3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4677-4683. doi:10.7150/jca.26461 This issue Cite
Research Paper
High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis
1. The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health, Xiangya Hospital, Central South University, Changsha, Hunan, China;
2. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute and School of Basic Medical Sciences, Central South University, Changsha, Hunan, China;
3. Institute of Reproductive and Stem Cell Engineering, School of Basic Medical Science, Central South University, Changsha, China;
4. Department of Plastic Surgery, Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, Third Xiangya Hospital, Central South University, Changsha, China;
5. Hunan Key Laboratory of Translational Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China;
6. Department of Oral and Maxillofacial Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan, China.
Received 2018-4-3; Accepted 2018-9-9; Published 2018-11-24
Abstract
Long non-coding RNAs (lncRNAs) are dysregulated in various cancers. However, the clinical relevance and functional roles of AFAP1-AS1 in colon cancer (CC) have not been clarified. We analyzed the lncRNA expression patterns in Gene Expression Omnibus (GEO) datasets and the Cancer Genome Atlas (TCGA) RNA-seq datasets, and found that the expression level of AFAP1-AS1 was significantly elevated in CC tissues. High levels of AFAP1-AS1 were associated with poor disease-free survival and overall survival in CC patients. In vitro experiments demonstrated that AFAP1-AS1 knockdown significantly inhibited the cell invasive and migration capability in CC cell line HT-29. AFAP1-AS1 knockdown also increased the expression of E-cadherin and ZO-1 while inhibited the expression of Vimentin, MMP9, ZEB1 and β-catenin, suggesting that AFAP1-AS1 is involved in the epithelial-mesenchymal transition (EMT) process of CC. Further studies confirmed that AFAP1-AS1 knockdown also affected the actin-cytokeratin signaling pathway. Thus, AFAP1-AS1 might be a potential novel diagnostic marker and therapeutic target for CC.
Keywords: long non-coding RNA (lncRNA), AFAP1 antisense RNA 1 (AFAP1-AS1), colon cancer (CC), metastasis, prognosis
Introduction
Colon cancer (CC) is a common digestive system's cancer that ranks fourth in cancer-associated mortality in the world and has increasing morbidity each year [1, 2]. Although encouraging progress for diagnosing and treating CC has been achieved, the underlying mechanisms of tumor development have yet to be fully elucidated.
Long non-coding RNAs (lncRNAs) are a kind of non-coding RNA longer than 200 nucleotides and lack significant open reading frames (ORFs) [3, 4]. Previous studies show that lncRNAs function in many biological processes, including cell differentiation, proliferation, and apoptosis [5, 6]. Recently, studies have shown that dysregulated lncRNAs also play important roles in human cancers [7-13]. Several lncRNAs are upregulated in CC tissues and cell lines relative to normal samples and therefore serve as oncogenes. BCAR4 promotes tumor growth and metastasis in CC by activating the Wnt/β-catenin signaling pathway [14]. HOTAIR is associated with the epithelial-mesenchymal transition (EMT) in CC and functions as a prognostic marker [15]. HNF1A-AS1 greatly enhances cell metastasis [16]. However, some lncRNAs, such as HOXB-AS3 [17] and CASC7 [18], are down regulated in CC, indicating they may act as tumor suppressers and potential targets for clinical diagnosis, prognosis and therapy.
In this study, we found a candidate CC biomarker, AFAP1-AS1, by analyzing lncRNA gene expression data in the TCGA RNA-Seq and GEO datasets. AFAP1-AS1 was significantly elevated in CC tissues relative to normal tissues. We aimed to determine the expression level and prognostic significance of AFAP1-AS1 in CC as well as investigate the underlying molecular mechanisms.
Materials and methods
Data mining and analysis
The lncRNA expression data and clinical data of CC tissues and normal tissues were obtained from the Gene Expression Omnibus (GEO) profile database (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE39582 [19] and GSE37364 [20]). The TCGA data of lncRNA levels (RNA-Seq dataset) in CC patients were obtained from the Cancer RNA-Seq Nexus database [21] (CRN, http://syslab4.nchu.edu.tw/index.jsp) and the clinical prognosis data were obtained from the GEPIA database [22] (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/).
Cell culture
The human CC cell lines SW480, SW620, HCT116 and HT-29 were maintained in our laboratory. And cells were cultured in RPMI 1640 medium (GIBCO, USA) supplemented with 10% fetal bovine serum (FBS, GIBCO, USA), 100 U/ml penicillin and 100 ug/ml streptomycin (Invitrogen, USA) at 37℃ in a humidified incubator air with 5% CO2.
siRNA transfection
The sequences of the two siRNAs that targeted AFAP1-AS1 were siRNA1, 5'-GGGCTTCAATTTACAAGCATT-3' and siRNA2, 5'-CCTATCTGGTCAACACGTATT-3'. Sequences of non-target negative controls (NC) were obtained from Life Technologies. The cells were cultured in 6-well plates over night. And then these cells were transfected with either 50 nM of siRNA1 or siRNA2 or NC using Lipofectamine RNAiMAX Reagent (Invitrogen, Breda, The Netherlands) in accordance with the manufacturer's instructions.
qRT-PCR
The total RNA was extracted using Trizol agent according to the manufacturer's instructions. SYBR_PremixExTaq II kit (Takara, Dalian, China) was used to carry out the reactions. The CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) was used to measure the relative expression levels of each gene. The gene-specific primers used were as follows: AFAP1-AS1, 5'-AATGGTGGTAGGAGGGAGGA-3' and 5'-CACACAGGGGAATGAAGAGG-3'; E-cadherin, 5'-GAGAACGCATTGCCACATACAC-3' and 5'-AAGAGCACCTTCCATGACAGAC-3'; Vimentin, 5'-TGCCAACCGGAACAACGAT-3' and 5'-AATTCTCTTCCATTTCACGCATC-3'; MMP-9, 5'-AAGGGCGTCGTGGTTCCAACTC-3' and 5'-AGCATTGCCGTCCTGGGTGTAG-3'; ZEB1, 5'-GATGATGAATGCGAGTCAGATGC-3' and 5'-CTGGTCCTCTTCAGGTGCC-3'; β-actin, 5'-TCACCAACTGGGACGACATG-3' and 5'-GTCACCGGAGTCCATCACGAT-3'. β-actin was used as an endogenous control.
Wound healing assay
We performed wound healing assays to assess the migration capacity of tumor cells as described previously [23-26]. Briefly, after transfection, the cells were seeded into 6-well plates (5×105 cells/well) and incubated for 24 h. When the cell cultures reached 95% confluency, a 10 μl tip was used to create a scratch in the cell monolayer. Images were captured at different time points after scratching (0 h, 24 h and 48 h) by microscopy to measure gap closure after wounding.
Τranswell invasion assays
Transwell Cell Culture Inserts (BD Biosciences, New Jersey, USA) were used to measure the invasive capacity [27-29]. In brief, HT-29 cells (1×105 cells in 100 μl of 2% FBS medium) were seeded on the top chamber and then placed into 24-well plates. The bottom well contained 800 µl of growth medium with 15% FBS. After the cells were incubated for 36 h at 37°C, the cells on the upper surface were removed, while the cells that invaded through the filter pores were fixed in 4% paraformaldehyde for 30 min and stained with gentian violet. Invaded cells were imaged using a microscope, and the cell number was counted in 5 randomly selected high-power fields.
Western blotting
The cells were harvested after 48h of treatment, and proteins were extracted in three decontamination lysis steps. Protein was quantified with a BCA protein assay kit, separated by 10% SDS-PAGE gels and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). We then followed standard procedures using the following anti-bodies: E-cadherin, β-catenin, Vimentin, MMP9, ZEB1 and ZO-1 (Cell Signaling Technology, Danvers, MA, USA); MPRIP, CTTN, RHOA, RHOC, RAB1B and β-actin (Proteintech, Wuhan, China). Immunoreactive bands were visualized by electrochemiluminescence. Visualization of β-actin expression was used as a loading control.
Statistical analysis
All the numerical data are expressed as the mean ± standard deviation (SD). Student's t-tests were used to analyze the data. Curve fitting analyses were performed with GraphPad Prism Software (GraphPad Software, San Diego, CA, USA), and the results of the analyses were considered significant if P< 0.05.
Results
LncRNA AFAP1-AS1 is upregulated in CC
To explore the clinical relevance of AFAP1-AS1, we first investigated its expression in two publicly available CC datasets (GSE39582 and GSE37364). AFAP1-AS1 was significantly upregulated in CC tissues compared to normal tissues (Figure 1A &1B). We next validated AFAP1-AS1 expression in an independent CC RNA-Seq dataset (TCGA-COAD). The results revealed that CC tissues have higher AFAP1-AS1 expression relative to normal tissues (Figure 1C).
Association between AFAP1-AS1 expression and CC patient survival
Based on the data obtained from GEPIA, we analyzed the prognostic value of AFAP1-AS1 in CC in the TCGA-COAD RNA-Seq dataset. The patients were classified into two groups according to the AFAP1-AS1 expression level: the low AFAP1-AS1-expressing group (n = 105), in which the expression of AFAP1-AS1 was lower than the median value, and the high AFAP1-AS1-expressing group (n = 135), in which the expression of AFAP1-AS1 was higher than the median value. Compared with the patients in the low expression group, patients expressing high levels of AFAP1-AS1 had significantly shorter overall survival (OS) and disease-free survival (DFS) (Figure 2A & 2B).
Knockdown of AFAP1-AS1 negatively regulates CC cell migration and invasion
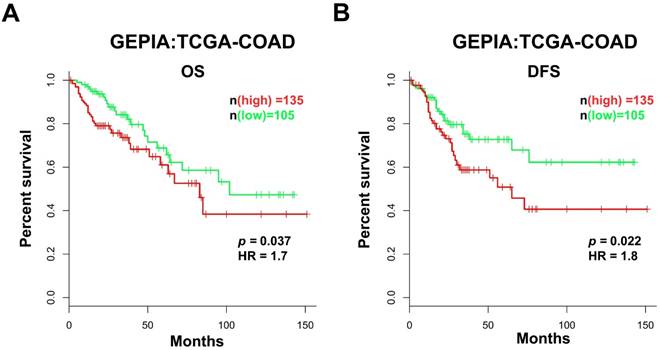
We measured the expression level of AFAP1-AS1 in 4 CC cell lines (SW480, SW620, HCT116, and HT-29) by qRT-PCR. Since HT-29 had the highest AFAP1-AS1 expression level (Figure 3A), we selected this cell line for further study. Two siRNAs targeting AFAP1-AS1 were transfected into HT-29 cells. The second siRNA (siRNA2) had a greater knockdown effect as measured by qRT-PCR (Fig. 3B). To elucidate the role of AFAP1-AS1 in CC, siRNA2 or a negative control (NC) siRNA was transfected into HT-29 cells. Migration studies revealed that the gap distance in the siRNA2-treated group was much longer than the NC group, suggesting that the knockdown of AFAP1-AS1 inhibited cell migration ability (Figure 3C & 3D). Transwell matrigel invasion assay was used to assess the effects of AFAP1-AS1 knockdown on cell invasion. As expected, the number of invading cells in the AFAP1-AS1 knockdown group was much lower than the number in the NC group (Figure 3E &3F).
Silencing AFAP1-AS1 influences the expression of EMT-related genes
To explore how AFAP1-AS1 regulates CC cell migration and invasion, we measured the expression level of EMT-related genes, E-cadherin, vimentin, MMP9, ZEB1, β-catenin and ZO-1 in CC cells after AFAP1-AS1 silencing. As shown by our qRT-PCR results, silencing AFAP1-AS1 upregulated the expression of E-cadherin while inhibiting the expression of vimentin, MMP9 and ZEB1 (Figure 4A). We also measured protein levels of EMT-marker proteins by western blotting. Similarly, silencing AFAP1-AS1 upregulated E-cadherin and ZO-1 protein while downregulating β-catenin, vimentin, MMP9 and ZEB1 (Figure 4B).
AFAP1-AS1 is elevated in CC tissues relative to normal tissues. The AFAP1-AS1 expression data in CC tissues were obtained from the GEO datasets in accession GSE39582 (A) and GSE37364 (B), Analysis revealed that CC tissues have a higher AFAP1-AS1 expression level than normal tissues. (C) Comparison of AFAP1-AS1 expression levels in CC tissues and normal tissues from the TCGA-COAD RNA-Seq dataset within the Cancer RNA-Seq Nexus database.
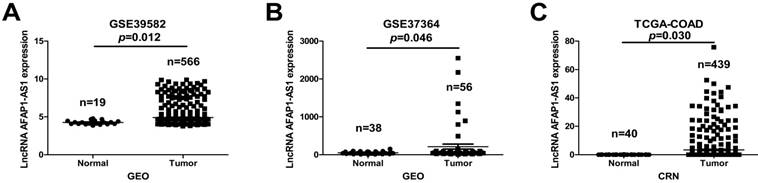
High AFAP1-AS1 is associated with poor prognosis in CC patients. CC patients with high AFAP1-AS1 have shorter overall survival (OS, A) and disease-free survival (DFS, B). Data were obtained from the TCGA-COAD RNA-Seq dataset within the GEPIA database. The Kaplan-Meier method was used to calculate curves.
Knockdown of AFAP1-AS1 influences the actin-cytokeratin signaling pathway
Previous studies have demonstrated that down regulation of AFAP1-AS1 suppressed tumor migration and invasion by regulating EMT-related genes. Future studies should investigate which signaling pathways are involved in tumor progression. In a previous study, we showed that RHOA, RAC2, RAB10 and RAB11A were significantly upregulated, while RHOC, RAB11B and LASP1 were down regulated after silencing AFAP1-AS1 in nasopharyngeal carcinoma cells [30]. We therefore measured these molecules in CC cells. Western blotting analysis showed that CTTN, RHOA and RAB1B were elevated, while MPRIP was lower after silencing AFAP1-AS1 (Figure 4C).
Discussion
Emerging evidence indicates that lncRNAs play important roles in multiple physiological and pathological processes, including tumor development [31-37]. AFAP1-AS1, also known as LOC84740, is located on chromosome 4p16.1 and acts as an antisense lncRNA regulator of the AFAP1 gene [30]. AFAP1-AS1 was originally identified in esophageal adenocarcinoma [38], is frequently upregulated in CC and serves a vital role in a variety of tumor types, including pancreatic cancer, hepatocellular carcinoma and lung cancer [39-42]. In liver cancer, AFAP1-AS1 is highly expressed and promotes cell growth by activating the RHOA/RAC2 pathway [43].
Previous studies confirmed via qRT-PCR that AFAP1-AS1 was frequently highly expressed in colorectal cancer (CRC) tissues and that AFAP1-AS1 is a potential diagnostic biomarker for CRC patients [44]. In the present study, we collected data from different platforms to determine the expression tendency of AFAP1-AS1 and its prognostic value in CC. Our analysis is consistent with that of previous studies; AFAP1-AS1 was elevated in CC tissues. Our findings also confirmed that AFAP1-AS1 expression was highly correlated with poor prognosis. These findings suggest that AFAP1-AS1 may act as an oncogene in CC. We found that HT-29 cells had a higher expression level of AFAP1-AS1. Therefore, HT-29 cells were used to further explore the role of AFAP1-AS1 in CC. We found AFAP1-AS1 knockdown significantly suppressed cell migration and invasion ability. We also determined that AFAP1-AS1 acts as a tumor promoter. EMT was characterized by cancer cell migration and invasion. E-cadherin suppression is the first step of EMT initiation, resulting in reduced cell adhesion. Our data suggest that AFAP1-AS1 knockdown causes the upregulation of E-cadherin while inhibiting Vimentin, MMP9 and ZEB1 mRNA expression. Furthermore, AFAP1-AS1 knockdown increases E-cadherin and ZO-1 protein levels while decreasing β-catenin, Vimentin, MMP9 and ZEB1. These findings provide evidence for the anti-metastasis action of AFAP1-AS1 silencing.
Cell migration and invasion are important aspects of cancer progression, and studies have highlighted the key signaling pathways that mediate tumor cell migration and invasion through regulation of the cytoskeleton assembly [45-47]. In a previous study, we confirmed that some molecules associated with the actin-cytokeratin signaling pathway were regulated by AFAP1-AS1 in nasopharyngeal carcinoma [30]. We also measured the molecules key to the actin-cytokeratin signaling pathway in the present study and found that CTTN, RHOA and RAB1B were elevated while MPRIP decreased after silencing AFAP1-AS1. These results suggest that AFAP1-AS1 knockdown might affect tumor cell invasion and migration through regulation of the actin-cytokeratin signaling pathway.
In conclusion, we found and confirmed that AFAP1-AS1 is upregulated in CC. AFAP1-AS1 knockdown significantly suppressed CC progression by regulating the actin-cytokeratin signaling pathway and EMT-related genes, indicating that AFAP1-AS1 may be a novel therapeutic target for CC.
AFAP1-AS1 knockdown inhibits cell migration and invasion. (A) AFAP1-AS1 expression levels across four CC cell lines were measured by qRT-PCR. (B) Expression levels of AFAP1-AS1 in HT-29 cells were measured by qRT-PCR after transfected with siRNAs targeting AFAP1-AS1 (siRNA1 and siRNA2), or a scrambled negative control (NC). qRT-PCR results indicated that the siRNA2 had the strongest inhibitory effect. (C) Representative images of cells that migrated into the scratched area and (D) the gap distance of each groups at 0 h, 24 h and 48 h. (E) Representative images of invasive cells transfected with NC or with siRNA2. (F) Measurement of the relative number of invaded cells in each group.*P< 0.05, **P < 0.01, ***P < 0.001.
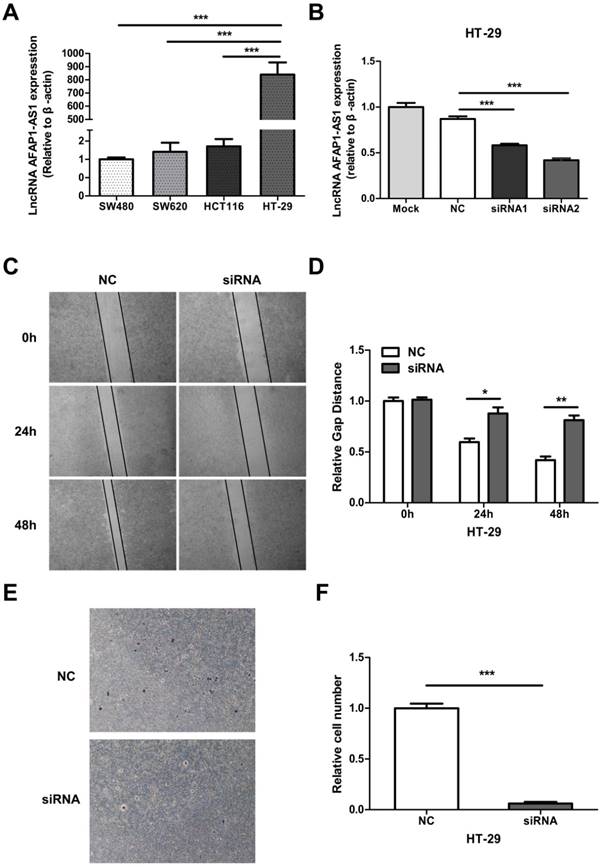
AFAP1-AS1 knockdown influences EMT-associated gene expression and the actin-cytokeratin signaling pathway. (A) qRT-PCR-measured mRNA expression of E-cadherin, vimentin, MMP9 and ZEB1 in siRNA2-transfected HT-29 cells. (B) The protein expression of E-cadherin, Vimentin, MMP9, ZEB1, β-catenin and ZO-1 were detected by Western blotting. (C) Western blotting analysis showed that CTTN, RHOA and RAB1B were elevated while MPRIP decreased after silencing AFAP1-AS1.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81572787, 81672683, 81772901, 81702907, 81772928 and 81803025), the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2015, 2018JJ3704, 2018JJ3815, 2018SK21210 and 2018SK21211), the Fundamental Research Funds for the Central Universities of Central South University (1053320171023) and the Special Fund of Clinical Medicine of Chinese Medical Association (17020280697).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
2. Wei F, Wu Y, Tang L, Xiong F, Guo C, Li X. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1271-5
3. Gong Z, Zhang S, Zhang W, Huang H, Li Q, Deng H. et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120-4
4. Wang JP, Tang YY, Fan CM, Guo C, Zhou YH, Li Z. et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2017;9:12487-502
5. Gong Z, Zhang S, Zeng Z, Wu H, Yang Q, Xiong F. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674
6. Yang L, Tang Y, Xiong F, He Y, Wei F, Zhang S. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2017;9:1426-43
7. He Y, Jing Y, Wei F, Tang Y, Yang L, Luo J. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235
8. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130
9. Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
10. Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683-95
11. Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X. et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545-54
12. Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y. et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134-42
13. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79
14. Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:92815-26
15. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S. et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395-402
16. Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B. et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50-62
17. Huang JZ, Chen M, Chen, Gao XC, Zhu S, Huang H. et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell. 2017;68:171-84 e6
18. Zhang Z, Fu C, Xu Q, Wei X. Long non-coding RNA CASC7 inhibits the proliferation and migration of colon cancer cells via inhibiting microRNA-21. Biomed Pharmacother. 2017;95:1644-53
19. Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453
20. Valcz G, Patai AV, Kalmar A, Peterfia B, Furi I, Wichmann B. et al. Myofibroblast-derived SFRP1 as potential inhibitor of colorectal carcinoma field effect. PLoS One. 2014;9:e106143
21. Li JR, Sun CH, Li W, Chao RF, Huang CC, Zhou XJ. et al. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016;44:D944-51
22. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
23. Wei F, Tang L, He Y, Wu Y, Shi L, Xiong F. et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9:432
24. Wei F, Wu Y, Tang L, He Y, Shi L, Xiong F. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer. 2018;118:233-47
25. Wang W, Yi M, Zhang R, Li J, Chen S, Cai J. et al. Vimentin is a crucial target for anti-metastasis therapy of nasopharyngeal carcinoma. Mol Cell Biochem. 2018;438:47-57
26. Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C. et al. Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens. J Virol. 2017:91 pii: e00301-17. doi: 10.1128/JVI.00301-17
27. Tu C, Zeng Z, Qi P, Li X, Guo C, Xiong F. et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole genome sequencing. Carcinogenesis. 2018 doi: 10.1093/carcin/bgy108. [Epub ahead of print]
28. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y. et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852-64
29. Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87
30. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
31. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46
32. He B, Li W, Wu Y, Wei F, Gong Z, Bo H. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353
33. Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
34. Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16:42
35. Wang Y, Mo Y, Yang X, Zhou R, Wu Z, He Y. et al. Long non-coding RNA AFAP1-AS1 is a novel biomarker in various cancers: a systematic review and meta-analysis based on the literature and GEO datasets. Oncotarget. 2017;8:102346-60
36. Yang L, Tang Y, He Y, Wang Y, Lian Y, Xiong F. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97-103
37. Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang Y. et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523-30
38. Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R. et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956-66 e4
39. Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother. 2015;75:8-11
40. Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L. et al. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699-707
41. Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y. et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137
42. Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729-37
43. Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY. et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590-8
44. Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152-9
45. Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171:5507-23
46. Tang Y, He Y, Zhang P, Wang J, Fan C, Yang L. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77
47. Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45
Author contact
![]() Corresponding authors: Fang Xiong, or Zhaojian Gong. Tel: 86-731-84805446; E-mail: xiongfedu.cn or gongzhaojian4458edu.cn
Corresponding authors: Fang Xiong, or Zhaojian Gong. Tel: 86-731-84805446; E-mail: xiongfedu.cn or gongzhaojian4458edu.cn

 Global reach, higher impact
Global reach, higher impact