Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(1):51-60. doi:10.7150/jca.26908 This issue Cite
Research Paper
Identification of Key Genes and Pathways in Female Lung Cancer Patients Who Never Smoked by a Bioinformatics Analysis
1. Department of Geriatrics, Clinical Laboratory, Xiangya Hospital of Central South University, Changsha, People's Republic of China
2. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital of Central South University, Changsha, People's Republic of China
3. Department of Pathology, the First Affiliated Hospital of Hunan University of Medicine, Huaihua, People's Republic of China
*These authors contributed equally to this study.
Received 2018-4-26; Accepted 2018-10-17; Published 2019-1-1
Abstract
Smoking is considered the major risk factor for lung cancer, but only a small portion of female lung adenocarcinoma patients are associated with smoking. Thus, identifying crucial genes and pathways related to nonsmoking female lung cancer patients is of great importance. Gene expression profiles were downloaded from the Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA) databases. The R software packages were applied to screen the differentially expressed genes (DEGs). GO term enrichment and KEGG pathway analyses were carried out using DAVID tools. The protein-protein interaction (PPI) network was constructed by Cytoscape software. In total, 487 downregulated and 199 upregulated DEGs were identified. The down-regulated DEGs were mainly enriched for behavior and response to wounding, and the upregulated DEGs were significantly enriched for multicellular organismal metabolic process and cell division. The KEGG pathway analysis revealed that the downregulated DEGs were significantly enriched for cell adhesion molecules and neuroactive ligand-receptor interaction, while the upregulated DEGs were mainly enriched for cell cycle and the p53 signaling pathway. The top 10 hub genes and top 3 gene interaction modules were selected from the PPI network. Of the ten hub genes, a high expression of five genes was related to a poor OS in female lung cancer patients who never smoked, including IL6, CXCR2, FPR2, PPBP and HBA1. However, a low expression of GNG11, LRRK2, CDH5, CAV1 and SELE was associated with a worse OS for the female lung cancer patients who never smoked. In conclusion, our study provides novel insight for a better understanding of the pathogenesis of nonsmoking female lung cancer, and these identified DEGs may serve as biomarkers for diagnostics and treatment.
Keywords: female lung cancer, never smoking, hub genes, bioinformatics
Introduction
Lung cancer is the most incident and lethal malignant tumor all over the world, with an estimated 1, 800, 000 new cases and 1, 600,000 deaths from this disease in 2012 [1]. Lung cancer is currently on the rise in female patients and represents the first cause of cancer-related death [2]. Cigarette smoking is considered the major risk factor for lung cancer, and it is reported that approximately 15% of all male lung cancer patients and 53% of female patients never smoked. In addition, it is estimated that 25% of all patients diagnosed with lung cancer worldwide are not attributable to smoking [3, 4]. Furthermore, lung cancer in non-smokers has been rising over time, showing a different carcinogenic pathway, clinicopathological characteristics, epidemiology and natural history [4, 5].
Currently, there is little information about the molecular mechanisms, biology markers and risk factors of lung cancer in non-smoking female lung cancer patients. Some recent studies report that several key genes are related to lung carcinoma in nonsmokers, such as the EGFR tyrosine kinase domain mutation [6], the TP53 mutation [7], ALK translocations [8], EML4-ALK fusion [9], and the loss of mRNA and protein expression of hMSH2 [10]. A variety of risk factors are considered to be involved in the initiation of cancer in non-smokers, including occupational exposure, second-hand smoke exposure, cooking oil fumes, indoor and outdoor air pollution, coal fumes, previous diseases and genetic factors [5, 11]. However, the molecular mechanisms of the tumorigenesis and development of lung cancer in nonsmoking women still remain unclear.
In the present study, the microarray data GSE19804 and GSE31210 were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) database. We identified never smoking female lung cancer patients associated differentially expressed genes (DEGs) between the cancerous and normal samples using a bioinformatics analysis. Subsequently, we performed GO term and KEGG pathway enrichment analyses, as well as a protein-protein interaction (PPI) analysis to screen the crucial genes and pathways that were closely associated with female lung cancer patients who never smoked. This study promotes our understanding the molecular mechanism of tumor development in nonsmoking women with lung cancer and provides potential target genes for diagnosis, prognosis, and drug design.
Methods and materials
Acquisition of the microarray data
Two gene expression profiles (GSE19804 and GSE31210) were obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) database, which were based on the Affymetrix Human Genome U133 Plus 2.0 Array. The GSE19804 dataset was submitted by Lu et al., including 60 nonsmoking female lung cancer tissue samples and 60 normal samples [12]. The microarray data of GSE31210 was submitted by Kohno T, which contained 246 samples, and consisted of 98 female lung cancer patients who never smoked and 4 normal female lung tissues [13].
Data preprocessing and DEGs identifying
The raw data were preprocessed and normalized by using the affy package (http://www.bioconductor.org/packages/release/bioc/html/affy.html) under the R environment (version 3.3.4, https://www.r-project.org/). The sva package (https://bioconductor.org/packages/release/bioc/html/sva.html) and pamr package (https://cran.r-project.org/web/packages/pamr/index.html) were used to remove the batch effects. The probe annotation files were downloaded from GEO. In order to handle the probes that corresponded to one gene, the average value was regarded as the gene expression level. Following preprocessing, the limma package (https://bioconductor.org/packages/release/bioc/html/limma. html) from Bioconductor was used to identify the DEGs based on Empirical Bayes method between the lung cancer samples and normal lung samples. All the P value less than 0.05 were considered significantly different.
GO and Functional enrichment analyses
The biological function of the DEGs was investigated by a GO enrichment analysis, which is comprised 3 terms, including biological process (BP), molecular function (MF) and cellular component (CC). The Kyoto Encyclopedia of Genes and Genomes database (www.genome.jp/kegg/) includes genomic, chemical, functional and metabolic information [14]. The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) is a powerful gene function analysis tool [15]. In the present study, the GO and KEGG pathway enrichment analysis were executed using DAVID online tools with a P value less than 0.05 and gene number more than 5.
PPI network construction and module analysis
The search Tool for the Retrieval of Interacting Genes (STRING, http://www.string-db.org/) database is an online tool for exploring protein-protein interactions. The PPI information from the DEGs was obtained from the STRING database and was further constructed by using Cytoscape 3.4.0 software. Subsequently, the plug-in Molecular Complex Detection (MCODE) in Cytoscape was applied to screen module with scores >3 and node counts >5. Moreover, the function and pathway enrichment analyses were performed using the plug-in Biological Network Gene Ontology tool (BiNGO). P less than 0.05 was treated as a significant difference.
Data validation
To validate the data reliability and reproducibility, we downloaded the RNA sequencing and clinical data of female lung cancer patients who never smoked from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/). The DEGs were screened by R packages. All the P value less than 0.05 were considered significant differences.
Results
Selecting the DEGs in female lung cancer patients who never smoked
The GSE19804 and GSE31210 datasets were downloaded from the GEO database. A total of 158 lung cancer samples and 64 normal lung samples were analysed. The software R was used to process these microarray data and detect the DEGs. Based on the R analysis, altogether 2844 DEGs were identified; of which, 686 DEGs met our selection criteria (P<0.05, fold change ≥2.0), including 199 up-regulated and 487 down-regulated genes (Figure 1). The expression level of the top 100 DEGs is shown in Figure 2.
GO analysis of the DEGs
To learn more about the function of the identified DEGs, we uploaded these genes to the online tool DAVID to determine the GO classifications and pathways. The results indicated that the down-regulated DEGs were mainly enriched in the biological processes (BP) associated behavior, response to wounding, regulation of cell proliferation, blood vessel morphogenesis, chemotaxis, taxis, and biological adhesion (Figure 3). The up-regulated DEGs were significantly enriched in multicellular organismal metabolic process, cell division, multicellular organismal macromolecule metabolic process and cell cycle phase (Figure 4). As for molecular function (MF), the downregulated DEGs were mainly enriched in cytokine activity, heparin binding, cytokine binding, glycosaminoglycan binding and pattern binding (Figure 3). The overexpressed DEGs were significantly enriched in metallopeptidase activity, extracellular matrix structural constituent, platelet-derived growth factor binding and enzyme inhibitor activity (Figure 4). Additionally, the results for the cellular component (CC) analysis showed that the downregulated DEGs were mainly enriched in extracellular matrix, proteinaceous extracellular matrix, integral to plasma membrane, intrinsic to plasma membrane and plasma membrane part (Figure 3), and the upregulated DEGs were notably enriched in spindle, spindle pole, fibrillar collagen, microtubule cytoskeleton and extracellular region (Figure 4).
Volcano plot of 2844 differentially expressed genes. Red: up-regulation with fold change of more than 2; green: down-regulation with a fold change of more than 2; blue: unchanged genes.
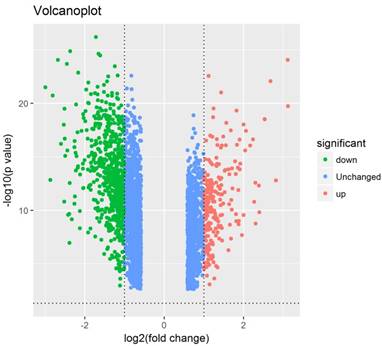
Heatmap of the top 100 differentially expressed genes with a fold change of more than 2. Red: up-regulation; green: down-regulation.
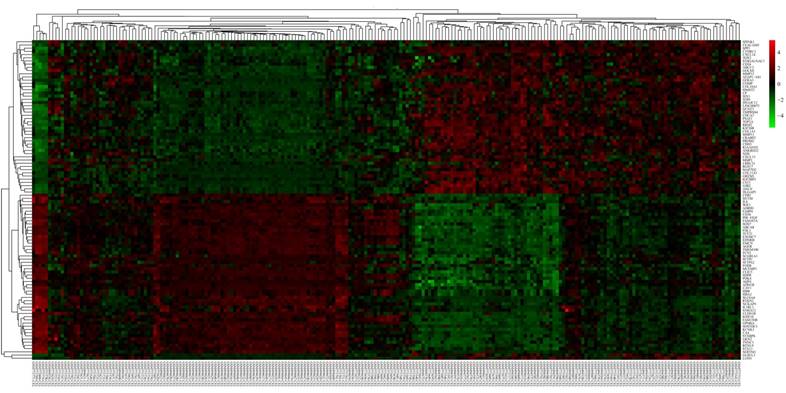
Gene ontology and KEGG pathway analyses results of the down-regulated differentially expressed genes with a fold change of more than 2. BP: biological process; CC: cellular component; MF: molecular function.
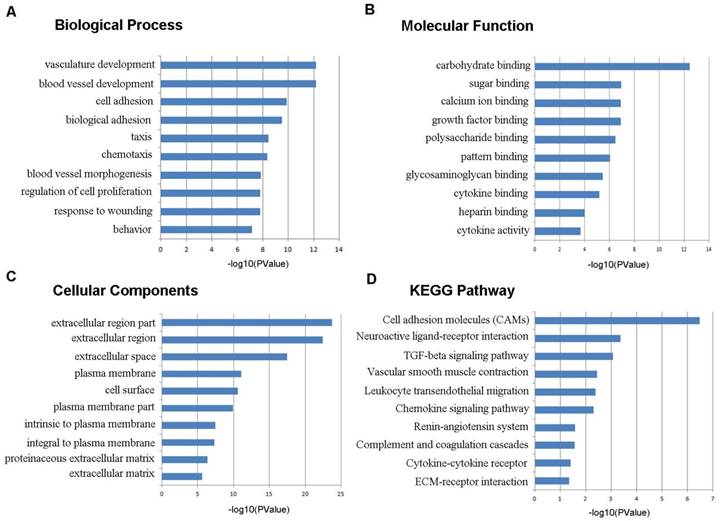
Gene ontology and KEGG pathway analyses results of the up-regulated differentially expressed genes with a fold change of more than 2. BP: biological process; CC: cellular component; MF: molecular function.
KEGG pathway analysis of the DEGs
As shown in Figure 3D, the downregulated DEGs were significantly enriched for cell adhesion molecules (CAMs), neuroactive ligand-receptor interaction, the TGF-beta signaling pathway, vascular smooth muscle contraction, leukocyte transendothelial migration and the chemokine signaling pathway. The upregulated DEGs were mostly enriched in ECM-receptor interaction, cell cycle, focal adhesion, the p53 signaling pathway and oocyte meiosis (Figure 4D).
Module screening from the PPI network
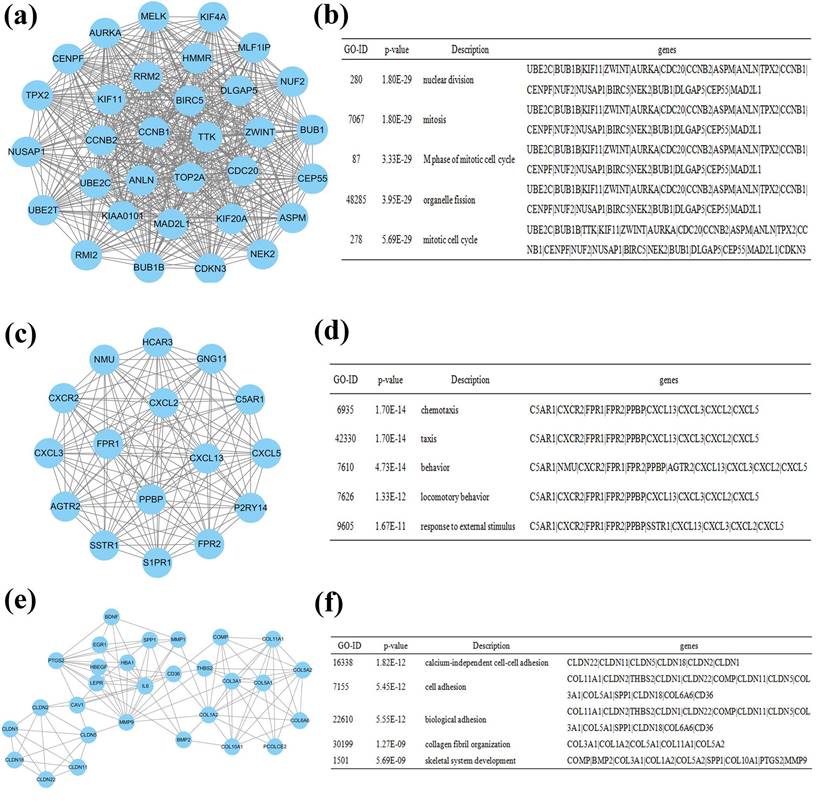
The PPI network was constructed using Cytoscape software, which included 641 nodes and 2255 edges. The hub genes were interleukin 6 (IL6), guanine nucleotide binding protein gamma 11 (GNG11), chemokine (C-X-C motif) receptor 2 (CXCR2), leucine-rich repeat kinase 2 (LRRK2), cadherin 5 (CDH5), formyl peptide receptor 2 (FPR2), pro-platelet basic protein (PPBP), caveolin 1 (CAV1), hemoglobin, alpha 1 (HBA1), and selectin E (SELE). These genes may play vital roles in the progression of lung cancer in females who never smoked. A total of 641 nodes were further analyzed by employing the plug-ins MODE and BiNGO to detect potential modules. The top 3 significant modules were acquired, and were enriched in nuclear division, chemotaxis and calcium-independent cell-cell adhesion, respectively (Figure 5).
Prognostic value of ten hub genes
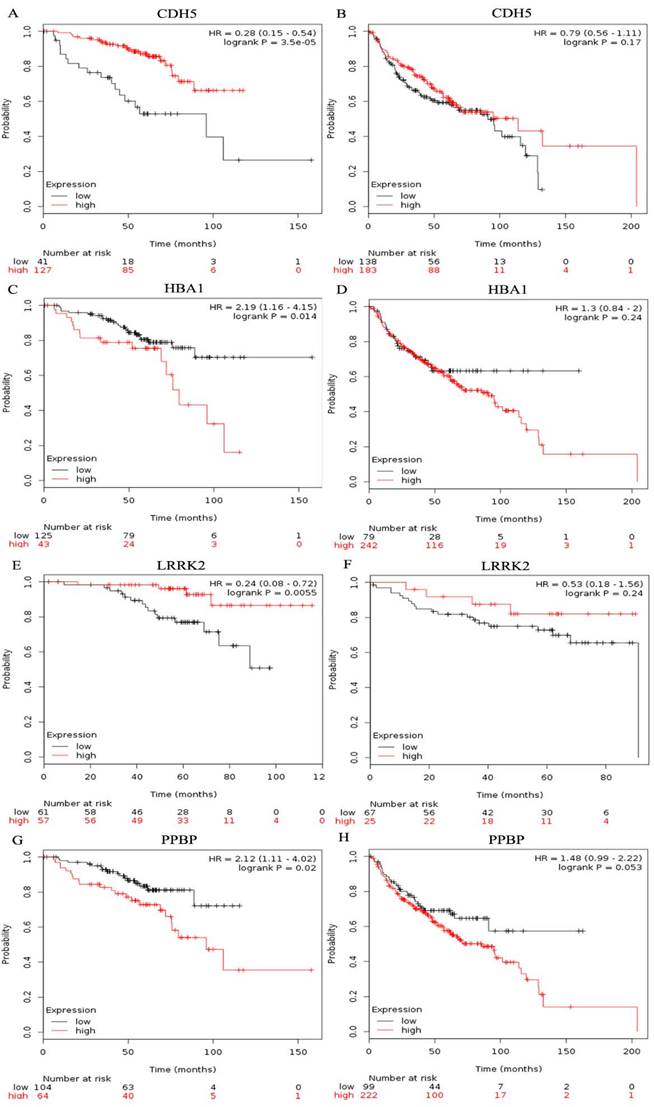
The prognostic value of ten hub genes in the PPI network was evaluated by using Kaplan-Meier plotter (www.kmplot.com). The overall survival(OS) for female lung cancer patients was calculated on the basis of the high and low expression of each gene. The results showed that a high expression of IL6 (HR=2.5, 95% CI: 1.32-4.73, P=0.0035) was negatively associated with OS in female lung cancer patients who never smoked, as well as CXCR2 (HR=3.38, 95% CI: 1.2-9.54, P=0.015), FPR2(HR=2.56, 95% CI: 1.36-4.82, P=0.0025), PPBP (HR=2.12, 95% CI: 1.11-4.02, P=0.02) and HBA1(HR=2.19, 95% CI: 1.16-4.15, P=0.014) (Table 1). However, low GNG11 (HR=0.49, 95% CI: 0.25-0.98, P=0.038), LRRK2(HR=0.24, 95% CI: 0.08-0.72, P=0.0055), CDH5(HR=0.28, 95% CI: 0.15-0.54, P=3.5e-05), CAV1(HR=0.22, 95% CI: 0.09-0.52, P=1.6e-04) and SELE(HR=0.34, 95% CI: 0.18-0.65, P=6e-04) expressions were related with a worse OS for female lung cancer patients who never smoked (Table 1). In addition, we also found that the expression of LRRK2, CDH5, PPBP and HBA1 showed no significant correlation with the OS of smoking female lung cancer patients (All P > 0.05, Figure 6).
Prognostic value of the ten hub genes in the female lung cancer patients who never smoked.
| Gene name | HR with 95% CI | P value |
|---|---|---|
| IL6 | 2.5(1.32, 4.73) | 3.5 e-03 |
| GNG11 | 0.49(0.25,0.98) | 0.038 |
| CXCR2 | 3.38(1.2-9.54) | 0.015 |
| LRRK2 | 0.24(0.08-0.72) | 5.5e-03 |
| CDH5 | 0.28(0.15-0.54) | 3.5e-05 |
| FPR2 | 2.56(1.36-4.82) | 2.5e-03 |
| PPBP | 2.12(1.11-4.02) | 0.02 |
| CAV1 | 0.22(0.09-0.52) | 1.6e-04 |
| HBA1 | 2.19(1.16-4.15) | 0.014 |
| SELE | 0.34(0.18-0.65) | 6e-04 |
HR: Hazard ratio; CI: Confidence interval.
Data validation
A total of 238 lung cancer samples and 24 normal lung samples were obtained from TCGA. After the R analysis, 3056 DEGs were identified according to our screening criteria (adj.P<0.05, |logFC|≥2.0), which consisted of 2223 up-regulated and 833 down-regulated genes. Among these DEGs, 1306 genes were classified as RNA genes or pseudogenes. In this study, we focused on the genes that had consistent expression patterns in the TCGA. When we compared the hub genes from the PPI network, we found that GNG11, LRRK2, CDH5, FPR2, PPBP, CAV1, and HBA1 showed similar expression profiles in the datasets from GEO (GSE19804 and GSE31210). Notably, although IL6, CXCR2 and SELE did not meet the screening criteria in our present work (all |logFC| < 2.0), they were also down-regulated in the lung cancer samples from TCGA.
Discussion
Lung cancer is the leading cause of death all around the world and brings about significant socioeconomic implications for patients, families and society as a whole. Smoking is strongly associated with the occurrence of lung cancer. However, most women with lung cancer were never smokers. Understanding the molecular mechanism of nonsmoking female lung cancer is of important clinical significance for diagnosis and treatment. In this study, we obtained gene expression data from the GSE19804 and GSE31210 datasets, and selected 199 upregulated and 487 downregulated DEGs between the lung cancer sample and normal lung tissues. The function and pathway analyses displayed that these DEGs were mainly enriched in the regulation of cell proliferation, multicellular organismal metabolic process, cell division, cell adhesion molecules, neuroactive ligand-receptor interaction, and ECM-receptor interaction. We also determined several crucial genes that might offer novel clues for diagnostic and treatment research in women with lung cancer who never smoked.
Module analysis of the PPI network. (a): module 1; (b): GO analysis of module 1; (c): module 2; (d): GO analysis of module 2; (e): module 3; (f): GO analysis of module 3.
A total of 158 female lung cancer samples and 64 normal lung samples were retrieved from the microarray data GSE19804 and GSE31210. All the data were extracted and analyzed using statistical software R. This research, altogether, identified 2844 DEGs, and among these, 686 DEGs showed a fold change of more than 2, and were comprised of 199 upregulated and 487 downregulated genes. Accumulating evidence shows that co-expressed genes are usually composed of a set of genes with similar expression profiles, which are also often involved in parallel biological processes[16, 17] . To further investigate the interactions of these DEGs, we carried out GO term and KEGG pathway analyses.
In our study, the GO term analysis results indicated that the downregulated DEGs were mainly enriched in response to wounding, regulation of cell proliferation, blood vessel morphogenesis, cytokine activity, and extracellular matrix, and the upregulated DEGs were significantly enriched in multicellular organismal metabolic process, cell division, cell cycle phase, metalloendopeptidase activity, and spindle. Response to wounding means a change in the state or activity of a cell or a stimulus indicating damage to the organism, which is closely related to the occurrence and development of various diseases [18-20]. Dysfunction of the regulation of cell proliferation, blood vessel morphogenesis, cell division and cell cycle are critical causes of tumorigenesis and cancer progression [21-23]. The KEGG pathways analysis suggested that the downregulated DEGs were significantly enriched in cell adhesion molecules, neuroactive ligand-receptor interaction, the TGF-beta signaling pathway, vascular smooth muscle contraction, leukocyte transendothelial migration and the chemokine signaling pathway. Cell adhesion molecules play important roles in tumorigenesis, progression, invasion and metastasis. Previous studies show that the loss of the function or the expression of some adhesion molecules could promote cell invasion and metastasis in lung cancer [24]. The endothelium is the first obstacle that leukocytes must overcome when they are recruited to the site of inflammation. Therefore, inhibiting leukocyte transendothelial migration is of much importance to the occurrence of a variety of inflammatory related diseases [25]. The up-regulated DEGs were mainly enriched for ECM-receptor interaction, cell cycle, focal adhesion, and the p53 signalling pathway. Recent studies show that the overexpression of some cell cycle-related genes predicts a poor prognosis as well as promotes tumor progression and drug resistance in lung cancer [19, 26]. Previous studies indicate that lung cancer cells and CCD32 normal cells interact by cell-cell contacts, which are strictly dependent on ECM components [27]. Furthermore, the relationship between a disturbance in the p53 signaling pathway and lung cancer is extensively researched [28, 29].
In addition, we also constructed a PPI network of the DEGs and identified the top 10 hub genes, including IL6, GNG11, CXCR2, LRRK2, CDH5, FPR2, PPBP, CAV1, HBA1 and SELE. IL6, with the highest degree of connectivity in the PPI network, is an important immunomodulatory cytokine that functions in inflammation and the maturation of B cells. IL6 promotes cachexia in lung adenocarcinoma, and high IL6 levels are associated with a worse prognosis in lung cancer patients [30, 31], which is consistent with our results. The second hub gene GNG11, as a crucial member of the gamma subunit family of G protein, plays an important role in the transmembrane signaling system. GNG11 is reported to induce cellular senescence in human fibroblasts, and inhibits SUSM-1 cell growth [32] but not HeLa cell growth [33]. However, the specific biological function of GNG11 in lung cancer still unknown.
FPR2 is a Gi-protein-coupled receptor that is mainly involved in antibacterial host defense and inflammation. Previous studies show that the overexpression of FPR2 participates in restraining the epithelial mesenchymal transition of A549 lung cancer cells [34].CXCR2, a key chemokine receptor, mediates the initiation and progression of various cancers including lung carcinoma, prostate cancer, breast cancer, and colorectal carcinoma [35]. Moreover, PPBP is a platelet-derived growth factor that belongs to the CXC chemokine family. Recent studies reveal that PPBP is significantly increased in lung cancer tissue and blood samples and serves as a novel diagnostic biomarker for lung carcinoma [36, 37]. Thus far, the biological functions of PPBP in lung cancer remain unclear. LRRK2 is a member of the leucine-rich repeat kinase family, and it is related to a variety of human diseases, such as Parkinson's disease, cancer, Crohn's disease and leprosy, but little know about its physiological effects [38]. Both CDH5 and SELE are adhesion molecules, but the former is a classical cadherin of the cadherin superfamily and the latter is the part of the selectin family. Cell adhesion molecules contribute significantly to tumorigenesis and tumor progression [24, 39]. In our present study, we identified that the expression of LRRK2 and CDH5 were decreased in female lung cancer patients who never smoked and was negatively associated with OS. However, there was no significant correlation between LRRK2 or CDH5 and OS in smoking female lung cancer patients. Therefore, we have reason to believe that LRRK2 and CDH5 play an important role in the pathogenesis of lung cancer in females who never smoked. CAV1, a main component of the caveolae plasma membranes, plays an important role in the Ras-ERK pathway and in promoting cell cycle progression. Furthermore, CAV1 is closely related to lung cancer drug resistance [40].
The module analysis of the PPI network revealed that the progression of non-smoking female lung cancer was associated with nuclear division, chemotaxis and calcium-independent cell-cell adhesion. DNA damage is the leading cause of cancer and nuclear division index is an important indicator of DNA damage [41]. Abnormal nuclear division is closely related to the occurrence of multiple cancers [41, 42]. Chemotaxis is reported to correlate with tumorigenesis. Mesenchymal stromal cells and macrophage chemotaxis are important drivers of the malignant phenotype for lung cancer [43]. In addition, calcium-independent cell-cell adhesion plays a critical role in the regulation of various cellular processes, such as polarization, cell adhesion, cell-cell contact formation, movement, survival, and proliferation. Adhesion molecules are implicated in the development and carcinogenesis of a wide variety of tumors [44, 45].
Prognostic value of four genes in female lung cancer patients. Prognostic value of CDH5(A), HBA1(C), LRRK2(E), and PPBP(G) in female lung cancer patients who never smoked. Prognostic value of CDH5(B), HBA1(D), LRRK2(F), and PPBP(H) in smoking female lung cancer patients.
In sum, we identified some crucial genes and pathways that were closely correlated with lung cancer tumorigenesis and cancer progression in females who never smoked by a bioinformatics analysis of the DEGs between the cancer tissues and normal lung tissues. These results provide more detailed clues for understanding the molecular mechanism of the pathogenesis of lung cancer in females who never smoked and to identify potential diagnostic biomarkers and therapeutic targets. However, further molecular biological experiments are needed to verify these findings.
Acknowledgements
This work was supported by funding from the Key Program of Hunan province Science and Technology Hall (2015SK20662).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7-30
2. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9-29
3. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74-108
4. Ziebarth NR. Lung cancer risk perception biases. Preventive medicine. 2018;110:16-23
5. Planchard D, Besse B. Lung cancer in never-smokers. The European respiratory journal. 2015;45:1214-7
6. Subramanian J, Govindan R. Molecular profile of lung cancer in never smokers. EJC supplements: EJC: official journal of EORTC, European Organization for Research and Treatment of Cancer [et al]. 2013;11:248-53
7. Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Human mutation. 2003;21:229-39
8. Dias M, Linhas R, Campainha S, Conde S, Barroso A. Lung cancer in never-smokers - what are the differences? Acta oncologica. 2017;56:931-5
9. Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC. et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723-33
10. Wang YC, Lu YP, Tseng RC, Lin RK, Chang JW, Chen JT. et al. Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. The Journal of clinical investigation. 2003;111:887-95
11. Torres-Duran M, Ruano-Ravina A, Kelsey KT, Parente-Lamelas I, Leiro-Fernandez V, Abdulkader I. et al. Environmental tobacco smoke exposure and EGFR and ALK alterations in never smokers' lung cancer. Results from the LCRINS study. Cancer letters. 2017;411:130-5
12. Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW. et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:2590-7
13. Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T. et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PloS one. 2012;7:e43923
14. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27-30
15. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44-57
16. Qiao F, Li N, Li W. Integrative Bioinformatics Analysis Reveals Potential Long Non-Coding RNA Biomarkers and Analysis of Function in Non-Smoking Females with Lung Cancer. Medical science monitor: international medical journal of experimental and clinical research. 2018;24:5771-8
17. Zhang L, Yang Y, Cheng L, Cheng Y, Zhou HH, Tan ZR. Identification of Common Genes Refers to Colorectal Carcinogenesis with Paired Cancer and Noncancer Samples. Disease markers. 2018;2018:3452739
18. Derrett-Smith EC, Martyanov V, Chighizola CB, Moinzadeh P, Campochiaro C, Khan K. et al. Limited cutaneous systemic sclerosis skin demonstrates distinct molecular subsets separated by a cardiovascular development gene expression signature. Arthritis Res Ther. 2017;19:156
19. Lee TG, Jeong EH, Min IJ, Kim SY, Kim HR, Kim CH. Altered expression of cellular proliferation, apoptosis and the cell cycle-related genes in lung cancer cells with acquired resistance to EGFR tyrosine kinase inhibitors. Oncology letters. 2017;14:2191-7
20. Wang Y, Guo Z, Zhao Y, Jin Y, An L, Wu B. et al. Genetic polymorphisms of lncRNA-p53 regulatory network genes are associated with concurrent chemoradiotherapy toxicities and efficacy in nasopharyngeal carcinoma patients. Scientific reports. 2017;7:8320
21. Herbert SP, Stainier DY. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nature reviews Molecular cell biology. 2011;12:551-64
22. Graham SV. The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review. Clin Sci (Lond). 2017;131:2201-21
23. Tan X, Zhang Z, Yao H, Shen L. Tim-4 promotes the growth of colorectal cancer by activating angiogenesis and recruiting tumor-associated macrophages via the PI3K/AKT/mTOR signaling pathway. Cancer letters. 2018;436:119-28
24. Charalabopoulos K, Gogali A, Kostoula OK, Constantopoulos SH. Cadherin superfamily of adhesion molecules in primary lung cancer. Experimental oncology. 2004;26:256-60
25. Weber EW, Han F, Tauseef M, Birnbaumer L, Mehta D, Muller WA. TRPC6 is the endothelial calcium channel that regulates leukocyte transendothelial migration during the inflammatory response. J Exp Med. 2015;212:1883-99
26. Fan C, Chen L, Huang Q, Shen T, Welsh EA, Teer JK. et al. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. British journal of cancer. 2015;113:1735-43
27. Cordes N, van Beuningen D. Cell adhesion to the extracellular matrix protein fibronectin modulates radiation-dependent G2 phase arrest involving integrin-linked kinase (ILK) and glycogen synthase kinase-3beta (GSK-3beta) in vitro. British journal of cancer. 2003;88:1470-9
28. Andriani F, Roz E, Caserini R, Conte D, Pastorino U, Sozzi G. et al. Inactivation of both FHIT and p53 cooperate in deregulating proliferation-related pathways in lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2012;7:631-42
29. Jiang J, Gu Y, Liu J, Wu R, Fu L, Zhao J. et al. Coexistence of p16/CDKN2A homozygous deletions and activating EGFR mutations in lung adenocarcinoma patients signifies a poor response to EGFR-TKIs. Lung cancer (Amsterdam, Netherlands). 2016;102:101-7
30. Miller A, McLeod L, Alhayyani S, Szczepny A, Watkins DN, Chen W. et al. Blockade of the IL-6 trans-signalling/STAT3 axis suppresses cachexia in Kras-induced lung adenocarcinoma. Oncogene. 2017;36:3059-66
31. Silva EM, Mariano VS, Pastrez PRA, Pinto MC, Castro AG, Syrjanen KJ. et al. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PloS one. 2017;12:e0181125
32. Hossain MN, Sakemura R, Fujii M, Ayusawa D. G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun. 2006;351:645-50
33. Takauji Y, Kudo I, En A, Matsuo R, Hossain MN, Nakabayashi K. et al. GNG11 (G-protein subunit gamma 11) suppresses cell growth with induction of reactive oxygen species and abnormal nuclear morphology in human SUSM-1 cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2017;95:517-23
34. Lee HJ, Park MK, Lee EJ, Lee CH. Resolvin D1 inhibits TGF-beta1-induced epithelial mesenchymal transition of A549 lung cancer cells via lipoxin A4 receptor/formyl peptide receptor 2 and GPR32. The international journal of biochemistry & cell biology. 2013;45:2801-7
35. Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T. et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61-71
36. Xiong D, Pan J, Zhang Q, Szabo E, Miller MS, Lubet RA. et al. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget. 2017;8:18885-900
37. Ulivi P, Mercatali L, Casoni GL, Scarpi E, Bucchi L, Silvestrini R. et al. Multiple marker detection in peripheral blood for NSCLC diagnosis. PloS one. 2013;8:e57401
38. Manzoni C. LRRK2 and autophagy: a common pathway for disease. Biochemical Society transactions. 2012;40:1147-51
39. Kourtidis A, Lu R, Pence LJ, Anastasiadis PZ. A central role for cadherin signaling in cancer. Experimental cell research. 2017;358:78-85
40. Halim H, Luanpitpong S, Chanvorachote P. Acquisition of anoikis resistance up-regulates caveolin-1 expression in human non-small cell lung cancer cells. Anticancer research. 2012;32:1649-58
41. El-Zein RA, Fenech M, Lopez MS, Spitz MR, Etzel CJ. Cytokinesis-blocked micronucleus cytome assay biomarkers identify lung cancer cases amongst smokers. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:1111-9
42. Ionescu ME, Ciocirlan M, Becheanu G, Nicolaie T, Ditescu C, Teiusanu AG. et al. Nuclear Division Index may Predict Neoplastic Colorectal Lesions. Maedica. 2011;6:173-8
43. Watts TL, Cui R, Szaniszlo P, Resto VA, Powell DW, Pinchuk IV. PDGF-AA mediates mesenchymal stromal cell chemotaxis to the head and neck squamous cell carcinoma tumor microenvironment. Journal of translational medicine. 2016;14:337
44. Rajwar YC, Jain N, Bhatia G, Sikka N, Garg B, Walia E. Expression and Significance of Cadherins and Its Subtypes in Development and Progression of Oral Cancers: A Review. Journal of clinical and diagnostic research: JCDR. 2015;9:ZE05-7
45. Yang Y, Li X, Du J, Yin Y, Li Y. Involvement of microRNAs-MMPs-E-cadherin in the migration and invasion of gastric cancer cells infected with Helicobacter pylori. Experimental cell research. 2018;367:196-204
Author contact
![]() Corresponding author: Wei Li, Email: xylw2015edu.cn
Corresponding author: Wei Li, Email: xylw2015edu.cn

 Global reach, higher impact
Global reach, higher impact