3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4684-4695. doi:10.7150/jca.27381 This issue Cite
Research Paper
OIP5 Promotes Growth, Metastasis and Chemoresistance to Cisplatin in Bladder Cancer Cells
1. Department of Urology, Shenzhen Second People's Hospital, Guangzhou Medical University, Guangdong, China.
2. Carson International Cancer Center, Shenzhen University School of Medicine, Shenzhen, China
3. College of pharmacy, Guangdong Pharmaceutical University, Guangdong, China.
4. Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China.
5. Guangdong Key Laboratory of Systems Biology and Synthetic Biology for Urogenital Tumors, Shenzhen, China.
* The three authors contributed to this work equally.
Received 2018-5-21; Accepted 2018-8-12; Published 2018-11-24
Abstract
Opa interacting protein 5 (OIP5) has previously been identified as a tumorigenesis gene. The purpose of this study is to explore the role of OIP5 in the progression of bladder cancer (BC). The OIP5 expression and clinical behaviors in bladder cancer were collected from lager database. Our study showed that OIP5 was highly expressed in bladder cancer tissues and cells. Overexpression of OIP5 in tumor patients predicted worse overall survival (OS) and higher histological grade. Vitro and vivo experiments demonstrated that knockdown of OIP5 significantly inhibited cell growth of BC. Scratch assay and transwell assay suggested that migration capacity of BC cells was decreased after knockdown of OIP5. Cisplatin sensitivity assay indicated that depletion of OIP5 increased the sensitivity of BC cells to cisplatin. Finally, we identified 38 overlapping differentially expressed genes (DEGs) between RNA-seq and TCGA analyses which were closely linked to OIP5. Bioinformatics analysis showed that these DEGs enriched in oocyte meiosis, fanconi anemia pathway, cell cycle, and microRNAs regulation. TOP2A, SPAG5, SKA1, EXO1, TK1 were confirmed to associated with bladder cancer development. Our study suggests that OIP5 may be a potential biomarker for growth, metastasis and drug-resistance in bladder cancer.
Keywords: OIP5, bladder cancer, metastasis, cisplatin-resistance
Introduction
Bladder cancer is the ninth most common malignancy worldwide and the most common cause of death in genitourinary tumor[1]. It was reported that 81, 190 new cases of bladder cancer and 17, 240 cancer-related deaths are predicted to occur in 2017 in the United States[2]. For non-muscle-invasive bladder cancer(NMIBC), the standard treatment is transurethral resection of bladder tumour (TURBT), followed with risk-based intravesical pharmacotherapy, but more than 70% of the patients tend to recrudescence and nearly 25% will develop to muscle-invasive bladder cancer (MIBC)[3]. MIBC is frequently managed with radical cystectomy with neoadjuvant chemotherapy[4]. Although BC patients can acquire benefits from multiple therapeutic interventions, involved in refining surgical techniques, continuously improving chemotherapy and radiotherapy regimens, even immunotherapy, targeted therapy, platinum-based systemic chemotherapy remains the cornerstone of agents for advanced disease[5]. However, patients with BC frequently suffer from its high recurrence and the 5-year survival rate for patients remains at 50-60% [6]. In addition, the clinical success of BC always compromised due to early metastasis and drug resistance. Consequently, extensive investigations of potential diagnosis biomarkers and overcoming drug resistance for BC patients are urgently needed.
Opa interacting protein 5 (OIP5), a protein coding gene that is located on chromosome 15, belongs to cancer/testis antigens (CTAs)[7]. OIP5 encodes a 25-kDa protein with a coiled-coil domain which was originally found as an Opa (Neisseria gonorrhoeae opacity associated) interacting protein by yeast two-hybrid analysis[8]. Previous studies have demonstrated that OIP5 was highly expressed in various cancers, involved in various biological processes of tumor[9, 10]. OIP5 was identified as a CTAs in gastric cancer, which indicated it could be a novel target for cancer specific immunotherapy [11]. Gong et al found that overexpression of OIP5 was inclined to get a shorter overall survival time in clear cell renal cell carcinoma [10]. Chun et al suggested that knockdown of OIP5 inhibited cell growth and induced apoptosis in colorectal and gastric cancers [12]. Although it has been reported that OIP5 participated in the proliferation, apoptosis, and cell cycle process of BC[13], the relationship between OIP5 and migration or drug resistance in BC is unknown.
Platinum-based chemotherapy remains regarded as the first-line chemotherapeutic regimens for ovarian, gastric, lung, testicular as well as bladder cancer[14]. However chemoresistance to cisplatin occurs frequently. The mechanisms that tumors development resistant to platinum drugs are multifactorial, including: 1)Reduced intracellular drug uptake. For example, copper transporter 1 (CTR1) has been found to play an important role in the uptake of the platinum drug. And downregulation of CTR1 could lead to poor therapeutic response to cisplatin; 2)Promoted drug efflux by increasing cellular glutathione(GSH). For example, Kotoh et al found that the content of GSH was significantly increased in the cisplatin-resistant BC cell line (T24/DDP7), compared with that of the T24 parental line; 3)Enhanced repair of DNA damage through reactivation of DNA repair pathway, such as nucletide excising repair (NER), and homologous recombination repair(HR); 4)Defects in apoptosis pathway, for example the loss expression of p53[15]. Moreover, the role of non-coding genes in the regulation of chemoresistance has attracted increasing attention. As a study found that overexpression of miR-218 was responsible for the enhancement of cisplatin sensitivity in T24 and EJ cells by reduced glucose uptake, cellular GSH content and enhanced ROS [16].
In the present study, we found that knockdown of OIP5 inhibited the proliferation, metastasis, and increased the sensitivity to cisplatin in BC. Our results demonstrated that OIP5 may be used as a potential therapeutic target to tackle with patients of bladder cancer.
Materials and Methods
Large database information mining and arrangement
The tissue expression and clinical information of OIP5 were collected from The Cancer Genome Atlas (TCGA) and Sequence Read Archive (SRA) database. Relevant mRNA expression and clinical data of bladder cancer in TCGA cohort (TCGA-BLCA) were download from UCSC Xena (https://xenabrowser. net/heatmap/#, up to October, 22, 2017). Data from Sequence Read Archive under accession SRA063495 was based on whole-genome sequencing of bladder cancer [17]. The mRNA expression of OIP5 in different urinary tract cancer cell linces was searched from Cancer Cell line Encyclopedia (CCLE)[18].
Cell culture
Human bladder transitional cell carcinoma cell lines (SW780, 5637), human normal bladder epithelial cell line(SVHUC-1) and human embryonic kidney 293 cell lines (293T) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The 5637 cells were cultured in RPMI-1640 (1640) (Gibco, USA), supplemented with10% fetal bovine serum (Gibco). The SW780 and 293T cells were cultured in Dulbecco's modified Eagle's medium(DMEM) (Gibco), supplemented with 10 % fetal bovine serum (Gibco). The SV-HUC-1 cells were cultured in F12K medium (Gibco), supplemented with 10 % fetal bovine serum(Gibco). All cells were maintained at 37°C with a humidified atmosphere of 5 % CO2 in incubator.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Quality of extracted total RNA was evaluated according to 260/280 absorbance ratio, measured by Nano Drop spectrometer (Thermo Scientific, Waltham, MA, USA). Revertra Ace qPCR RT Kit (Toyobo, Japan) was used to transfor mRNA to cDNA. The mRNA expression levels of OIP5 were measured by using SYBR® Premix Ex TaqTM (TaKaRa, Japan) according to the user's manuals. The qPCR primers are listed in Table 1. The PCR mixtures were prepared according to the manufacturer's protocols with PCR conditions of 40 cycles of 15 sec at 95°C, 20 sec at 55°C, and 30 sec at 70°C on a ABI PRISM 7300 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). The 2-△△ct method was used to calculate the relative amount of OIP5. All experiments were carried out at least three repetitions.
Relative primers used in this research.
| Name | Sequences |
|---|---|
| OIP5-F | CTTGTGGGATTCCCGTTGGTT |
| OIP5-R | TGTGCGTTAGCACTATCTTCTCT |
| GAPDH-F | CGCTCTCTGCTCCTCCTGTTC |
| GAPDH-R | ATCCGTTGACTCCGACCTTCAC |
Note: F, forward primer; R, reverse primer
Stable transfected cells construction
A lentivirus-mediated miRNA targeting OIP5 was designed and synthesized by SyngenTech (BeiJing, China). The sequence of miR-OIP5 is: TGCTGTAGAAGTCAACATTAAGCCCAGTTTTGGCCACTGACTGACTGGGCTTAGTTGACTTCTACAGG. The sequence of miRNA-NC is: TGCTGCATACGGCCCACAGGTATTTCGTTTTGGCCACTGACTGACGAAATACGTGGGCCGTATGCAGG. The 293T cells were transfected with shuttle vectors combined with helper plamids of pMD2. G and psPAX. 2 by Lipofectamine 3000 following the manufacturer's protocol (Invitrogen, USA). The supernatant was harvested after 48 h, 72h, and then filtered through 0. 45μm PVDF filter. Then, the lentivirus supernatant was concentrated by lentivirus concentration solution (1:5, BioGeek) at 4℃ overnight. After concentration, the lentivirus particles were collected and resuspend. The target cells were infected by the viral liquid with polybrene (8ug/ml, HanBio). After 48 h of infection, the stably transduced cells were selected with puromycin. Finally, the selected clones were expanded for further studies.
Western Blot Assays
Cells were collected and lysed in RIPA buffer (Beyotime, ShangHai, China) with protease inhibitor on ice for 30 min, followed by centrifugation at 14, 000g for 10 minutes at 4°C. BCA protein assay kit (Pierce, USA) was used to quantify the protein concentration. The protein was diluted in 4× loading buffer (TransGen, BeiJing), and then boiled for 5 min at 100℃. Equal amounts of protein was loaded onto SDS-PAGE (10%) gels for electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, USA). The membranes were blocked with 5% Albumin Bovine (BSA; Biotechnology, Lot# 0164C419) for 1 hour at room temperature and then incubated with the following primary antibodies at 4℃ overnight: mouse monoclonal anti-β-actin (1:1000; Beyotime, ShangHai), rabbit monoclonal anti-OIP5 (1:200; Sigma, St. Louis, MO USA). After washing 3 times, the membranes were incubated with a secondary antibody (1:5000 dilution; Abcam) for 2 hours. Then, the membranes were visualized after development with an enhanced chemiluminescence (ECL) substrate (Thermo, USA). Chromogenic reaction was performed by Image Lab 5. 2 software (GelBoc, USA). Finally, Adobe Photoshop software(Adobe Systems, USA) was used to analyze the protein bands.
Cell proliferation assay
Cell Counting Kit-8, CCK-8 (TransGen, Beijing, China) and 5-ethynyl-20-deoxyuridine (EdU) assay kit (Ribobio, Guangzhou, China) were used for cell proliferation according to the manufacturer's instructions. For CCK-8 assay, the stably transfected cells of miR-OIP5 and miR-NC group were seeded in a 96-well plates, and cultured in normal medium. At 24, 48 or 72 h, 96h, 10 μl CCK-8 reagent was added to each well and cultured for 1 h. The absorbance at 450 nm was detected by automatic microplate reader (Bio-Rad, Hercules, CA, USA).
For EdU assay, stably transfected cells were seeded in a 12-well flat-bottomed plate, and incubated with 100μl of 50μM EdU per well for 2h. Then, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After removing the buffer, the cells were incubated with 200μl of 2mg/ml glycine for 5min followed by washing with 200μl of PBS. The cells in each well were then treated with 0. 5% Triton X-100 for 20 min at room temperature for permeabilization. After washing with PBS three times, cells were added with 200μl of 1X Apollo solution for 30 min at room temperature in the dark. After that, cells were incubated with 200μl of Hoechst33342 for 30 min at room temperature in the dark followed by washing with PBS. The cells were then observed using fluorescence microscopy. All experiments were performed three times.
Tumor xenograft model and tumorigenicity assay
SW780 cells stably transfected with OIP5 knockdown vector (miRNA-OIP5) and its negative control (miRNA-NC) were employed to the tumorigenicity assay. Five female BALB/c athymic nude mice (Five-six weeks old, 16-20g) were purchased from Beijing Vital River Laboratory Animal Technology Co, Ltd. All mice were housed in a climate-control SPF (Specific Pathogen Free) facility. Cell suspension (1× 107 cells in 200 μl PBS per mouse) was subcutaneously injected into the flanks of the nude mice. And, the miRNA-OIP5 cell line was injected on the right side, and the miRNA-NC cell line was injected on the left side, respectively. The tumor size, volume and mice weight were measured twice a week. The tumor volume was calculated using the formula: length × width2 × 1/2. After inoculation 4 weeks, the mice were euthanized and the tumors were removed and weighed.
Cell migration assay
Cell migration ability was measured by scratch assay. Cells were seeded in 6-well plates and incubated in an incubator to get 100% confluence. Clear lines in the wells were generated using a sterile 200 μl pipette tip. A digital camera system was used to take pictures from each well quickly. 24 hours later, pictures were taken again. Migration distance was measured at the time of 0 h and 24 h. The wound gaps were digital quantified using Image-Pro Plus software (Version 5. 1, Media Cybernetics, Inc. ).
Transwell assay also be adopted to further confirm the result. Transwell insert with 8. 0μm pores (COStor, Alfred Road, Kennebunk ME, USA) were placed in a 24-well plate. 5x105 cells suspended with 200μl FBS-free medium were added to the top chamber and 500μl medium supplemented with 10% FBS was added to the bottom chamber. After 24 hour of incubation at 37℃, cells that migrated through the chamber membrane were fixed in 4% paraformaldehyde and then stained with 0. 1% crystal violet (Sigma-Aldrich). After dyed for 20 min, the redundant dyes adhered to the upper surface of the polycarbonate membrane was removed by cotton swab. The invaded cells were observed and photographed using microscopy. Then, the chambers were soaked into 1ml 3% glacial acetic acid to wash out the crystal violet with the invaded cells. After 10 min, 100μl elutropic crystal violet was added into per well of 96-well plates, and the absorbance was measured at a wavelength of 570nm using a microplate reader (Bio-Rad, Hercules, CA, USA). Each experiment was carried out at least three times.
Drug sensitivity assay
The CCK-8 assay(TransGen, Beijing, China) was used to measure the cell viability and half maximal inhibitory concentration (IC50). Cells were seeded in 96-well plates at a concentration of 8x103 cells per well and cultured overnight. When the cells reached about 60% confluence, different concentrations of cisplatin was added to the wells. After treated with 72 h, 10 μl of CCK-8 reagent with 90 μl of culture medium was added into each well and incubated for 3 hours. Then, optical density (OD) value at the wavelength of 450nm was measured using a microplate reader (Thermo). The cell viability and IC50 was calculated using GraphPad Prism 5.0 software. The half maximal inhibitory concentration (IC50) was measured by linear regression. The concentrations of cisplatin was as follows:SW780 cells (0, 0.794, 1.587, 3.175, 6.35, 12.7, 25.4, 50.8, 101.6, and 203.2 μM/mL), 5637 cells (0, 0.287, 0.574, 1.148, 2.295, 4.59, 9.18, 18.36, 36.72, and 73.44 μM/mL). Each experiment was repeated three times.
RNA sequencing and Bioinformatics analyses
RNA was isolated from OIP5 stable knockdown of SW780 cell lines (miR-OIP5) and its negative control (miR-NC). The differentially expressed genes (DEGs) in both samples were screened using Illumina's Genome Analyzer II platform. Pearson test was used to search for genes related to OIP5 in TCGA cohort (TCGA-BLCA). The candidate genes which absolute value of Pearson's r>0.6 was the included object. Then, we sought intersection of mRNAs between the RNA-seq and TCGA cohort. To assess the potential function of the consensus target genes, Database for Annotation, Visualization and Integrated Discovery (DAVID) [19] was used to enrich the pathway in Kyoto Encyclopedia of Genes and Genomes (KEGG). The P value < 0.05 and gene count >2 generated by DAVID was defined as the cutoff criterion. And the assessment and integration of protein-protein interactions were processed by STRING v10.5 database [20] and modeled by Cytoscape v3. 5. 1. The minimum required interaction score generated by STRING were set in medium confidence (0. 400).
Statistical analyses
All the experiments were independently repeated at least three times and presented as mean ± standard deviation (SD). SPSS 20.0 (IBM, Armonk, NY) was used to perform the statistical analysis. All experimental data were analyzed by Student's t-test or ANOVA and P< 0.05 was considered statistically significant. In addition, GraphPad Prism 6 (La Jolla, CA, USA) was used to deal with all data.
Result
OIP5 was upregulated in bladder cancer, and its clinicopathologic characteristics
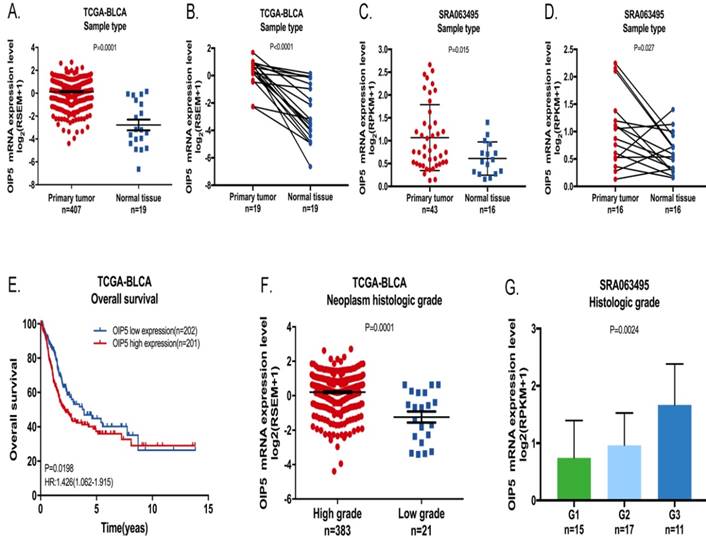
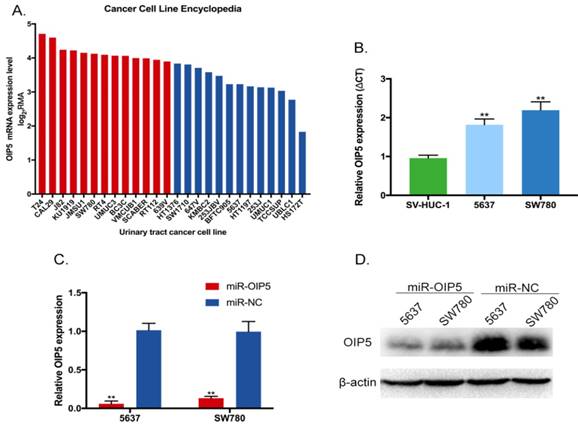
We explored the tissue expression and clinical characteristics of OIP5 through large database mining. Initially, data from the TCGA-BLCA (Fig. 1A, 1B) suggested that OIP5 had a higher expression in tumor tissues(n=407) than that in matched normal peritumoral tissues(n=19). Likewise, compared with para-cancer tissues(n=16), the OIP5 expression was significantly upregulated in tumor tissues(n=43) based on SRA database (Fig. 1C, 1D). Fig. 2A exhibited the OIP5 expression level in various bladder cancer cell lines searched from Cancer Cell line Encyclopedia (CCLE). Then, the relative expression level of OIP5 was detected using real-time qPCR in bladder cancer and normal urothelial cell lines. As shown in Fig. 2B, OIP5 was significantly upregulated in 5637 (P<0. 01) and SW780 (P<0. 01) compared to SV-HUC1 cell line. Meanwhile, overexpression of OIP5 was correlated with poor overall survival(OS) (P = 0. 0198) (Fig. 1E) and advanced histological grade(p=0. 0001) ( Fig. 1F) for BC patients in TCGA database. In addition, the results based on the SRA063495 database further verified that the differential expression of OIP5 was correlated with tumor grade (Fig. 1G). These results manifested that OIP5 was overexpressed in bladder cancer and contributed to patients prognosis.
Efficiency of OIP5 was suppressed by lentivirus-mediated miRNA infection in SW780 and 5637 cells
To assess the phenotypes of OIP5 in BC, 5637 and SW780 cell lines were stably transfected with lentiviral vector (miRNA-OIP5), as well as negative control (miRNA-NC). The knockdown efficiency of OIP5 was determined by RT-Qpcr and western blotting (WB). And, the expression levels of OIP5 were obviously down-regulated in cells infected by miRNA-OIP5 compared with miR-NC in mRNA (Fig. 2C) and protein (Fig. 2D) levels.
Knockdown of OIP5 inhibited cell growth in vitro and vivo
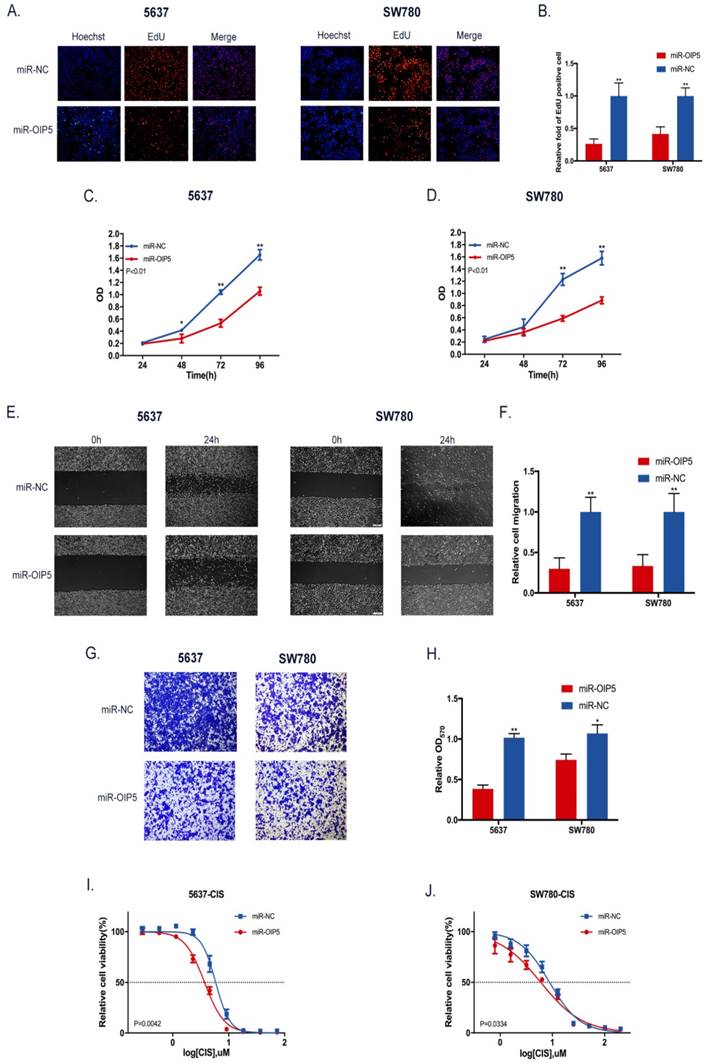
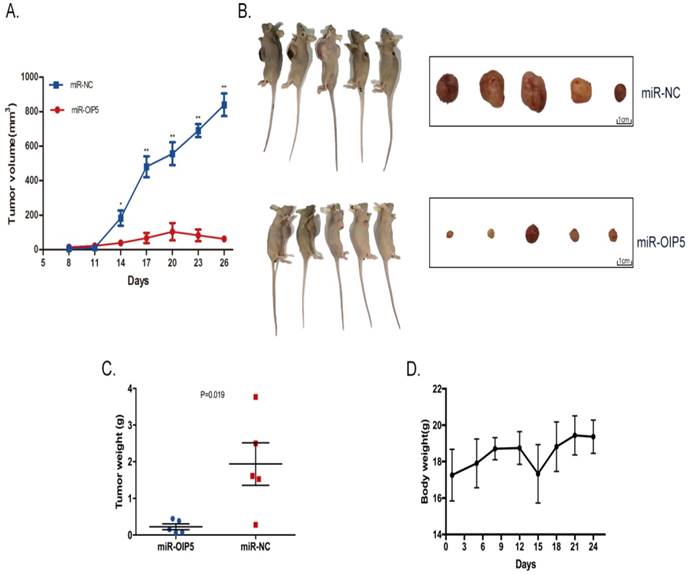
CCK-8 and EdU assays were utilized to determine whether OIP5 promotes cell proliferation in bladder cancer. As detected in CCK-8 (Fig. 3C, 3D), stably knockdown of OIP5 remarkably inhibited cell proliferation in miRNA-OIP5 cells compared with miR-NC cells. Then EdU assay proved that the rate of EdU positive of 5637 and SW780 cells were significantly decreased in miR-OIP5 group compared with miR-NC group(Fig. 3A). Meanwhile, the quantitative results suggested that the number of EdU positive cells in miR-OIP5 group was approximately decreased by 73. 7% in 5637 cells (P < 0. 01) and 58. 45% in SW780 cells (P < 0. 01) (Fig. 3B). Furthermore, we also employed xenograft assay to verify the results in vivo. Xenograft experiment showed that tumors formed by miR-OIP5 cells exhibited a smaller volume (Fig. 4A, 4B) and lighter weight( Fig. 4C) relative to control group. Fig. 4D showed the mouse weight fluctuation during the experiment. These results indicated that knockdown of OIP5 significantly inhibited cell growth of bladder cancer.
The OIP5 expression levels and clinical data in BC based on large database. OIP5 expression was higher in bladder cancer tissues than those in para-cancer tissues analyzed by TCGA-BLCA (A, B) and SRA063495(C, D). Correlation between OIP5 expression and OS based on TCGA-BLCA cohort (E). Correlation between OIP5 expression and histologic stage of bladder cancer patients based on TCGA database(F) and SRA063495(G).
OIP5 was up-regulated in BC cells and inhibited by miRNA-OIP5 lentivirus infection. (A) The OIP5 expression levels in various bladder cancer cell lines searched from Cancer Cell line Encyclopedia(CCLE). (B) The relative mRNA expression of OIP5 was higher in bladder cancer cell lines (SW780, 5637) than those in normal urothelial cell line(SV-HUC1). (C, D) The expression levels of OIP5 were knockdown in mRNA and protein levels by infected with miRNA-OIP5. Data are shown as mean ± SD, (**P<0. 01).
Knockdown of OIP5 inhibited cell migration of bladder cancer
To investigate the role of OIP5 in the cell migration and invasion in BC, the cell scratch and transwell assay were adopted. Scratch assay illustrated that migration area of miR-NC group was obviously greater than that of miR-OIP5 group in two cell lines (Fig. 3E). The ratio of the relative migration in the miRNA-OIP5 group was reduced by 70. 14% in 5637 (P <0. 01) and decreased by 66. 68% in T24 (P <0. 01) (Fig. 3F). Likewise, the transwell assay showed that invaded cells were obviously decreased after miR-OIP5 vector treatment compared to the miR-NC group. (Fig. 3G, 3H). These results indicated that knockdown of OIP5 suppresses cell migration capacity of bladder cancer.
Knockdown of OIP5 sensitized SW780 and 5637 cells to cisplatin
We explored the association between OIP5 expression and cisplatin chemoresistance in BC cells using drug sensitivity assay. In the cell viability assay, OIP5 downregulation markedly increased the sensitivity to cisplatin both in 5637 and SW780 cells, compared with the negative control cells (Fig. 3I, 3J). The IC50 values of cisplatin for OIP5 down-regulated cells was approximately reduced by 1. 58 times in 5637 (P = 0. 0042) and by 1. 47 times in SW780 (P = 0. 0334), compared with that of control cells. These result confirmed that knockdown of OIP5 both in 5637 and SW780 cells by lentivirus-mediated miRNA rendered the cells more sensitive to cisplatin compared with the negative control cells. Our results indicated that OIP5 may play significant roles in chemotherapy resistance in BC.
Differential expression genes profiles and pathway
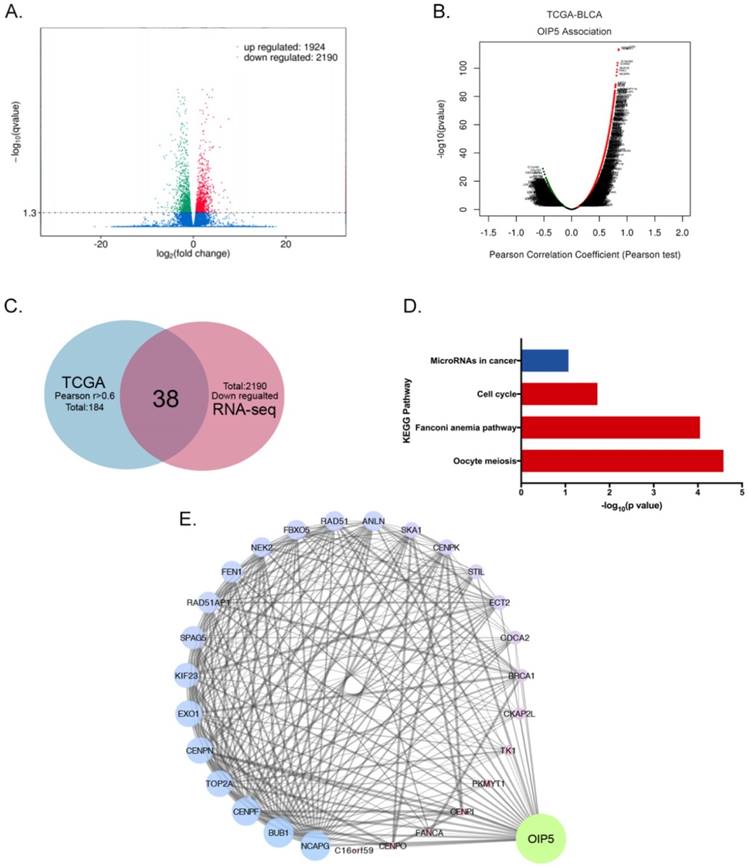
RNA sequencing combined with TCGA database and bioinformatics analyses were performed to access the biological mechanism of OIP5. The volcano plot showed 1924 up-regulated and 2190 down-regulated DEGs were identified between miR-OIP5 group and miR-NC group (Fig. 5A). Fig. 5B exhibited the genes associated with OIP5 in TCGA cohort(TCGA-BLCA) according to pearson test. In TCGA data analysis, there were 184 positive correlation genes (Pearson's r >0. 6), but the Pearson's r maximum absolute value of negative correlation genes was only 0. 52. Then we identified 38 intersection genes between and down-regulated DEGs (2190) and the positive correlation genes(184) (Fig. 5C). However, the overlap of up-regulated and negative correlation genes was small. KEGG pathway analysis indicated that the signaling pathway of the overlapping genes were enriched in oocyte meiosis, fanconi anemia pathway, cell cycle, microRNAs in cancer (Fig. 5D). As the interaction network indicated that TK1, TOP2A, SPAG5, SKA1, EXO1, RAD51AP1, PKMYT1 et al were closely linked to OIP5 (Fig. 5E).
The influence of OIP5 downregulation on cells malignant phenotypes. (A) EdU experiment was adopted to determine the cells growth ability. (B) The quantitative measurement of EdU positive cells. (C, D) Cell proliferation was detected in miR-OIP5 and miR-NC cells by CCK-8 assays. (E) Cell migration arrest was observed in stable transfected cells in cell scratch assays. (F) The quantitative measurement of migration areas. (G)Transwell assay was used to affirm the cell migration ability. (H)The quantitative test of invasive cells. Data are shown as mean ± SD. (*p<0. 05, **P<0. 01). (I, J)Stable transfected cells were exposed to increasing doses of cisplatin for 72h. Knockdown of OIP5 enhances sensitivity to cisplatin in miR-OIP5 group cells compared with miR-NC group.
Knockdown of OIP5 suppresses tumor growth in vivo. (A)The tumour volume in miR-OIP5 and miR-NC mice. (B)Photographs of the mice and dissected tumors that were injected with stably transfected SW780 cells for 4 weeks. (C)The tumor weight of the mice were measured at 4 weeks after injection. (D)Body weight fluctuation of the nude mice during the 4 weeks. Data are presented as mean ± SD; *P < 0. 05, **P<0. 01.
Discussion
Recurrence, drug-resistance, and metastasis continue to be the challenges to the bladder cancer management. Mounting published evidence demonstrated that OIP5 could induce tumorigenesis in various tumors, including bladder cancer[13]. Koinuma et al reported that knockdown of OIP5 by siRNA inhibited cell growth of lung cancer and esophageal cancer. They also found that OIP5 may affect the aggressive phenotype of by interacting with Raf1 [9]. Raf1 acts as a trigger of a cascade of cell signaling responses, involved in cell proliferation and survival, and widely expressed in tumor[21]. The stability of OIP5 protein is likely to be regulated by its interaction with Raf1. Li et al proved that OIP5 regulates the breast cancer proliferation through miR-139-5p/Notch1 pathway[22]. In hepatocellular carcinoma(HCC), OIP5 activates AKT through the mTORC2 and p38/PTEN signaling pathways, which contributed to tumor cell growth and metastasis[23]. A recent study suggested that OIP5 acts as a downstream gene of E2F1, and maintains the stability of E2F1 signaling pathway which promotes tumorigenesis and metastasis in glioblastoma. Our results further affirmed that OIP5 was present at high levels and OIP5 knockdown inhibited the growth of BC cells both in vitro and vivo, promoting OIP5 as a potential access point for therapeutic targets. Moreover, we first found that OIP5 suppression impeded BC cells invasion, indicating that OIP5 is substantially related to metastasis and invasion in BC. In addition, data from TCGA-BLCA data and SRA063495 [17] suggested that high expression of OIP5 predicted poor overall survival and advanced grade, which means it may be a prognostic biomarker for BC patients.
We explored the biological mechanism of OIP5 in BC through sequencing analysis and mining of large data information. In this study, 38 overlapping down-regulated DEGs were linked to OIP5 and bladder cancer, involved in oocyte meiosis, cell cycle, microRNAs in cancer. Among these DEGs, TK1, TOP2A, SPAG5, SKA1, EXO1, RAD51AP1, PKMYT1 were closely associated to bladder cancer development. A microarray analysis has suggested that the expression level of TK1 is elevated in primary bladder tumors [24]. Thymidine kinase 1 (TK1), a salvage pathway enzyme, located on chromosomes 17q23. 2- q25. 3, involved in DNA synthesis and repair[25, 26]. Rausch et al reported that TK1 is highly expressed in Muscle-invasive bladder cancer, and recognized as proliferation and prognostic marker[27]. Interestingly, serum concentration of TK1 were decreased after bladder tumor ectomy, indicating it acted as a role of monitoring the effectiveness of surgery[28]. Topoisomerase-II alpha (TopoIIA) encodes a DNA topoisomerase, an enzyme that contributed to DNA processes of DNA replication, transcription and cell cycle regulation[29]. This gene also functions as the target for several anticancer agents and its mutation have been associated with the development of drug resistance[30]. JaeKim et al found that deregulated TOP2A activity attenuates the cytotoxic of chemotherapeutic agents by promoting drug efflux through increasing cellular glutathione(GSH) levels in the doxorubicin -resistant BC cell line (5637/DR50) [31]. Sperm-associated antigen 5 (SPAG5) plays an important role in dynamic regulation of mitotic spindles[32]. In prostate cancer, SPAG5 is a direct target of miR-539, linked to tumor growth and metastasis, and invasion[33]. A study published in The Lancet Oncology suggested that SPAG5 is confirmed as prognostic biomarkers for combination cytotoxic chemotherapy sensitivity in breast cancer[34]. In addition, Yuan et described that SPAG5 downregulation alter sensitivity to taxol via the mTOR signaling pathway which activity is regulated depend on taxol dose in hela cells[35]. Spindle and kinetochore-associated protein 1 (SKA1) is a component of the kinetochore-microtubule interface and associates with cell cycle regulation[36]. In bladder cancer, depletion of SKA1 induces cell cycle arrest at S phase and impaired cell growth ability through down-regulation of CDK4 and Cyclin D1, and alleviated the activations of ERK2 and AKT[37]. In general, these DEGs influence diverse biological processes of bladder cancer, including cell cycle, proliferation, invasion, and drug response regulation. In our study, the role of OIP5 in tumor devolvement was consistent with these DEGs, but further validation is required to verify its molecular mechanism.
The most significant finding of our study was inhibition of OIP5 promoted sensitivity to cisplatin in BC cells. Cis-disamminedichloroplatinum (Ⅱ), also known as cisplatin or DDP, falls into a class of DNA-damaging agents [38]. The cytotoxic activity of cisplatin comprises a variety of cellular mechanisms, one of which is its interaction with DNA [39]. It has been widely accepted that cisplatin induced tumor apoptosis via influencing DNA replication and inhibiting mitosis [40]. Cisplatin induces DNA lesion through formation of intra- and inter-strand platinum-DNA covalent adducts, such as 1, 2-intrastrand, 1, 3- and longer range intrastrand and protein- DNA crosslinks. An HMG-domain protein binds to the cisplatin-DNA adduct, blocking its repair [38]. It has been established that repair of these DNA-crosslinking and enhancement of DNA repair play an integral role in the process of drug-resistance [41]. Nucletide excising repair (NER), a DNA repair pathway is responsible for removing DNA-crosslinking [42]. As already studied, 1, 2-and 1, 3 intrastrand crosslinks are repaired by the NER pathway[43]. Cells deficient in NER are hypersensitive to cisplatin, and the sensitive to cisplatin will recover when the NER function returns to normal. For instance, cisplatin is an excellent agent for the treatment of testicular tumors due to its deficient in NER[44]. Other mechanisms attributed to DNA repair process include inter-crosslinking repair (ICR), homologous recombination repair(HR), Homologous mismatch (HM), and mismatch repair (MMR)[45].
Based on the above discussion, there is strong evidence to ascertain that the principal target of cisplatin is DNA. As above mentioned, OIP5 is a coding gene, localizes to centromeres, we hypothesized it may play a role in cisplatin resistance. OIP5 is also named as Mis18beta and LINT-25, which is essential for the recruitment of CENP-A to the centromere through the mediator Holiday junction recognition protein [46]. CENP-A (centromere protein A) is a histone H3 variant and essential for centromere structure and function [47]. LINT-25 has been suggested to interact with Lamina-associated polypeptide (LAP2a), which was accumulated during G1 phase in proliferating cells [46]. This protein also interacts with the A-type lamins and retinoblastoma protein, and regulates cell cycle progression via the E2F-Rb pathway[48]. A recent study reported that OIP5 expression was linked with the mitotic proliferation of HCC, and knockdown of OIP5 elicited cell cycle arrest in the G2/M phase [23]. He et al found that suppression of OIP5 increased the cell population of the G1 phase in 5637 cell lines of BC [13]. Additionally, OIP5 repressed docetaxel-induced mitochondrial damage by modulating the mitophagy pathway[49]. These studies robustly suggested that OIP5 play an important role in cells mitosis, involved in DNA replication, chromosome maintenance. And cisplatin belongs to a cell cycle non-specific drug, forming DNA adducts in cell nucleus, so as to inhibit cell mitosis and DNA replication. These associations indicate that OIP5 may serve as a promising target for cisplatin resistant in BC. However, the detailed mechanisms that cisplatin-resistant result from OIP5 need further elucidation.
Differential expression genes profiles and pathway based on RNA sequencing and TCGA database analyses. (A)The volcano plot of DEGs between miR-OIP5 and miR-NC cells. The screening threshold is set to Qvalue < 0. 05 by default (if Qvalue < 0. 05, means screening differential genes is too small, pvalue < 0. 05 is used for differential screening). (B)The mRNAs associated with OIP5 TCGA cohort(TCGA-BLCA) according to pearson test. Pearson Correlation Coefficient(Pearson's r), a measure of the linear correlation between two variables X and Y. Its value ranges from -1 to 1, r<0: negative linear correlation, r=0:no linear correlation, r>0: positive linear correlation. (C)The shared genes(38) between down-regulated DEGs(2190) the positive correlation mRNA(184). The positive correlation cohort range was restricted as r>0. 6. (D) The pathway enrichment of the overlapping genes through KEGG pathway analysis. (E)The interaction network of DEGs and OIP5. The degree value is denoted by the size and color of map nodes. The degree value renders a gradual process accompanied by the size. The small size with a low degree in red, large size with a high degree in blue and transition in purple. The sizes of map edges are determined by the combine score.
In conclusion, the present study showed that OIP5 upregulation promotes the proliferation, metastasis, and drug-resistance progression of bladder cancer. In the future, OIP5 may be a potential therapeutic target and prognosis biomarker for bladder cancer.
Acknowledgements
We are indebted to the donors whose names were not included in the author list, but who participated in this program. This work was supported by the National Key Basic Research Program of China (973 Program) (2014CB745201), National Natural Science Foundation of China (81772737, 31570115), National Science Foundation Projects of Guangdong Province (2017B030301015), the Shenzhen Municipal Government of China (JCYJ20170413161749433, JSGG20160301161836370), the Sanming Project of Shenzhen Health and Family Planning Commission, SZSM201412018, SZSM201512037, and the high level university's medical discipline construction 2016031638.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. European urology. 2016Jun28;71(1):96-108
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018Jan;68(1):7-30
3. Kamat AM, Sylvester RJ, Bohle A, Palou J, Lamm DL, Brausi M. et al. Definitions, End Points, and Clinical Trial Designs for Non-Muscle-Invasive Bladder Cancer: Recommendations From the International Bladder Cancer Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016Jun1;34(16):1935-44
4. Sonpavde G, Lockhart ME, Nix JW. Chemotherapy for Muscle-Invasive Bladder Cancer: Better Late Than Never?. J Clin Oncol. 2016:34 (8)
5. Abufaraj M, Dalbagni G, Daneshmand S, Horenblas S, Kamat AM, Kanzaki R. et al. The Role of Surgery in Metastatic Bladder Cancer: A Systematic Review. European urology. 2017;17:308400
6. Kluth LA, Black PC, Bochner BH, Catto J, Lerner SP, Stenzl A. et al. Prognostic and Prediction Tools in Bladder Cancer: A Comprehensive Review of the Literature. European urology. 2015Aug;68(2):238-53
7. Afsharpad M, Nowroozi MR, Mobasheri MB, Ayati M, Nekoohesh L, Saffari M. et al. Cancer-Testis Antigens as New Candidate Diagnostic Biomarkers for Transitional Cell Carcinoma of Bladder. Pathology oncology research: POR. 2017 Oct 20
8. Williams JM CG, Zhu L, Rest RF. Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth. Mol Microbiol. 1998;27(1):171-86
9. Koinuma J, Akiyama H, Fujita M, Hosokawa M, Tsuchiya E, Kondo S. et al. Characterization of an Opa interacting protein 5 involved in lung and esophageal carcinogenesis. Cancer science. 2012Mar;103(3):577-86
10. Gong M, Xu Y, Dong W, Guo G, Ni W, Wang Y. et al. Expression of Opa interacting protein 5 (OIP5) is associated with tumor stage and prognosis of clear cell renal cell carcinoma. Acta histochemica. 2013Oct;115(8):810-5
11. Nakamura Y, Tanaka F, Nagahara H, Ieta K, Haraguchi N, Mimori K. et al. Opa interacting protein 5 (OIP5) is a novel cancer-testis specific gene in gastric cancer. Annals of surgical oncology. 2007Feb;14(2):885-92
12. Chun HK CK, Kim HC, Kang JE, Kang MA, Kim JT, Choi EH, Jung KE, Kim MH, Song EY, Kim SY, Won M, Lee HG. OIP5 is a highly expressed potential therapeutic target for colorectal and gastric cancers. BMB Rep. 2010;43(5):349-54
13. He X, Hou J, Ping J, Wen D, He J. Opa interacting protein 5 acts as an oncogene in bladder cancer. Journal of cancer research and clinical oncology. 2017Nov;143(11):2221-33
14. Oliver RT. Testicular cancer. Curr Opin Oncol. 1996;8:252-8
15. Dilruba S, Kalayda GV. Platinum-based drugs: past, present and future. Cancer chemotherapy and pharmacology. 2016Jun;77(6):1103-24
16. Li P, Yang X, Cheng Y, Zhang X, Yang C, Deng X. et al. MicroRNA-218 Increases the Sensitivity of Bladder Cancer to Cisplatin by Targeting Glut1. Cellular Physiology and Biochemistry. 2017;41(3):921-32
17. Guo G, Sun X, Chen C, Wu S, Huang P, Li Z. et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nature genetics. 2013Dec;45(12):1459-63
18. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012Mar28;483(7391):603-7
19. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44-57
20. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015Jan;43(Database issue):D447-52
21. Yuryev A, Wennogle LP. Novel raf kinase protein-protein interactions found by an exhaustive yeast two-hybrid analysis. Genomics. 2003;81(2):112-25
22. Li HC, Chen YF, Feng W, Cai H, Mei Y, Jiang YM. et al. Loss of the Opa interacting protein 5 inhibits breast cancer proliferation through miR-139-5p/NOTCH1 pathway. Gene. 2017Mar1;603:1-8
23. Li H ZJ, Lee MJ, Yu GR, Han X, Kim DG. OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget. 2017;8(11):18129-44
24. Doherty SC, McKeown SR, Lopez JA, Walsh IK, McKelvey-Martin VJ. Gene expression in normal urothelium depends on location within the bladder: a possible link to bladder carcinogenesis. European urology. 2006Aug;50(2):290-301
25. Li HX, Zhang S, Lei DS, Wang XQ, Skog S, He Q. Serum thymidine kinase 1 is a prognostic and monitoring factor in patients with non-small cell lung cancer. Oncology reports. 2005;13(1):145-9
26. Aufderklamm S, Todenhofer T, Gakis G, Kruck S, Hennenlotter J, Stenzl A. et al. Thymidine kinase and cancer monitoring. Cancer Lett. 2012Mar;316(1):6-10
27. Rausch S, Hennenlotter J, Teepe K, Kuehs U, Aufderklamm S, Bier S. et al. Muscle-invasive bladder cancer is characterized by overexpression of thymidine kinase 1. Urologic oncology. 2015Oct;33(10):426 e21-9
28. Zhang J JQ, Zou S, Zhang P, Zhang X, Skog S, Luo P, Zhang W, He Q. Thymidine kinase 1: A proliferation marker for determining prognosis and monitoring the surgical outcome of primary bladder carcinoma patients. Oncology reports. 2006;15(2):455-61
29. Strausfeld U, Richter A. Simultaneous purification of DNA topoisomerase I and II from eukaryotic cells. Preparative biochemistry. 1989;19(1):37-48
30. Tubbs R, Barlow WE, Budd GT, Swain E, Porter P, Gown A. et al. Outcome of patients with early-stage breast cancer treated with doxorubicin-based adjuvant chemotherapy as a function of HER2 and TOP2A status. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009Aug20;27(24):3881-6
31. Wun-JaeKim YK, Osamu Yoshida. Muhifactorial Involvement of Multidrug Resistance Protein, DNA Topoisomerase I1 and Clutathione/Clutathione-S-Transferase in NonPClycoprotein-Mediated Multidrug Resistance in Human Bladder Cancer Cells. Int J Urol. 1997;4:583-90
32. Gruber J HJ, Schnabel J, Weber K, Hatzfeld M. The mitotic-spindle-associated protein astrin is essential for progression through mitosis. J Cell Sci. 2002;115(Pt 21):4053-9
33. Zhang H, Li S, Yang X, Qiao B, Zhang Z, Xu Y. miR-539 inhibits prostate cancer progression by directly targeting SPAG5. Journal of experimental & clinical cancer research: CR. 2016Apr1;35:60
34. Abdel-Fatah TMA, Agarwal D, Liu D-X, Russell R, Rueda OM, Liu K. et al. SPAG5 as a prognostic biomarker and chemotherapy sensitivity predictor in breast cancer: a retrospective, integrated genomic, transcriptomic, and protein analysis. The Lancet Oncology. 2016;17(7):1004-18
35. Yuan LJ, Li JD, Zhang L, Wang JH, Wan T, Zhou Y. et al. SPAG5 upregulation predicts poor prognosis in cervical cancer patients and alters sensitivity to taxol treatment via the mTOR signaling pathway. Cell death & disease. 2014May22;5:e1247
36. Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Developmental cell. 2009Mar;16(3):374-85
37. Tian F, Xing X, Xu F, Cheng W, Zhang Z, Gao J. et al. Downregulation of SKA1 Gene Expression Inhibits Cell Growth in Human Bladder Cancer. Cancer biotherapy & radiopharmaceuticals. 2015Sep;30(7):271-7
38. Zamble DB LS. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem Sci. 1995:20 (10)
39. Raghavan D, Koczwara B, Javle M. Evolving Strategies of Cytotoxic Chemotherapy for Advanced Prostate Cancer. European joumal of cancer. 1997;33(4):566
40. Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R. et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nature Communications. 2013:4
41. Reed E OR, Tarone R, Yuspa SH, Poirier MC. Platinum-DNA adducts in leukocyte DNA correlate with disease response in ovarian cancer patients receiving platinum-based chemotherapy. Proc Natl Acad Sci U S A. 1987;84(14):5024-8
42. Berra CM, de Oliveira CS, Garcia CCM, Rocha CRR, Lerner LK, Lima LCdA. et al. Nucleotide excision repair activity on DNA damage induced by photoactivated methylene blue. Free Radical Biology and Medicine. 2013;61:343-56
43. Huang JC ZD, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci U S A. 1994;91(22):10394-8
44. Welsh C, Day R, McGurk C, Masters JRW, Wood RD, Köberle B. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. International Journal of Cancer. 2004;110(3):352-61
45. Bohm L. Inhibition of homologous recombination repair with Pentoxifylline targets G2 cells generated by radiotherapy and induces major enhancements of the toxicity of cisplatin and melphalan given after irradiation. Radiat Oncol. 2006May3;1(1):12
46. Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C. et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Developmental cell. 2007Jan;12(1):17-30
47. Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Current Opinion in Cell Biology. 2008;20(1):91-100
48. Naetar N HS, Dorner D, Dechat T, Korbei B, Gotzmann J, Beug H, Foisner R. LAP2-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci. 2007;120(Pt5):737-47
49. Kim TW, Lee SJ, Park YJ, Park SY, Oh BM, Park YS. et al. Opa-interacting protein 5 modulates docetaxel-induced cell death via regulation of mitophagy in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017Oct;39(10):1010428317733985
Author contact
![]() Corresponding authors: pony8980com (WH); liuyuchenmdcgcom (YL)
Corresponding authors: pony8980com (WH); liuyuchenmdcgcom (YL)

 Global reach, higher impact
Global reach, higher impact