Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(1):277-286. doi:10.7150/jca.27536 This issue Cite
Research Paper
The Significance Role of microRNA-200c as a Prognostic Factor in Various Human Solid Malignant Neoplasms: A Meta-Analysis
1. Department of Urology, The Affiliated Huaian No.1 People's Hospital of Nanjing Medical University, Huai'an, 223300, China.
2. Department of Urology, Nanjing First Hospital, Nanjing Medical University, Nanjing, 210006, China.
3. Department of Ultrasound, Jiangsu Province Hospital of TCM, Affiliated Hospital of Nanjing University of TCM, Nanjing, 210029, China.
4. Department of Urology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, China.
5. Department of Oncology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, China.
* Feng Wang, Lei Zhang and Haoxiang Xu contributed equally to this work.
Received 2018-5-29; Accepted 2018-8-19; Published 2019-1-1
Abstract
Objective: The aim of this study was to conduct a meta-analysis of 49 relevant studies to evaluate the prognostic value of miRNA-200c in various human malignant neoplasms.
Methods: All relevant studies were identified by searching PubMed, Embase and Web of Science until August 15st, 2018. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) of miRNA-200c for overall survival (OS) and progression-free survival (PFS)/recurrence-free survival (RFS)/disease-free survival (DFS) were calculated to investigate such associations.
Results: Overall, 49 eligible studies were included in this meta-analysis. Our results showed that high expression of miRNA-200c was significantly correlated with a poor OS in cancer (pooled HR = 1.32, 95% CI: 1.06-1.65), but was not significantly correlated with PFS/RFS/DFS in cancer (pooled HR=1.05, 95% CI: 0.84-1.23). In our subgroup analysis, high miRNA-200c expression predicted a significantly worse OS (pooled HR = 1.50, 95% CI: 1.12-2.01) only in Caucasians. Moreover, high miRNA-200c expression even showed significantly poor OS (pooled HR = 1.88, 95% CI: 1.39-2.54) in blood samples. In addition, a significantly unfavorable OS (pooled HR = 2.69, 95% CI: 1.49-4.85) and (pooled HR = 2.66, 95% CI: 1.07-6.59) associated with up-regulated miRNA-200c expression were observed in breast cancer and endometrial cancer, respectively. Besides, high miRNA-200c expression also showed significantly poor PFS/RFS/DFS (pooled HR=1.66, 95% CI: 1.03-2.67) in breast cancer.
Conclusions: Our findings indicated that high miRNA-200c expression was a promising biomarker for patient survival and disease progression in malignant tumors, especially in breast cancer and endometrial cancer. Considering the insufficient evidence, further large-scale researches and clinical studies were needed to verify these results.
Keywords: malignant neoplasms, miRNA-200c, prognosis, overall survival, progression-free survival
Introduction
MicroRNAs (miRNAs) are a class of single stranded, highly conserved, endogenous, small non-coding RNAs (18-25 nucleotides in length) [1, 2]. Mature miRNAs negatively regulate target genes expression at the post-transcriptional level by binding to the 3' untranslated region (3'UTR) of messenger RNA (mRNA), resulting in suppression of translation or degradation of mRNAs. Therefore, miRNAs play vital roles in diverse biological processes, including cell cycle, apoptosis, differentiation and growth [3-6]. Due to the aberrant expression discrepancy between tumor tissues and normal tissues, emerging studies have demonstrated that miRNAs were willing to considered as diagnostic or prognostic biomarkers for multiple human cancers [7-9]. In addition, miRNAs can be classified into two categories, including protective miRNAs which are associated with favorable prognosis, and hazardous miRNAs which are known as poor survival predictors [10, 11]. As a member of miRNAs, miRNA-200c has been studied relatively intensively and thoroughly for the prognosis of cancer.
The miRNA-200 family consists of five members: miRNA-200a, miRNA-200b, miRNA-200c, miRNA-429, and miRNA-141, is closely associated with human health and disease [12, 13]. Recent studies have shown that the miRNA-200 family members, especially miRNA-200c, played a vital role in the process of epithelial-mesenchymal transition (EMT), which was important for embryonic development, cancer, and other diseases [12-15]. MiRNA-200c is highly enriched in epithelial tissues, and is deemed to repress the expression of E-cadherin transcription inhibitors ZEB1 and ZEB2, and has a direct influence on EMT. In recent times, increasing evidences have demonstrated that miRNA-200c was associated with patient prognosis and clinicopathology significance, such as ovarian cancer, gastric cancer, pancreatic cancer, colorectal cancer, esophageal cancer, and so on [16-22]. What's more, miRNA-200c expression was also related to tumor drug resistance [23, 24]. In addition, some studies have reported that miRNA-200c could be detectable in blood samples and had the potential to be new biomarkers in patients with cancer [25, 26]. Therefore, it is necessary and timely to perform a meta-analysis to evaluate the prognostic value and clinicopathology significance of miRNA-200c expression in patients with cancer.
To investigate whether the miRNA-200c expression could serve as a prognostic or clinical biomarker for cancer, we performed a systematic review and meta-analysis by extracting summary statistics of the published literature for survival endpoints.
Materials and Methods
Search Strategy
We systematically and carefully searched online databases, including PubMed, Embase and Web of Science for original articles analyzing the prognostic value of miRNA-200c in various cancers until August 15st, 2018. The relevant studies were selected according to varying combinations of the following keywords: 'cancer', 'carcinoma', 'neoplasm', 'tumour', 'tumor', 'microRNA-200c', 'microrna-200c', 'miRNA-200c', 'miR-200c', 'overall survival', 'recurrence', and 'prognosis'. Besides, the following criteria should be considered to select the literatures: (1) the studies exploring various human malignancies; (2) the relationship between miRNA200c and cancer prognosis. In order to supplement our literature search, the bibliographies of all eligible studies were reviewed for additional relevant publications. When more than one study had been published using the same series of study subjects, only the most recent and the most complete study was chosen in this meta-analysis.
Quality assessment
This meta-analysis was strictly performed according to a critical review checklist of the Dutch Cochrane Centre proposed by MOOSE. The key points of the quality assessment included clear definitions of the following: (1) clear origin of country and description of study population; (2) clear definition of type of carcinoma; (3) clear explanation of study design; (4) clear detection of samples; (5) clear description of outcome assessment; (6) sufficient follow-up period. Studies without meeting all of the above criteria were excluded to maintain the quality of the meta-analysis. In addition, sensitivity analyses and published bias were also performed to promote the quality of this meta-analysis.
Data extraction
Two investigators (Feng Wang and Lei Zhang) independently identified all eligible studies, and uncertain data were reassessed by Haoxiang Xu. The extracted data elements included the following: (1) first author's name and year of publication; (2) study population, ethnicity and nationality; (3) study design, malignant types and detected samples; (4) HRs along with their 95% CIs associated with high miRNA-200c for overall survival (OS) and progression-free survival (PFS)/recurrence-free survival (RFS)/disease-free survival (DFS) were collected. In addition, only Kaplan-Meier curves was provided in some studies, the HRs and their 95% CIs were extracted from graphical survival plots using Engauge Digitizer version 4.1 [27, 28].
Statistical analysis
Heterogeneity test for pooled HRs was calculated by Cochran Q-test and Higgins I-squared statistic (I2). P < 0.05 was considered statistically significance. According to the heterogeneity among the pooled investigations, the fixed-effects model (Mantel- Haenszel method) or the random-effects model (DerSimonianLaird method) were chosen in this meta-analysis. If the heterogeneity was observed (P < 0.05 or I2 > 50%), a random-effects model was utilized; Otherwise, a fixed-effects model was conducted [29]. What's more, subgroup analysis upon similar characteristics were performed to minimize the influence of heterogeneity. To examine the stability and reliability of the overall meta-analysis results, sensitivity analysis was performed by excluding one single study one by one and recalculating their HRs. Publication bias was assessed by Begg's funnel plot and Egger linear regression test [30, 31]. All statistical analyses were conducted by using Stata version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Summary of enrolled studies
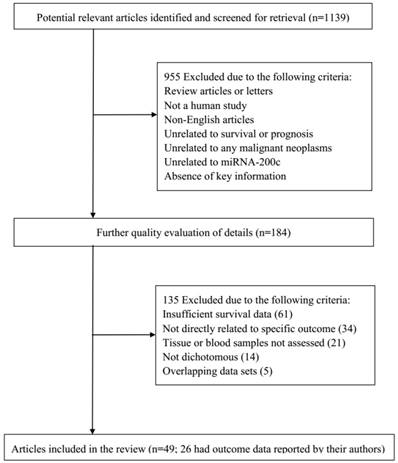
According to the study selection process, a total of 1,139 studies from PubMed, Embase, and the Web of Science were collected to focus on the relationship between miRNA-200c and the prognosis of cancer. After a primary evaluation of titles and abstracts, 955 studies were excluded if they were letters or review articles, were not human studies, were not related to prognosis or outcomes, and did not have relationship between miRNA-200c and malignancies. For further quality evaluation of remaining studies, 184 studies were excluded because of insufficient survival data, indirectly concern with specific outcomes, unrelated to tumor tissues and blood samples, incomplete or repeated data. Finally, 49 studies were considered applicable to this meta-analysis [16-22, 25, 26, 32-71]. The flowchart of literature search and selection procedure is shown in Figure 1.
Of the 49 eligible studies, 45 studies focused on OS, and 27 studies reported PFS/RFS/DFS. During these studies, 26 studies related to Asian population while 26 studies reported Caucasians. Meanwhile, 36 studies focused on tissues and other 17 studies investigated blood samples. What's more, the malignant neoplasms consisted of pancreatic cancer, ovarian cancer, bladder cancer, gastric cancer, endometrial cancer, lung cancer, breast cancer, colorectal cancer. All the analyses were retrospective studies with a maximum follow-up of 13-192 months. In this meta-analysis, all of the baseline characteristics of the studies associated with the prognosis of cancer are comprehensively listed in Table 1.
OS associated with miRNA-200c expression
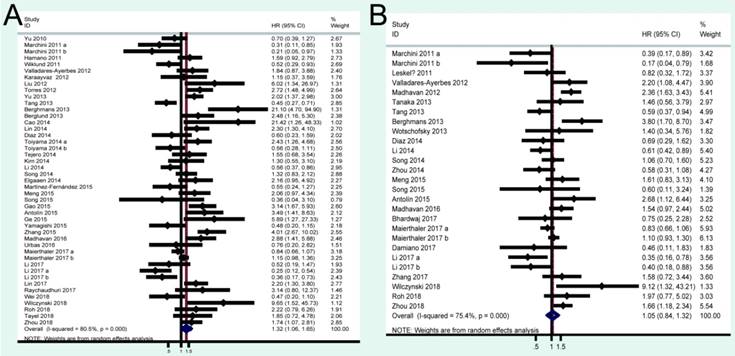
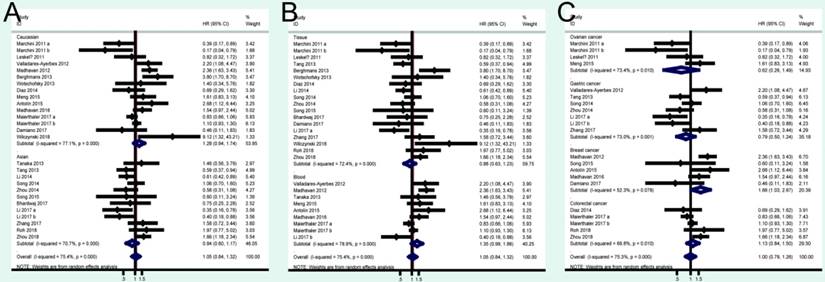
A total of 41 studies were included in OS analysis with significant heterogeneity (P < 0.001, I2 = 80.5%). Thus, a random model was used to calculate a pooled HRs and 95% CIs. Our results found that high miRNA-200c expression was a significant predictor of poor OS (pooled HR = 1.32, 95% CI: 1.06-1.65) (Figure 2A). We conducted three subtotal analyses stratified to avoid the influence of heterogeneity, including dominant ethnicity, categories of detected samples, and malignant diseases. First of all, 22 studies in Caucasians demonstrated that patients with elevated miRNA-200c expression had a significantly poorer OS (pooled HR = 1.50, 95% CI: 1.12-2.01) by a random- effects model, because of significant heterogeneity among pooled studies. However, high miRNA-200c expression was not significantly associated with enhancing OS (pooled HR = 1.17, 95% CI: 0.81-1.69) by a random model in Asian populations (Figure 3A). Secondly, in subtotal analysis stratified by the category of detected samples, high miRNA-200c expression showed a significant relationship with poor OS by a random-effects model, mainly in blood samples (pooled HR = 1.88, 95% CI: 1.39-2.54) but not in tissue (Figure 3B). Thirdly, when stratified by malignant types, pooled analysis in the breast cancer subgroup with the fixed-effects model (pooled HR = 2.69, 95% CI: 1.49-4.85) and endometrial cancer subgroup with the random-effects model (pooled HR = 2.66, 95% CI: 1.07-6.59), exhibited a significant association between up-regulated expression of miR-200c. However, no other subgroups, including ovarian cancer, gastric cancer, lung cancer and colorectal cancer discovered any significant results, as shown by stratified analyses (Figure 3C).
Flowchart of literature search and selection process.
Main characteristics of studies included in the meta-analysis.
| First author | Publication year | Case nationality | Dominant ethnicity | Study design | Number of cases | Malignant disease | Detected sample | Survival analysis | Source of HR | Maximum months of follow-up |
|---|---|---|---|---|---|---|---|---|---|---|
| Yu | 2010 | Japan | Asian | R | 99 | Pancreatic cancer | Tissue | OS | Reported | 101 |
| Marchini a | 2011 | Italy | Caucasian | R | 89 | Ovarian cancer | Tissue | OS,PFS | Reported | 139.2 |
| Marchini b | 2011 | Italy | Caucasian | R | 55 | Ovarian cancer | Tissue | OS,PFS | Reported | 139.2 |
| Hamano | 2011 | Japan | Asian | R | 98 | Esophageal cancer | Tissue | OS | Estimated | 96.7 |
| Leskelä | 2011 | Spain | Caucasian | R | 72 | Ovarian cancer | Tissue | PFS | Reported | 125 |
| Wiklund | 2011 | Denmark | Caucasian | R | 100 | Bladder cancer | Tissue | OS | Estimated | >40 |
| Valladares-Ayerbes | 2012 | Spain | Caucasian | R | 52 | Gastric cancer | Blood | OS,PFS | Estimated | 60 |
| Karaayvaz | 2012 | America | Caucasian | R | 34 | Endometrial cancer | Tissue | OS | Reported | 125 |
| Liu | 2012 | China | Asian | R | 70 | Lung cancer | Tissue | OS | Reported | 30 |
| Madhavan | 2012 | Germany | Caucasian | R | 164 | Breast cancer | Blood | PFS | Estimated | 24 |
| Torres | 2012 | Poland | Caucasian | R | 73 | Endometrioid endometrial cancer | Blood | OS | Estimated | 150 |
| Yu | 2013 | China | Asian | R | 157 | Esophageal cancer | Blood | OS | Reported | 50 |
| Tanaka | 2013 | Japan | Asian | R | 64 | Esophageal cancer | Blood | PFS | Reported | 42 |
| Tang | 2013 | China | Asian | R | 126 | Gastric cancer | Tissue | OS,DFS | Estimated | 60 |
| Wotschofsky | 2013 | Germany | Caucasian | R | 89 | Renal cell carcinoma | Tissue | PFS | Reported | 80 |
| Berghmans | 2013 | Belgium | Caucasian | R | 38 | Lung cancer | Tissue | OS | Reported | 60 |
| Berglund | 2013 | Sweden | Caucasian | R | 61 | Diffuse large B-cell lymphoma | Tissue | OS | Estimated | 192 |
| Cao | 2014 | China | Asian | R | 100 | Ovarian cancer | Tissue | OS | Reported | 56 |
| Lin | 2014 | Australia | Caucasian | R | 97 | Prostate cancer | Blood | OS | Reported | 62 |
| Diaz | 2014 | Spain | Caucasian | R | 127 | Colorectal cancer | Tissue | OS,DFS | Estimated | 120 |
| Toiyama a | 2014 | Japan | Asian | R | 156 | Colorectal cancer | Blood | OS | Reported | 70 |
| Toiyama b | 2014 | Japan | Asian | R | 182 | Colorectal cancer | Tissue | OS | Reported | 70 |
| Tejero | 2014 | Spain | Caucasian | R | 155 | Lung cancer | Tissue | OS | Estimated | |
| Kim | 2014 | SouthKorea | Asian | R | 72 | Lung cancer | Tissue | OS | Reported | 135 |
| Li | 2014 | China | Asian | R | 150 | Lung cancer | Tissue | OS,PFS | Reported | 20.6 |
| Song | 2014 | china | Asian | R | 385 | Gastric cancer | Tissue | OS,PFS | Estimated | 112 |
| Elgaaen | 2014 | Norway | Caucasian | R | 35 | Ovarian cancer | Tissue | OS | Estimated | >100 |
| Zhou | 2014 | China | Asian | R | 64 | Gastric cancer | Tissue | DFS | Estimated | >30 |
| Martínez-Fernández | 2015 | Spain | Caucasian | R | 87 | Bladder cancer | Tissue | OS | Estimated | 133.3 |
| Meng | 2015 | Germany | Caucasian | R | 163 | Ovarian cancer | Blood | OS,PFS | Estimated | 136 |
| Song | 2015 | China | Asian | R | 134 | Breast cancer | Tissue | OS,PFS | Estimated | 100 |
| Gao | 2015 | China | Asian | R | 93 | Ovarian cancer | Blood | OS | Reported | >100 |
| Antolín | 2015 | Spain | Caucasian | R | 57 | Breast cancer | Blood | OS,PFS | Reported | 310.8 |
| Ge | 2015 | China | Asian | R | 163 | Renal cell carcinoma | Tissue | OS | Reported | 24.3 |
| Yamagishi | 2015 | Japan | Asian | R | 83 | Malignant lymphoma | Tissue | OS | Estimated | >70 |
| Zhang | 2015 | China | Asian | R | 98 | Gastric cancer | Blood | OS | Reported | 60 |
| Madhavan | 2016 | Germany | Caucasian | R | 225 | Breast cancer | Blood | OS,PFS | Estimated | >30 |
| Urbas | 2016 | Austria | Caucasian | R | 78 | Biliary tract cancer | Tissue | OS | Reported | 140 |
| Bhardwaj | 2017 | india | Asian | R | 42 | Eyelid sebaceousgland carcinoma | Tissue | DFS | Estimated | 44 |
| Maierthaler a | 2017 | Germany | Caucasian | R | 308 | Colorectal cancer | Blood | OS,RFS | Reported | 72 |
| Maierthaler b | 2017 | Germany | Caucasian | R | 219 | Colorectal cancer | Blood | OS,RFS | Reported | 72 |
| Damiano | 2017 | Italy | Caucasian | R | 51 | Breast cancer | Tissue | DFS | Estimated | 92 |
| Li | 2017 | China | Asian | R | 48 | Colorectal cancer | Tissue | OS | Estimated | >60 |
| Li a | 2017 | China | Asian | R | 51 | Gastric cancer | Tissue | OS,PFS | Reported | 13 |
| Li b | 2017 | China | Asian | R | 51 | Gastric cancer | Blood | OS,PFS | Reported | 13 |
| Lin | 2017 | Australia | Caucasian | R | 89 | Prostate cancer | Blood | OS | Reported | >40 |
| Raychaudhuri | 2017 | Germany | Caucasian | R | 42 | Breast cancer | Tissue | OS | Estimated | 120 |
| Zhang | 2017 | China | Asian | R | 169 | Gastric cancer | Tissue | DFS | Reported | 65.4 |
| Wei | 2018 | China | Asian | R | 60 | Lung cancer | Tissue | OS | Estimated | 60 |
| Wilczynski | 2018 | Polish | Caucasian | R | 119 | Endometrioid endometrial cancer | Tissue | OS,DFS | Reported | 119 |
| Roh | 2018 | Korea | Asian | R | 93 | Colorectal cancer | Tissue | OS,RFS | Reported | 93 |
| Tayel | 2018 | Egypt | African | R | 25 | Colorectal cancer | Blood | OS | Estimated | 25 |
| Zhou | 2018 | China | Asian | R | 108 | Colorectal cancer | Tissue | OS,RFS | Reported | 108 |
Study design is described as prospective (P) or retrospective (R).
OS, overall survival; PFS, progression-free survival; DFS, disease-free survival; RFS, relapse-free survival; NM, not mentioned.
Recurrence associated with miRNA-200c expression
A total of 24 studies focused on the PFS/RFS/ DFS analysis revealed a protective role of high miRNA-200c expression (pooled HR=1.05, 95% CI: 0.84-1.32) by a random-effects model (Figure 2B). In the stratified analyses, increased miRNA-200c expression correlated with favorable PFS/RFS/DFS in the Asian subgroup (pooled HR=0.84, 95% CI: 0.60-1.17; Figure 4A). In detected samples subgroup, the results suggested that high miRNA-200c expression was not a significant association with enhanced PFS/RFS/DFS in blood sample (pooled HR=1.35, 95% CI: 0.99-1.86; Figure 4B), and in tissue samples (pooled HR=0.88, 95% CI: 0.63-1.23; Figure 4B). What's more, up-regulated miRNA-200c expression was found to be significantly associated with worse PFS/RFS/DFS, only in breast cancer (pooled HR=1.66, 95% CI: 1.03-2.67; Figure 4C), but no significant relevance was observed in other subgroup of ovarian cancer (pooled HR=0.62, 95% CI: 0.26-1.49; Figure 4C), gastric cancer (pooled HR=0.79, 95% CI: 0.50-1.24; Figure 4C), and colorectal cancer (pooled HR=1.13, 95% CI: 0.84-1.50; Figure 4C).
Sensitivity analyses
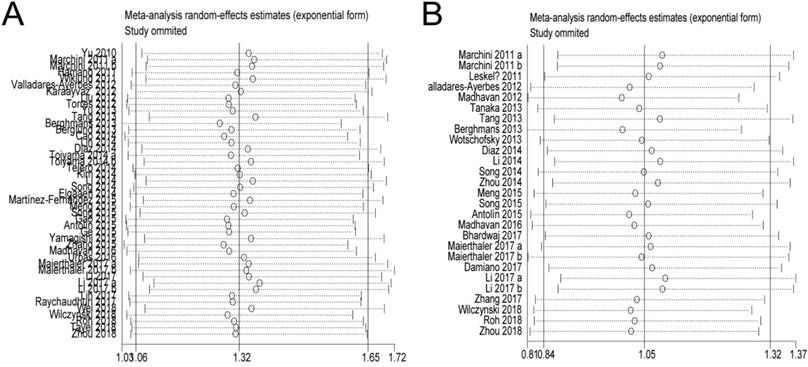
In the OS and PFS/DFS/DSS studies, sensitivity analyses did not indicate alterations in the results if any individual study was excluded (Figure 5), which suggested that no single study significantly influenced the pooled HRs or the 95% CIs.
Publication bias
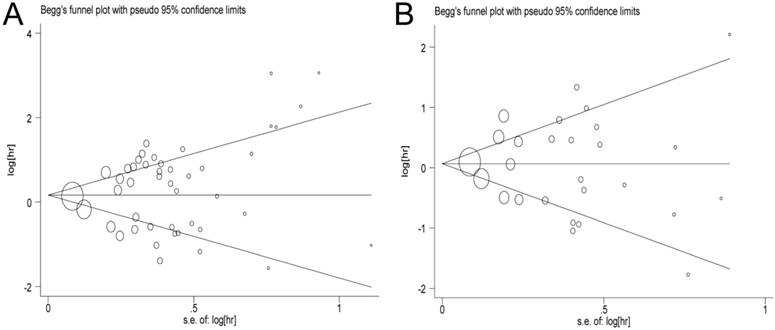
In this meta-analysis, publication bias was performed by Begg's tests and funnel plots (OS: P = 0.469, PFS/DFS/DSS: P = 0.917, respectively). As expected, the funnel plots were symmetrical, and the P values of Begg's tests were 0.897 for OS and 0.615 for PFS/DFS/DSS, suggesting that no significant publication bias was observed in the meta-analysis (Figure 6).
Forest plots of merged analyses of OS and PFS/RFS/DFS in association with miRNA-200c expression.
Forest plots of merged analyses of OS in association with miRNA-200c expression for the different subgroups. A: stratified by dominant ethnicities; B: stratified by detected sample; C: stratified by malignant diseases.
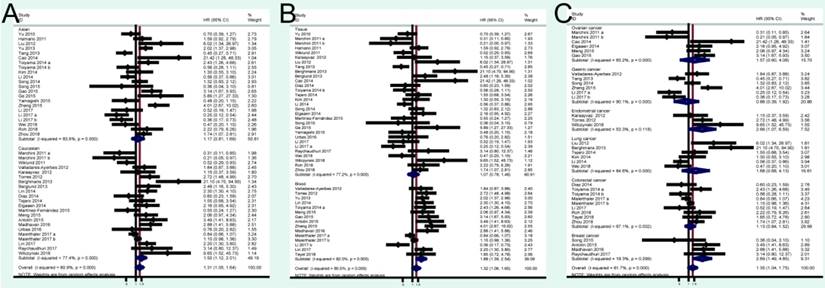
Forest plots of merged analyses of PFS/RFS/DFS in association with miRNA-200c expression for the different subgroups. A: stratified by dominant ethnicities; B: stratified by detected sample; C: stratified by malignant diseases.
Sensitivity analysis. A: Pooled HR for OS under Random-effort model; B: Pooled HR for PFS/RFS/DFS under Random-effort model.
Begg's funnel plot of publication bias test. A: Pooled HR for OS under Random-effort model; B: Pooled HR for PFS/RFS/DFS under Random-effort model.
The pooled HRs, 95% CIs, p values and I2 of OS and PFS/DFS stratified by ethnicity, detected samples
| OS | PFS/DFS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| N | pHR (95% CI) | p Value | I2 | N | pHR (95% CI) | p Value | I2 | ||
| All studies | 40 | 1.29 (1.02,1.63)a | 0.000 | 81.5% | 24 | 0.97(0.77,1.23)a | 0.000 | 74.7% | |
| Race | |||||||||
| Caucasian | 21 | 1.44(1.08,1.92)a | 0.000 | 77.1% | 14 | 1.22(0.90,1.65)a | 0.000 | 76.7% | |
| Asian | 19 | 1.16(0.77,1.76)a | 0.000 | 85.2% | 10 | 0.71(0.53,0.94)b | 0.056 | 45.7% | |
| Detected sample | |||||||||
| Tissue | 26 | 1.00(0.71,1.40)a | 0.000 | 76.6% | 15 | 0.74(0.54,1.01)a | 0.001 | 60.3% | |
| Blood | 14 | 1.88(1.38,2.57)a | 0.000 | 83.2% | 9 | 1.19(0.99,1.86)a | 0.000 | 78.9% | |
| Disease type | |||||||||
| Ovarian cancer | 6 | 1.57(0.60,4.08)a | 0.000 | 83.2% | 4 | 0.62(0.26,1.49)a | 0.010 | 73.4% | |
| Gastric cancer | 6 | 0.86(0.39,1.92)a | 0.000 | 90.1% | 7 | 0.79(0.50,1.24)a | 0.001 | 73.0% | |
| Lung cancer | 5 | 1.68(0.68,4.13)a | 0.000 | 84.6% | - | - | - | - | |
| Colorectal cancer | 6 | 0.95(0.68,1.32)a | 0.005 | 70.5% | 3 | 0.95(0.75,1.21)b | 0.124 | 52.1% | |
| Breast cancer | 4 | 2.69(1.49,4.85)b | 0.299 | 18.3% | 5 | 1.66(1.03,2.67)b | 0.078 | 52.3% | |
a, The HRs and 95% CIs of the enrolled studies were pooled using the random-effects model if the p Value for the heterogeneity test was less than 0.05 or I2 was greater than 50%. b, The HRs and 95% CIs of the enrolled studies were pooled using the fixed-effects model.
pHR, pooled HR; OS, overall survival; PFS, progression-free survival; DFS, disease-free survival; N, number of studies.
Discussion
MiRNA-200c is a member of the miRNA-200 family, which inhibits epithelialto-mesenchymal transition (EMT) by targeting the transcriptional repressor of cadherin 1 (CDH1), zinc fnger E-box binding homeobox 1 (ZEB1), and survival of motor neuron protein interacting protein 1 (SIP1), leading to prevent tumor progression and metastasis in various malignancies [13, 14, 72, 73]. Growing evidences have demonstrated that the dysregulation of miRNA-200c was involved in several malignancies. MiRNA-200c is up-regulated in several cancers, such as ovarian cancer, bile duct cancer, nasopharyngeal carcinoma, lung cancer, colorectal cancer, gastric cancer [18, 35, 74-79]; whereas miRNA-200c is down-regulated in bladder cancer, spinal cord injury, breast cancer and so on [52, 80, 81]. Meanwhile, the prognostic role of miRNA-200c in human malignancies was still controversial. Jurmeister S et al. [82] found that miRNA-200c repressed migration and invasion of breast cancer cells by targeting actin-regulatory proteins formin homology 2 domain containing 1 (FHOD1) and protein phosphatase, Mg2/Mn2 dependent, 1F (PPM1F). Song et al. [52] showed that miRNA-200c inhibited breast cancer proliferation by targeting KRAS. However, some studies found that miRNA-200c was considered as a driver of biological aggressiveness in ovarian cancer and colon cancer [83, 84]. What's more, miRNA-200c expression was related with tumor drug resistance. For instance, it has been reported that miRNA-200c was a marker of chemoresistance and aggressiveness in female reproductive cancers [23, 24]. Cochrane DR et al. [24] found that miRNA-200c suppressed invasiveness and restored sensitivity to microtubule-targeting chemotherapeutic agents in breast and ovarian cancer cells. Shimono Y et al. [85] showed that down-regulation of miRNA-200c linked breast cancer stem cells with normal stem cells. Thus, miRNA-200c was undeniably an promising biomarker of human malignancies and played important roles in tumour initiation and progression. In this meta-analysis, we aimed to explore the association between miRNA-200c expression and the prognosis and clinicopathology of cancer.
Meta-analysis is a powerful tool and can provide more sufficient results compared to a single study especially in analyzing unexplained studies. As a result, we suggested that meta-analysis had some stronger advantages to prove the prognostic role of microRNA-200c expression in multiple human malignancies. In our meta-analysis, high expression of miRNA-200c was significantly correlated with a poor OS in cancer, but was not significantly correlated with PFS/RFS/DFS in cancer. These inconsistent pooled outcomes might imply that patient survival or tumor progression was influenced by dissimilar potential mechanisms. These inconsistent outcomes might hint at dissimilar potential mechanisms that affected patient survival or tumor progression.
Furthermore, in subgroup analyses based on dominant ethnicity, categories of detected samples, and malignant diseases, we successfully drew some valuable conclusions for clinical application. Our study found that increased miRNA-200c expression predicted a significantly worse OS in Caucasians, but there was no statistical significance in Asians. Besides, the high expression of miRNA-200c could predict a significantly favorable PFS/RFS/DFS in Asians. These discrepancies might result from different hereditary backgrounds and environmental exposure. Extensive researchers have discovered the predictive values of miRNAs and diverse expression levels in different ethnic groups [86-88]. Recent evidences have suggested that miRNAs were considered as potential biomarkers in various cancers by detected in blood, including hepatocellular carcinoma [89], esophageal squamous cell cancer [90], non-small cell lung cancer [91] and breast cancer [92]. Valladares-Ayerbes M et al. reported that circulating miRNA-200c could be considered as a diagnostic and prognostic biomarker for gastric cancer [25]. In our subgroup analysis of detected samples, we found that high expression of miRNA-200c significantly related to a poor OS in blood samples, but not in tissue. Consistent with previous results, up-regulated miRNA-200c expression was found to be significantly associated with enhanced PFS/RFS/DFS in blood subgroup, but failed to find a significant consequence in tissue. These results might demonstrate that miRNA-200c might serve as a blood biomarker for cancer. Meanwhile, compared with tissue sample, detection by blood samples was faster and convenient, which could effectively evaluate survival prognosis and recurrence risk at any time point. Hence, it might be efficacious for dynamically monitoring the prognosis and therapeutic effects in cancer patients by detecting blood miRNA-200c during follow-up. What's more, in subgroup analyses based on different malignant diseases, we observed statistically significant outcomes in both the OS of the breast cancer subgroup and PFS/RFS/DFS of the breast cancer subgroup. Therefore, we suggested that high miRNA-200c expression might be a promising risk biomarker for poor prognosis in breast cancer.
These results suggested that miRNA-200c was a promising biomarker, to be used to predict cancer prognosis. However, there were still some limitations in our meta-analysis. Firstly, the number of studies included was not sufficient, which weakened the reliability of our results and led to the relative insufficiency of studies in subgroup analyses. The subgroups based on detected samples and tumor types were not fully elucidated due to the insufficient studies. Secondly, no independent study in Africans was included in the meta-analysis, which might hinder the comprehensive investigation of the relationship between miRNA-200c expression and cancer prognosis. What's more, only articles in English were included in our meta-analysis, which might lead to a possibility of language bias. Thirdly, cut-off values were different among these eligible studies; it was difficult for us to set up a baseline referring to high miRNA-200c expression. Thus, the pooled outcome could be higher or lower than the actual value, which might have caused a bias in the results. In addition, the heterogeneity existed in the total OS and PFS/RFS/DFS analyses. The source of heterogeneity was likely because of different characteristics of the patients, including race, age, pathological type, disease type, and cut-off value and so on. Moreover, as a cancer biomarker, detection of miR-200c in blood samples was faster, more convenient and more acceptable for patients to dynamically monitor their prognosis and therapeutic effects through their lifetime. Therefore, more studies were needed to evaluate further the prognostic role of miRNA-200c in various human malignant diseases.
Conclusions
In summary, this meta-analysis demonstrated that miRNA-200c over-expression could significantly predict poor prognostic outcomes in diverse human neoplasms, particularly in breast cancer and endometrial cancer. Moreover, further large-scale researches and clinical studies were needed to investigate the relationship between high miRNA-200c expression and the prognosis of cancer.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642-655
2. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854
3. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102-114
4. Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3:140-155
5. Hulf T, Sibbritt T, Wiklund ED. et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene. 2013;32:2891-2899
6. Markou A, Yousef GM, Stathopoulos E. et al. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014;60:197-205
7. Lu J, Getz G, Miska EA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838
8. Karakatsanis A, Papaconstantinou I, Gazouli M. et al. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297-303
9. Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572-594
10. Campayo M, Navarro A, Vinolas N. et al. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J. 2013;41:1172-1178
11. Lee HK, Bier A, Cazacu S. et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652
12. Burk U, Schubert J, Wellner U. et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582-589
13. Gregory PA, Bert AG, Paterson EL. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601
14. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910-14914
15. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-890
16. Leskela S, Leandro-Garcia LJ, Mendiola M. et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2011;18:85-95
17. Marchini S, Cavalieri D, Fruscio R. et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12:273-285
18. Tang H, Deng M, Tang Y. et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602-5612
19. Yu J, Ohuchida K, Mizumoto K. et al. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169
20. Diaz T, Tejero R, Moreno I. et al. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol. 2014;109:676-683
21. Tanaka K, Miyata H, Yamasaki M. et al. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20(Suppl 3):S607-S615
22. Madhavan D, Zucknick M, Wallwiener M. et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18:5972-5982
23. Cochrane DR, Spoelstra NS, Howe EN. et al. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055-1066
24. Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J Oncol. 2010;2010:821717
25. Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V. et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186
26. Li M, Gu K, Liu W. et al. MicroRNA-200c as a prognostic and sensitivity marker for platinum chemotherapy in advanced gastric cancer. Oncotarget. 2017
27. Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21:3337-3351
28. Tierney JF, Stewart LA, Ghersi D. et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
29. DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105-114
30. Egger M, Smith GD. Bias in location and selection of studies. BMJ. 1998;316:61-66
31. Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634
32. Hamano R, Miyata H, Yamasaki M. et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029-3038
33. Wiklund ED, Bramsen JB, Hulf T. et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327-1334
34. Karaayvaz M, Zhang C, Liang S. et al. Prognostic significance of miR-205 in endometrial cancer. PLoS One. 2012;7:e35158
35. Liu XG, Zhu WY, Huang YY. et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618-626
36. Torres A, Torres K, Pesci A. et al. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer. 2013;132:1633-1645
37. Yu H, Duan B, Jiang L. et al. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res. 2013;6:71-77
38. Wotschofsky Z, Busch J, Jung M. et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin Chim Acta. 2013;416:5-10
39. Berghmans T, Ameye L, Willems L. et al. Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer. 2013;82:340-345
40. Berglund M, Hedstrom G, Amini RM. et al. High expression of microRNA-200c predicts poor clinical outcome in diffuse large B-cell lymphoma. Oncol Rep. 2013;29:720-724
41. Cao Q, Lu K, Dai S. et al. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol. 2014;7:2392-2401
42. Lin HM, Castillo L, Mahon KL. et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014;110:2462-2471
43. Toiyama Y, Hur K, Tanaka K. et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735-743
44. Tejero R, Navarro A, Campayo M. et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899
45. Kim MK, Jung SB, Kim JS. et al. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch. 2014;465:463-471
46. Li J, Li X, Ren S. et al. miR-200c overexpression is associated with better efficacy of EGFR-TKIs in non-small cell lung cancer patients with EGFR wild-type. Oncotarget. 2014;5:7902-7916
47. Song F, Yang D, Liu B. et al. Integrated microRNA network analyses identify a poor-prognosis subtype of gastric cancer characterized by the miR-200 family. Clin Cancer Res. 2014;20:878-889
48. Vilming EB, Olstad OK, Haug KB. et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. 2014;14:80
49. Zhou X, Wang Y, Shan B. et al. The downregulation of miR-200c/141 promotes ZEB1/2 expression and gastric cancer progression. Med Oncol. 2015;32:428
50. Martinez-Fernandez M, Duenas M, Feber A. et al. A Polycomb-mir200 loop regulates clinical outcome in bladder cancer. Oncotarget. 2015;6:42258-42275
51. Meng X, Muller V, Milde-Langosch K. et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7:16923-16935
52. Song C, Liu LZ, Pei XQ. et al. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6:34968-34978
53. Gao YC, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36:4843-4850
54. Antolin S, Calvo L, Blanco-Calvo M. et al. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer. 2015;15:297
55. Ge YZ, Xu LW, Xu Z. et al. Expression Profiles and Clinical Significance of MicroRNAs in Papillary Renal Cell Carcinoma: A STROBE-Compliant Observational Study. Medicine (Baltimore). 2015;94:e767
56. Yamagishi M, Katano H, Hishima T. et al. Coordinated loss of microRNA group causes defenseless signaling in malignant lymphoma. Sci Rep. 2015;5:17868
57. Zhang HP, Sun FB, Li SJ. Serum miR-200c expression level as a prognostic biomarker for gastric cancer. Genet Mol Res. 2015;14:15913-15920
58. Madhavan D, Peng C, Wallwiener M. et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37:461-470
59. Urbas R, Mayr C, Klieser E. et al. Relevance of MicroRNA200 Family and MicroRNA205 for Epithelial to Mesenchymal Transition and Clinical Outcome in Biliary Tract Cancer Patients. Int J Mol Sci. 2016:17
60. Bhardwaj M, Sen S, Chosdol K. et al. miRNA-200c and miRNA-141 as potential prognostic biomarkers and regulators of epithelial-mesenchymal transition in eyelid sebaceous gland carcinoma. Br J Ophthalmol. 2017;101:536-542
61. Maierthaler M, Benner A, Hoffmeister M. et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140:176-187
62. Damiano V, Brisotto G, Borgna S. et al. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes Chromosomes Cancer. 2017;56:147-158
63. Li W, Chang J, Tong D. et al. Differential microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Oncotarget. 2017;8:35783-35791
64. Lin HM, Mahon KL, Spielman C. et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br J Cancer. 2017;116:1002-1011
65. Raychaudhuri M, Bronger H, Buchner T. et al. MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2017;162:511-521
66. Zhang L, Huang Z, Zhang H. et al. Prognostic value of candidate microRNAs in gastric cancer: A validation study. Cancer Biomark. 2017;18:221-230
67. Lei W, Kang W, Nan Y. et al. The downregulation of miR-200c promotes lactate dehydrogenase A expression and non-small cell lung cancer progression. ONCOL RES. 2018;2018:01-10
68. Wilczynski M, Danielska J, Domanska-Senderowska D, Dzieniecka M, Szymanska B, Malinowski A. Association of microRNA-200c expression levels with clinicopathological factors and prognosis in endometrioid endometrial cancer. Acta Obstet Gynecol Scand. 2018;97(5):560-9
69. Roh MS, Lee HW, Jung SB. et al. Expression of miR-200c and its clinicopathological significance in patients with colorectal cancer. PATHOL RES PRACT. 2018;214(3):350-5
70. Tayel SI, Fouda E, Gohar SF, Elshayeb EI, El-Sayed EH, El-Kousy SM. Potential role of MicroRNA 200c gene expression in assessment of colorectal cancer. ARCH BIOCHEM BIOPHYS. 2018;647:41-6
71. Lei Z, Xiaomin Y, He H, Jian C, Xiaowu X. Nicotine downregulates microRNA-200c to promote metastasis and the epithelial-mesenchymal transition in human colorectal cancer cells. J CELL PHYSIOL. 2018;2018:08-04
72. Bracken CP, Gregory PA, Kolesnikoff N. et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846-7854
73. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894-907
74. Iorio MV, Visone R, Di Leva G. et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699-8707
75. Lee JW, Choi CH, Choi JJ. et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535-2542
76. Meng F, Henson R, Lang M. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113-2129
77. Zhang L, Deng T, Li X. et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559-566
78. Zhang GJ, Zhou T, Liu ZL. et al. Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol Clin Oncol. 2013;1:379-384
79. Cristobal I, Rincon R, Manso R. et al. Deregulation of miR-200b, miR-200c and miR-429 indicates its potential relevant role in patients with colorectal cancer liver metastasis. J Surg Oncol. 2014;110:484-485
80. Liu L, Qiu M, Tan G. et al. miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med. 2014;12:305
81. Yu DS, Lv G, Mei XF. et al. MiR-200c regulates ROS-induced apoptosis in murine BV-2 cells by targeting FAP-1. Spinal Cord. 2014
82. Jurmeister S, Baumann M, Balwierz A. et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633-651
83. Chen J, Wang W, Zhang Y. et al. The roles of miR-200c in colon cancer and associated molecular mechanisms. Tumour Biol. 2014;35:6475-6483
84. Nash MA, Deavers MT, Freedman RS. The expression of decorin in human ovarian tumors. Clin Cancer Res. 2002;8:1754-1760
85. Shimono Y, Zabala M, Cho RW. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592-603
86. Yazici H, Zipprich J, Peng T. et al. Investigation of the miR16-1 (C > T) + 7 Substitution in Seven Different Types of Cancer from Three Ethnic Groups. J Oncol. 2009;2009:827532
87. Bovell LC, Shanmugam C, Putcha BD. et al. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19:3955-3965
88. Huang RS, Gamazon ER, Ziliak D. et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692-701
89. Tomimaru Y, Eguchi H, Nagano H. et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175
90. Komatsu S, Ichikawa D, Takeshita H. et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104-111
91. Heegaard NH, Schetter AJ, Welsh JA. et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130:1378-1386
92. Asaga S, Kuo C, Nguyen T. et al. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84-91
Author contact
![]() Corresponding authors: Bing Zhong, Department of Urology, Huai'an First People's Hospital affiliated with Nanjing Medical University, Huai'an, 223300, China. E-mail: 15152569186com; TEL: +08615152569186 and Zhiqiang Qin, Department of Urology, Nanjing First Hospital, Nanjing Medical University, Nanjing, 210006, China. E-mail: qinzq_urologycom; TEL: +08615950523702
Corresponding authors: Bing Zhong, Department of Urology, Huai'an First People's Hospital affiliated with Nanjing Medical University, Huai'an, 223300, China. E-mail: 15152569186com; TEL: +08615152569186 and Zhiqiang Qin, Department of Urology, Nanjing First Hospital, Nanjing Medical University, Nanjing, 210006, China. E-mail: qinzq_urologycom; TEL: +08615950523702

 Global reach, higher impact
Global reach, higher impact