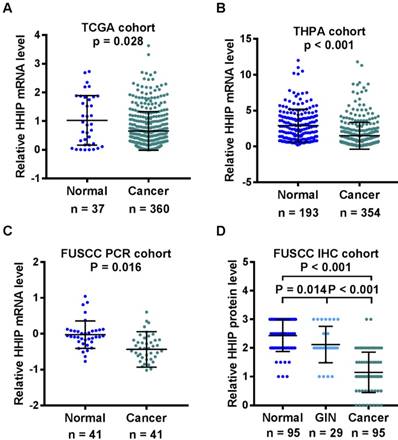
We first searched the TCGA and HPA database to analyze the mRNA expression of HHIP. The data of HHIP mRNA expression from the TCGA gastric datasets showed that the levels of HHIP mRNA expression were significantly lower in the GC tissue samples (0.657±0.673) compared with the normal gastric tissues (1.030±0.865) (Figure 1A). Consistently, the data of HHIP mRNA expression from the HPA database indicated that HHIP mRNA was also decreased in the GC tissue samples (1.500±1.873) compared with the normal tissue samples (2.913±2.281, P < 0.001; Figure 1B).
HHIP transcripts were then detected in 41 pairs of the primary GC tissues and the adjacent gastric mucosa. In comparison with these matched normal (1.546±2.229) samples, HHIP mRNA levels were significantly decreased in the cancer tissues (0.604±0.725, Figure 1C). To detected the HHIP protein expression in GC, a tissue microarray with 95 pairs of GC and normal mucosa samples and 29 cases of GIN were selected for HHIP IHC staining. Statistically significant score differences were observed between the GC (1.149±0.702) and normal gastric mucosa (2.432±0.554, P<0.001). In addition, HHIP immunostaining in the GIN (2.121±0.636) tissues was reduced compared with the normal tissue samples (2.432±0.554, P = 0.014) and increased compared with the GC tissue samples (1.149±0.702, P < 0.001; Figure 1D).
HHIP subcellular localization and immunostaining
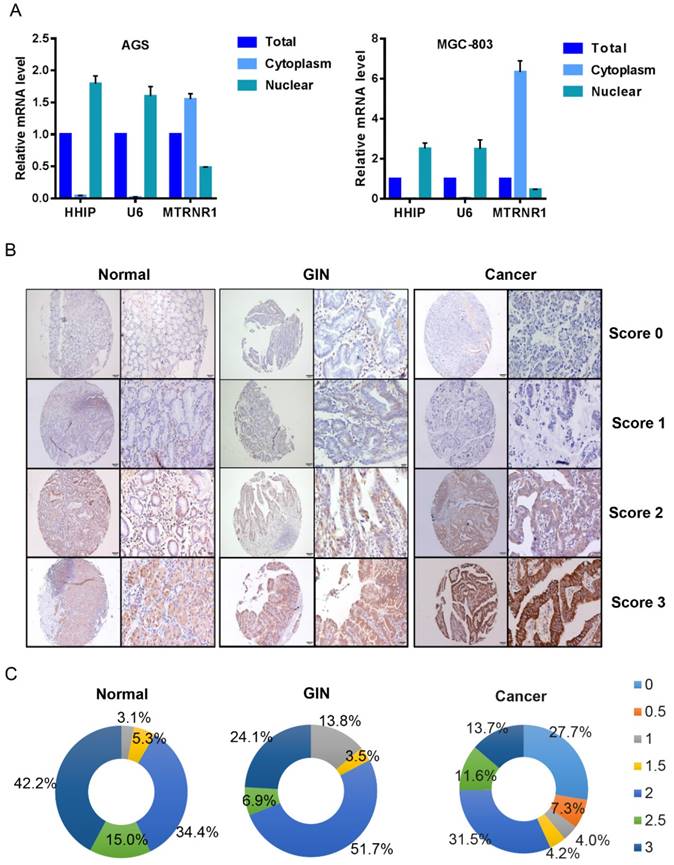
To determine the subcellular localization of HHIP mRNA in GC, we used a nucleoplasm separation method to detect the subcellular location of HHIP in AGS and MGC-803 cells; MT RNR1 was used as a cytoplasmic reference and U6 was used as a nucleus reference, and the results showed that HHIP was mainly located in GC cell nuclei (Figure 2A). By IHC, positive signals of HHIP immunostaining were observed in the cytoplasm and cell membrane of benign and malignant gastric mucosa cells (Figures 2B). A low expression (score 0-1.5) was observed in 41 GC cases, 5 IN cases and 8 normal cases; a high expression (score 2-3) was observed in 54 GC cases, 24 IN cases and 87 normal cases (Figure 2C). An interesting finding was that no normal gastric mucosa and GIN samples were scored as 0-0.5, whereas 35% of the gastric cancer samples were scored as 0-0.5 (Figure 2C).
Association of HHIP protein expression with GC patient clinicopathological features and prognosis
By dividing patients into the high HHIP mRNA level group and low HHIP mRNA level group using the mean value of HHIP mRNA expression in 41 cases of GC tissue samples, we analyzed the relationship between the HHIP mRNA expression levels and clinicopathological features of GC patients. However, no positive correlation was found (all P > 0.05; Table 1). We also analyzed the relationship between the HHIP protein expression levels and clinicopathological features of 95 cases of GC patients who were enrolled for IHC analysis. Chi-squared analysis demonstrated that HHIP protein expression was correlated with lymphatic metastasis (P = 0.041), nervous invasion (P=0.001) and pTNM stage (P = 0.007), while there was no significant correlation between HHIP protein expression and any other clinicopathological feature (all P > 0.05; Table 1).
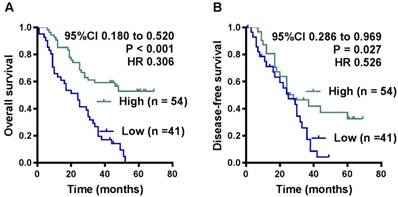
OS and DFS curves were plotted according to HHIP protein expression levels using the Kaplan-Meier method. As presented in Figure 3, the patients with high HHIP protein expression exhibited a significantly better OS (P < 0.001) and DFS (P = 0.027) than those with low HHIP expression. Univariate analysis of OS revealed that the HHIP expression level (P < 0.001), tumor size (P = 0.020), tumor depth (P = 0.008), nervous invasion (P = 0.021), lymphatic metastasis (P = 0.017), peritoneal metastasis (P < 0.001) and tumor stage (P < 0.001) were prognostic indicators for GC (Table 2). Further multivariate analysis revealed that HHIP expression (P < 0.001) and peritoneal metastasis (P < 0.001) were independent prognostic predictors for OS in GC patients (Table 2), while HHIP protein expression was not an independent prognostic indicator for DFS in GC patients (data not shown).
HHIP suppresses GC cell migration and invasion
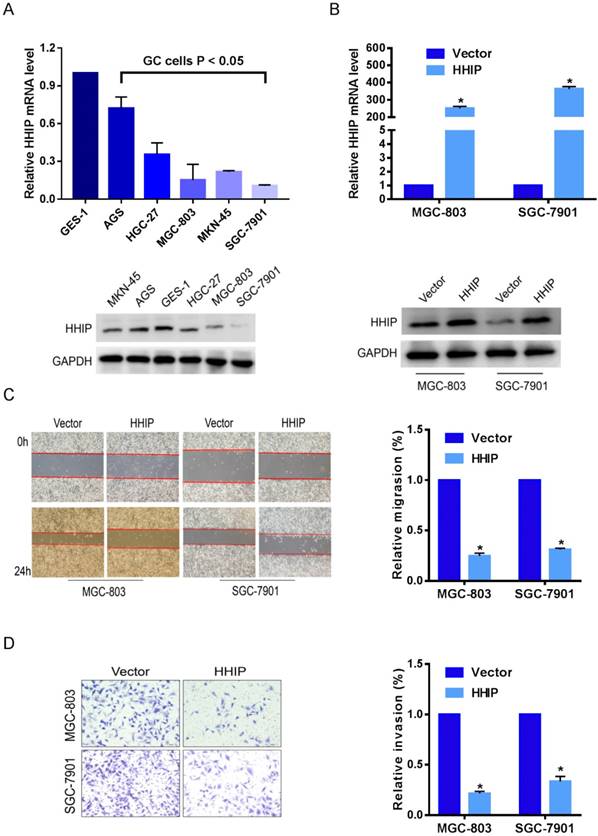
To investigate the biological effects of HHIP in GC progression, we examined the HHIP baseline expression levels in 5 gastric cancer cell lines (AGS, HGC-27, MGC-803, SGC-7901, MKN-45) and one normal gastric epithelial cell line (GES-1). The level of HHIP in gastric cancer cells was found to be significantly lower than that in GES-1 cells (Figure 4A). Then, SGC-7901 and MGC-803 cells were selected for overexpression experiments, and the efficiency of overexpression was validated by qRT-PCR and Western blotting (Figure 4B).
Because high HHIP expression levels were tightly correlated with lymph node metastasis and nervous invasion, we speculated that HHIP can suppress the metastatic ability of GC cells. With further in vitro experimental assays, we found that upregulation of HHIP resulted in attenuated wound-healing rates (P < 0.05; Figure 4C) and a remarkably decreased number of invaded cells (P < 0.05; Figure 4D) in both SGC-7901 and MGC-803 cells. These findings suggest that HHIP may be a metastatic-related suppressor in GC.
HHIP had lower expression in gastric cancer than normal tissues. A. Analysis of the TCGA GC dataset indicated that HHIP expression was decreased in the gastric cancers (n = 360) compared with normal gastric tissues (n = 37). (mean±SD, with Mann-Whitney test). B. Analysis of HPA GC dataset indicated there was decreased HHIP mRNA expression in the 354 GC samples compared to the normal tissue (n = 193). (mean±SD, with Mann-Whitney test). C. Analysis of the FUSCC cohort by RT-qPCR indicated that HHIP mRNA expression was significantly downregulated in the gastric cancer samples compared to the normal controls (n = 41, mean±SD, with paired t-test). D. Analysis of the FUSCC cohort by Western blotting indicated HHIP protein expression in 95 pairs of GC and normal tissues, as well as in 29 GIN samples. (mean±SD, with one-way ANOVA).
HHIP subcellular localization and immunostaining. A. HHIP mRNA subcellular localization in GC AGS and MGC-803 cell lines. B. Representative images of HHIP expression in gastric cancer, GIN and normal tissues, as visualized at 40× and 200× magnifications. C. Distribution of Immunoreactive Score (IRS) in TMA of gastric tissues and in TMA of indicated tissues.
Discussion
In human normal tissues, HHIP mRNA is mainly expressed in the adult lung, brain and stomach; however, it is weakly expressed in the esophagus, heart, skin, placenta and pancreas [8]. As a structural decoy receptor for vertebrate Hedgehog, HHIP acts as an antagonist of hedgehog signaling [9]. Low expression of HHIP has been reported in human malignancies such as hepatic [10-12], pancreatic [13], gastrointestinal [14], or medulloblastoma cancers [15]. In gastrointestinal cancer, including esophageal cancer, gastric cancer and pancreatic cancer, Hedgehog signaling is frequently activated due to epigenetic silencing of the HHIP1/HHIP gene, and HHIP is considered a Hedgehog inhibitor [5], whereas L Yang et al. reported that HHIP was highly expressed in esophageal squamous cell carcinomas (ESCC) tissue samples, suggest that silencing of HHIP is not a major mechanism for HH signaling activation in ESCC [16].
Clinical characteristics of gastric cancer patients
| Clinicopathologic feature | HHIP mRNA expression (RT-qPCR group) | Pa | HHIP protein expression (IHC group) | Pa | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Age (years) | 0.623 | 0.838 | ||||
| < 60 | 16 | 4 | 21 | 29 | ||
| ≥ 60 | 17 | 4 | 20 | 25 | ||
| Gender | 0.310 | 0.803 | ||||
| male | 26 | 8 | 31 | 43 | ||
| female | 7 | 0 | 10 | 11 | ||
| Tumor mass size | 0.697 | 0.830 | ||||
| < 5 cm | 16 | 5 | 25 | 35 | ||
| ≥ 5 cm | 17 | 3 | 16 | 19 | ||
| Histologic grade | 0.573 | 0.815 | ||||
| Good or moderate | 26 | 6 | 30 | 38 | ||
| Poor or undifferentiated | 7 | 2 | 10 | 15 | ||
| Tumor depth | 0.522 | 0.141 | ||||
| T1+T2 | 4 | 0 | 3 | 10 | ||
| T3+T4 | 29 | 8 | 38 | 44 | ||
| Lymphatic metastasis | 0.722 | 0.041 | ||||
| Absent | 6 | 1 | 3 | 13 | ||
| Present | 27 | 7 | 38 | 41 | ||
| Peritoneal metastasis | 0.412 | 0.488 | ||||
| Absent | 23 | 7 | 28 | 41 | ||
| Present | 10 | 1 | 13 | 13 | ||
| Vascular invasion | 0.627 | 0.828 | ||||
| Absent | 12 | 3 | 13 | 19 | ||
| Present | 21 | 5 | 28 | 35 | ||
| Nervous invasion | 0.692 | 0.001 | ||||
| Absent | 10 | 3 | 7 | 27 | ||
| Present | 23 | 5 | 34 | 27 | ||
| pTNM stage | 0.569 | 0.007 | ||||
| I+II | 10 | 2 | 4 | 19 | ||
| III+IV | 23 | 6 | 37 | 35 | ||
a All statistical tests were 2-sided. Significance level: p < 0.05.
Univariate and multivariate analyses of clinicopathological factors for overall survival in gastric cancer patients (IHC cohort)
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95 % CI) | pa | HR (95 % CI) | pa | |
| Age (≥ 60/< 60) | 0.882 (0.534-1.456) | 0.624 | ||
| Gender (Male/Female) | 1.089 (0.608-1.950) | 0.774 | ||
| Tumor size (≥ 5/< 5) | 1.823 (1.101-3.018) | 0.020 | ||
| Histologic grade (Poor, other/Good, mod) | 0.894 (0.504-1.587) | 0.703 | ||
| Depth of tumor (T3, T4/T1, T2) | 4.898 (1.527-15.709) | 0.008 | ||
| Vascular invasion (Present/Absent) | 1.746 (0.984-3.099) | 0.057 | ||
| Nervous invasion (Present/Absent) | 1.943 (1.105-3.419) | 0.021 | ||
| Lymphatic metastasis (Present/Absent) | 2.806 (1.205-6.553) | 0.017 | ||
| Peritoneal metastasis (Present/Absent) | 3.062 (1.803-5.200) | < 0.001 | 2.792 (1.617-4.821) | < 0.001 |
| TNM stage (III+IV/I+II) | 4.313 (1.945-9.564) | < 0.001 | ||
| HHIP protein expression (High/Low) | 0.294 (0.175-0.495) | < 0.001 | 0.327 (0.192-0.555) | < 0.001 |
HR: Hazard ratio; CI: confidence interval;
a All statistical tests were 2-sided. Significance level: p < 0.05.
Influence of HHIP protein expression patterns on overall survival and disease-free survival by Kaplan-Meier analyses in the FUSCC cohort. A. The Kaplan-Meier survival curve with log-rank analysis of OS showed statistical significance between the curves of patients with HHIP high-expression and low-expression (log-rank test). B. The Kaplan-Meier survival curve with log-rank analysis of DFS showed statistical significance between curves of patients with HHIP high-expression and low-expression (log-rank test).
In the present study, we first studied the expression of HHIP in the TCGA and HPA databases and found HHIP mRNA were downregulated in GC. These results were confirmed in the validation cohort, and HHIP mRNA expression levels in tumor tissues from 41 GC cases were significantly lower than the adjacent normal gastric tissue samples. HHIP immunostaining, both in the cytoplasm and cell membrane, also showed lower expression in the GC tissues compared with the normal gastric tissue samples. No previous studies had focused on HHIP expression in GIN, the precancerous lesion of GC. By IHC, our results revealed that HHIP immunostaining in the GIN tissues was reduced compared with the normal gastric tissue samples and increased in comparison to the GC tissue samples. Moreover, the negative and weak positive expressions were more common in the gastric cancer tissues samples. These results suggested that HHIP may act as a tumor suppressor. In addition, HHIP protein expression is an independent factor for predicting GC overall survival. All these data suggest that HHIP is a potential biomarker for GC diagnosis and prognosis.
HHIP downexpression is associated with a high tumor burden in human cancer [14, 17]. A previous study showed that HHIP downexpression is correlated with lymph node metastasis in colorectal cancer tissues and TNM stage in gastric cancers with strong Shh expression [14]. Consistently, our study discovered a significant association of lymphatic metastasis, nervous invasion and advanced pTNM stage with HHIP protein expression in GC. However, there was no correlation between HHIP mRNA and lymph node metastasis in our PCR cohort. In addition, two previous studies by Song Y et al. showed inconsistent results. They found no significant relationship between HHIP mRNA or protein expression with clinical features in GC [18, 19]. These discrepancies may be due to the differences in sample size, the detection methods, the use of different antibody clones and scoring methods. Since phenotypes prompt the underlying function, we further conducted in vitro experiment investigations and confirmed that HHIP may be a metastatic-related suppressor in GC.
In conclusion, by analyzing the TCGA and HPA databases and confirming the results in a cohort of in-house patients, we showed that both mRNA and protein levels of HHIP were significantly downexpressed in GC. Decreased HHIP expression during GC tumorigenesis and development and its prognostic correlation suggested that HHIP might have a potential role in GC prognostication. Combined with the functional experimental results that overexpression of HHIP decreased the metastatic ability of GC cells, our study provides evidence supporting HHIP as a promising suppressive target.
Overexpression of HHIP inhibits the metastasis of GC cells. A. HHIP baseline mRNA and protein expression levels in 5 gastric cancer cell lines and one normal gastric epithelial cell line (GES-1) by RT-qPCR (upper panel) and Western blotting (down panel); (mean±SD, with one-way ANOVA). B. The efficiency of HHIP overexpression were validated by qRT-PCR (upper panel) and Western blotting (down panel) in SGC-7901 and MGC-803 cells (mean±SD, with a paired t-test); *P<0.01. C. The representative pictures (left panel) and quantifications (right panel) of cell migration were analyzed using a wound-healing assay; images of SGC-7901 and MGC-803 cells were taken at 0 and 24 hours postscratch test (mean±SD, with paired t-test); *P<0.01. D. The cell invasion potential in SGC-7901 and MGC-803 cells was assessed using a transwell assay. The cell number was counted in 5 random view fields (mean±SD, with a paired t-test); *P < 0.01.
Acknowledgements
This study was supported by the National Human Genetic Resources Sharing Service Platform (2005DKA21300), National Natural Science Foundation of China (81602078, 81472220, 81802367, 81802361), Shanghai Science and Technology Development Fund (18ZR1408000, 17ZR1406500 and 15ZR1407400), and Hospital Foundation of Fudan University Shanghai Cancer Center (YJ201704).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7-30
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115-32
3. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059-87
4. Kobune M, Iyama S, Kikuchi S, Horiguchi H, Sato T, Murase K. et al. Stromal cells expressing hedgehog-interacting protein regulate the proliferation of myeloid neoplasms. Blood cancer journal. 2012;2:e87
5. Lin EH, Kao YR, Lin CA, Kuo TY, Yang SP, Hsu CF. et al. Hedgehog pathway maintains cell survival under stress conditions, and drives drug resistance in lung adenocarcinoma. Oncotarget. 2016;7:24179-93
6. Shahi MH, Rey JA, Castresana JS. The sonic hedgehog-GLI1 signaling pathway in brain tumor development. Expert opinion on therapeutic targets. 2012;16:1227-38
7. Zuo Y, Lv Y, Qian X, Wang S, Chen Z, Jiang Q. et al. Inhibition of HHIP Promoter Methylation Suppresses Human Gastric Cancer Cell Proliferation and Migration. Cell Physiol Biochem. 2018;45:1840-50
8. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & cellular proteomics: MCP. 2014;13:397-406
9. Bosanac I, Maun HR, Scales SJ, Wen X, Lingel A, Bazan JF. et al. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nature structural & molecular biology. 2009;16:691-7
10. Tada M, Kanai F, Tanaka Y, Tateishi K, Ohta M, Asaoka Y. et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14:3768-76
11. Eichenmuller M, Gruner I, Hagl B, Haberle B, Muller-Hocker J, von Schweinitz D. et al. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology (Baltimore, Md). 2009;49:482-90
12. Wang F, Anderson PW, Salem N, Kuang Y, Tennant BC, Lee Z. Gene expression studies of hepatitis virus-induced woodchuck hepatocellular carcinoma in correlation with human results. International journal of oncology. 2007;30:33-44
13. Martin ST, Sato N, Dhara S, Chang R, Hustinx SR, Abe T. et al. Aberrant methylation of the Human Hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer biology & therapy. 2005;4:728-33
14. Taniguchi H, Yamamoto H, Akutsu N, Nosho K, Adachi Y, Imai K. et al. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. The Journal of pathology. 2007;213:131-9
15. Shahi MH, Afzal M, Sinha S, Eberhart CG, Rey JA, Fan X. et al. Human hedgehog interacting protein expression and promoter methylation in medulloblastoma cell lines and primary tumor samples. Journal of neuro-oncology. 2011;103:287-96
16. Yang L, Bian Y, Huang S, Ma X, Zhang C, Su X. et al. Identification of signature genes for detecting hedgehog pathway activation in esophageal cancer. Pathology oncology research: POR. 2011;17:387-91
17. Chang L, Zhang P, Zhao D, Liu H, Wang Q, Li C. et al. The hedgehog antagonist HHIP as a favorable prognosticator in glioblastoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:3979-86
18. Song Y, Tian Y, Zuo Y, Tu JC, Feng YF, Qu CJ. Altered expression of PTCH and HHIP in gastric cancer through their gene promoter methylation: novel targets for gastric cancer. Molecular medicine reports. 2013;7:1159-68
19. Song Y, Zuo Y. Occurrence of HHIP gene CpG island methylation in gastric cancer. Oncol Lett. 2014;8:2340-4
20. Wittekind C, Compton CC, Greene FL, Sobin LH. TNM residual tumor classification revisited. Cancer. 2002;94:2511
21. Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q. et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2017;23:2071
22. Xu MD, Dong L, Qi P, Weng WW, Shen XH, Ni SJ. et al. Pituitary tumor-transforming gene-1 serves as an independent prognostic biomarker for gastric cancer. Gastric Cancer. 2016;19:107-15
Author contact
![]() Corresponding authors: Weiqi Sheng. M.D, PhD. Department of Pathology, Fudan University Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, People's Republic of China. E-mail: shengweiqi2006com; Tel: +86-21-64175590; Fax: +86-21-64174774. & Mi-die Xu, PhD. Department of Pathology, Fudan University Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, People's Republic of China. E-mail: xumd27202003com; Tel: +86-21-64175590; Fax: +86-21-64174774.
Corresponding authors: Weiqi Sheng. M.D, PhD. Department of Pathology, Fudan University Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, People's Republic of China. E-mail: shengweiqi2006com; Tel: +86-21-64175590; Fax: +86-21-64174774. & Mi-die Xu, PhD. Department of Pathology, Fudan University Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, People's Republic of China. E-mail: xumd27202003com; Tel: +86-21-64175590; Fax: +86-21-64174774.

 Global reach, higher impact
Global reach, higher impact