Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(4):843-852. doi:10.7150/jca.27856 This issue Cite
Research Paper
Tripartite motif-containing 15 overexpression in non-small cell lung cancer is associated with poor patient prognoses
1. The Institute of Oncology, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Jinan, 250021 P.R. China
2. Department of Oncology, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Jinan, 250021 P.R. China
3. Department of Thoracic Surgery, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Jinan, 250021 P.R. China
*These authors contributed equally to this work.
Received 2018-6-13; Accepted 2019-1-1; Published 2019-1-29
Abstract
Purpose: This study aimed to comprehensively investigate the differential expression and prognostic indicators of the tripartite motif-containing (TRIM) gene family in non-small cell lung cancer (NSCLC).
Methods: The Cancer Genome Atlas (TCGA) Research Network and three datasets from Gene Expression Omnibus (GEO) database were used to assess TRIM gene family expression patterns in NSCLC. Quantitative real-time PCR and immunohistochemistry (IHC) were conducted to confirm differentially expressed genes (DEGs). Kaplan-Meier survival analysis and univariate Cox regression analysis were carried out to analyze the association between TRIM gene expression and NSCLC prognoses. Gene set enrichment analysis (GSEA) was carried on for the predict the biological processes.
Results: Of the 78 TRIM family members measured, TRIM15 was selected due to the DEGs and the prognostic value regarding NSCLC. In lung squamous cell carcinoma (LUSC), the Log2 fold change (Log2FC) of TRIM15 was 5.16 (p= 0.00575), whereas in lung adenocarcinoma (LUAD), it was 6.37 (p =6.78E-07). TRIM15 upregulation was related to poor prognoses in both LUSC (HR 1.353; 95%CI 1.023-1.789; p =0.034) and LUAD (HR 1.560; 95%CI 1.159-2.101; p =0.003). Using immunohistochemistry, TRIM15 expression was significantly higher in NSCLC tissues compared with that of matched normal tissues (p =0.0009), and similar findings were generated with tissue microarray analysis (p<0.0001).
Conclusion: TRIM15 could act as a diagnostic predictor or therapeutic target for lung cancer treatments.
Keywords: Non-small cell lung cancer, E3 ubiquitin ligases, survival analysis, TRIM family genes, TRIM15
Introduction
Lung cancer is one of the most common human tumors, and is the leading cause of all cancer-related deaths, accounting for 27% of all cancers [1]. Non-small cell lung cancer (NSCLC) represents 80%-85% of lung cancers, and lung squamous cell carcinoma (LUSC) and lung adenocarcinomas (LUAD) represent the largest subgroups of NSCLC [2]. Tumor progression is closely related to many fundamental biological processes, including post-translational modifications for which ubiquitination is one of the most critical modifications [3].
The ubiquitin system exerts essential effects in many degradation processes of regulatory proteins involved in cell-cycle regulation, signal transduction, receptor regulation, and apoptosis [4]. The ubiquitination process is mediated by E1 ubiquitin-activating enzymes (E1s), E2 ubiquitin-conjugating enzymes (E2s), and E3 ubiquitin ligases (E3s). E1s activate ubiquitin (Ub) and transfer Ub to E2s to form a thioester bond with the C terminus of Ub, and E3s mediate the final step of Ub transfer by interacting with the E2-Ub complex and a specific substrate [5]. The ubiquitination process plays crucial roles in tumor development by regulating the degradation of oncogene products and tumor suppressors [6-9]. The E3s contain three families including the homologous to E6-AP COOH terminus (HECT) family, a really interesting new gene (RING) finger-containing protein family, and the ring between ring fingers (RBR) family [10]. Up to now, more than 600 different RING-domain-containing E3s have been discovered, such as the c-CBL family, the MDM family, and others [11]. The tripartite motif-containing (TRIM) proteins contain more than 80 members in humans, and approximately 10% are RING-Domain-containing E3s [12]. Although not all TRIM family members were confirmed to have E3 Ubiquitin ligase activity and only a few have been functionally validated. TRIM gene families have attracted a lot of attention in recent years especially related to the E3 research field.
TRIM family proteins are closely related to tumor progression. TRIM24 mediated tumor cell proliferation, and when upregulated, can predict disease recurrence in prostate [13] and breast [14] cancers. In liver cancer, TRIM24 suppressed tumorigenesis in a murine model [15]. TRIM29 is another prominent member that has been studied in various tumors, including gastric, bladder, esophageal, and pancreatic cancers, which indicated that TRIM29 upregulation was a poorer prognostic indicator [16-21]. In another study, TRIM29 suppressed breast cancer cells by inhibiting TWIST1 and suppressing the epithelial to mesenchymal transition (EMT) [22, 23]. Moreover, the expression and the function of TRIM family members are tissue-restricted, and it is of great significance to comprehensively investigate this family in relation to neoplastic conditions.
The TRIM family proteins play essential roles in tumor progression and tumor suppression, which varies in different cancer types. However, due to the large number of TRIM family members and the incomplete understanding of the complex mechanisms associated with the different TRIM proteins, many molecular mechanisms and how they relate to tumors is not clear. According to our observations, studies relating to the TRIM family of proteins and lung cancer are very limited. And, to better understand the effects of the TRIM family members and their prognostic significance associated with lung cancer, we analyzed gene expression of all TRIM family members and performed prognosis analyses using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. Of the 78 TRIM family members measured, TRIM15 was selected according to its differential expression and prognostic significance regarding NSCLC. We believe that TRIM15 could eventually be used as a prognostic predictor for diagnoses, or as a therapeutic target for patients with NSCLC.
Materials and Methods
Expression analysis
The RNA sequence data and the clinical data for LUSC and LUAD were downloaded from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). A total of 551 LUSC samples and 592 LUAD samples were available in TCGA database, and the LUSC samples consisted of 502 tumor tissues and 49 normal tissues, while the LUAD samples consisted of 533 tumor tissues and 59 normal tissues.
Before analyses were performed, original data was filtered and standardized through DEseq with the R Software. Then, we analyzed the differential expression of the TRIM family members in the LUSC and LUAD samples, by comparing the RNA-seq data of the tumor and normal tissues. Results had to satisfy the following two conditions, simultaneously, which were to have a log2 fold change ≥1 and an adjusted p-value<0.05 (to be considered statistically significant). Heat maps were generated by HemI (Heatmap Illustrator, version 1.0) to support the results.
The Series Matrix Files of the GSE75037 database (containing 83 LUAD tissues and 83 matched adjacent non-malignant lung tissues), the GSE19804 database (containing 60 non-smoking female lung cancer tissues and 60 matched adjacent non-malignant lung tissues), and the GSE43458 database (containing 80 lung adenocarcinoma tissues and 30 normal lung tissues) were downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), and the probes were transformed to the gene symbols based on GPL6884 platform[24]. The GES75037, GSE19804, and GSE43458 were selected according to the following criteria: (I) the samples were from human patients; and (II) that LUSC or LUAD patients were in the datasets. Figures were generated through GraphPad Prism7.
The R2 platform: Genomics Analysis and Visualization Platform (http://r2.amc.nl) was used to obtain data, and the histograms and the box-plots were used to test the differential gene expression.
The cBioPortal for Cancer Genomics was used to select samples with well-defined mutation types, such as EGFR, KRAS, LKB1, and p53. One hundred and thirty LUAD patients and 177 LUSC patients were selected. Of the selected LUAD samples, 33,76, 43, and 107 samples had EGFR, KRAS, LKB1, and p53 mutations, respectively. In selected LUSC samples, 128 samples had p53 mutations.
Prognosis analysis
We analyzed the relationship between gene expression and prognoses using the matching results between gene expression and patient clinical data downloaded from the TCGA. When selecting data, repeated samples, samples without survival times, survival times of less than 30 days, or cases without defined pathologic stages were excluded. The final selection included 467 cases of LUSC and 482 cases of LUAD (the selection process is shown in Supplementary Figures 1A and 1B). Prognoses were considered significant if a p-value of  0.05 was obtained.
0.05 was obtained.
The Kaplan Meier-plotter (http://kmplot.com/analysis/) was used to assess the prognostic value of the significant genes using Cox regression univariate analysis to auto-select the best cutoffs [25].
Cell culture, gene silencing, and reagents
H520 cells were maintained in RPMI and 10% FBS (Biological Industries) in a humidified environment at 5% CO2 and 37oC. In this study, siRNA transfection experiments were performed when cell densities reached 30%-40% using jetPRIME transfection kits (Polyplus company). SiRNA is synthesized by Shanghai Gemma gene and the sequence is as follows: negative control 5'-UUCUCCGAACGUGUCACGUTT-3'; TRIM15-1 5'-UGAGUCUCGAACAGCGGUUTTAACCHCUGUUCGAGACUCATT-3'; TRIM15-2 5'-GCAGCAGAUUUGGAAGGAGTTCUCCUUCCAAAUCUGCUGCTT-3'; TRIM15-3 5'-UCAAGAAGACCAGAAGCUUTTAAGCUUCUGGUCUUCUUGATT-3'; TRIM15-4 5'-CUCCUGACCUUGUCAAGAATTUUCUUGACAAGGUCAGGAGTT-3'.
The cell cycle assay
A Cell cycle detection kit was purchased from BD company. Adherent cells were digested with 0.25% trypsin and then collected into centrifuge tubes and washed three times with pre-cooled PBS to completely remove the trypsin. Cells were collected using a centrifuge at a 300g centrifugal force and 4 °C to avoid cell damage. The cells were resuspended by adding 500µl binding buffer and were then transferred to flow tubes. While vortexing (to loosen the pellet), 5 mL of cold 70 to 80% ethanol was added drop-by-drop into the cell pellet, and the cells were incubated at -20°C for a minimum of 2 hours. After the incubation, cells were washed twice to remove the ethanol. The washing was in 1X PBS. The cells were centrifuged for 10 minutes at 1,000 to 1,500 rpm, and then the supernatant was aspirated. The cells were incubated for 15 minutes at room temperature, and the cell cycle assay was performed with the Muse Cell Cycle assay kit, and the results were analyzed with flowjo_v10 software.
Patients, RNA extractions, and quantitative real-time PCR
Seventeen pairs of primary NSCLC and matched normal tissues were obtained from patients at the Shandong Provincial Hospital Affiliated with Shandong University and informed consents were obtained from all the patients. All tissue samples were from untreated patients undergoing surgery, and all clinicopathologic information (age, gender, pathology, differentiation, invasion depth, lymph node metastasis) was available. The study was approved by the Shandong Provincial Hospitals' Ethical Review Committee. All samples were snap frozen in liquid nitrogen and stored at -80 ◦C until the extraction of RNA.
Total RNA in tumor samples and the normal lung tissues were prepared with RNA TRIzol (Shanghai Excell biology, Inc). RNA quality was confirmed in an Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA synthesis was conducted with the High Capacity RNA-to-cDNA Kit. Quantitative real-time PCR was performed using SYBR Green PCR master mix on Roche LightCycler 480 System as follows: 1 cycle of 95 °C for 30s, 55 cycles of 95 °C for 5 s, 55 °C for 30 s and 72°C for 30s. A dissociation step was performed to generate a melting curve to confirm the specificity of the amplification. Real-time PCR assay kits were purchased from Takara Bio (Dalian, China). GAPDH was used as an internal reference gene. The fold change of gene expression was calculated by the 2- ΔΔ Ct method. Experiments were repeated in triplicate. The primer sequences are as follows: TRIM15 forward: 5'-AGGAAGGGAGAGATGGGACT-3', TRIM15 reverse: 5'-GAAGGTGAAGATGGGCTCCT-3'; GAPDH forward: 5'-CATGAGAAGTATGACAACAGCCT-3', reverse: 5'-AGTCCTTCCACGATACCAAAGT-3').
Immunohistochemistry
Tissue microarrays were purchased from Iwill Biological Technology (Wuhan, China), which contained 35 lung cancer tissues with the matched normal tissues. The TRIM15 antibody was purchased from the Proteintech Group, USA. Immunohistochemistry was performed as described previously [26]. Briefly, tissue sections underwent deparaffinization in xylene followed by rehydration through graded ethanol at room temperature and then were heated in a 1 mmol/L ethylenediaminetetraacetic acid (EDTA) buffer (water bath, 96-98 °C) for 15 min for antigen retrieval. Peroxidase activity was quenched with 0.3% hydrogen peroxide, and nonspecific binding was blocked with 10% normal goat serum. The primary anti-TRIM15 antibody was used at a dilution of 1:300. Normal serum was used as a negative control. After washing with PBS, slides were incubated with a biotin-labeled secondary antibody for 30 min and streptavidin-horseradish peroxidase for 30 min. Diaminobenzidine was used as the chromagen and slides were counterstained with hematoxylin.
To quantitate immunohistochemical staining, the following criteria were followed; (1) 0-score, absence of staining or the extent of staining is less than 10%; (2) 1-score, weak staining, or extent of staining is more than 10% and less than 30%, or both; (3) 2-score, moderate staining, or the extent of staining is more than 30% and less than 50%, or both; (4) 3-score, intense staining and the extent of staining is more than 50%, and less than 70%; (5) 4-score, intense staining and the extent of staining is more than 70%. Results were assessed by two pathologists, independently.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was used to analyze the correlations between TRIM15 expression and cancer-related pathways, using the above mentioned LUAD datasets downloaded from TCGA. The detailed protocol for GSEA is available on the Broad Institute Gene Set Enrichment Analysis website (http://www.broad.mit.edu/gsea). Datasets and phenotype label files were created and loaded onto GSEA software (v2.0.13; Broad Institute, Cambridge, USA). The gene sets were downloaded from the Broad Institute website. The phenotype label was TRIM15-high versus TRIM15-low. The number of permutations was set to 1000.
Statistical analysis
The Kaplan-Meier analysis and the univariable Cox proportional hazard model were performed to make the survival curve and determine the hazard ratio to analyze the prognostic effect of gene expression differences on survival using SPSS Statistics 22. Receiver operating characteristic (ROC) curves were used to identify the optimal cutoff points. To avoid the emergence of the bias, a running log-rank test was used at intervals between the 5th and 95th percentiles of the normalized expression of each gene. The cutoff value of each gene was defined when the log-rank statistical value was maximal. For all analysis, a p-value of <0.05 was considered to be statistically significant.
Results
Differential gene expression analysis of TRIM gene family members
Nineteen members in LUSC (TRIM29, TRIM22, TRIM16L, TRIM59, TRIM7, TRIM16, TRIM2, TRIM28, TRIM71, TRIM45, TRIM9, MEFV, TRIM17, TRIM15, TRIM43, TRIM72, TRIM58, TRIM63, and TRIM49C) and fourteen members in LUAD (TRIM2, TRIM15, TRIM16L, TRIM54, TRIM59, TRIM31, TRIM17, TRIM46, TRIM72, TRIM9, TRIM58, MEFV, TRIM45, and TRIM6) (supplementary Table 1) showed significant differential expression, an ten members (TRIM2, TRIM15, TRIM16L, TRIM59, TRIM17, TRIM72, TRIM9, TRIM58, MEFV, and TRIM45) showed significant differential expression in both LUSC and LUAD. Gene expression comparisons between normal and tumor tissues in LUSC and LUAD tissues are shown in Fig. 1A and Fig. 1B, respectively. Gene expression was ranked compared with normal expression from the highest to most similar to lowest, which meant that the top and bottom were the least similar to normal.
The prognostic value of the TRIM gene family members
We made the K-M survival curves for the members whose differential expressions were significant in LUSC and LUAD, respectively (Supplementary Fig. 2 and Supplementary Fig. 3). Three members (TRIM2, TRIM15, and TRIM58) showed significant prognostic value in both LUSC and LUAD (Fig. 2). TRIM58 expression was lower in tumor tissues compared with normal tissues, but among tumor tissues, lower expression was associated with better prognoses. These results of expression and prognosis were mutually exclusive, which were not easily explained. Fold change in TRIM2 expression was much less than TRIM15, and the relation between expression and prognosis was also mutually exclusive. TRIM15 was overexpressed in tumor tissues compared with normal tissues, and high expression was associated with poor prognoses. We selected TRIM15, as a diagnostic and prognostic indicator, with significance in differential expression and prognosis analyses regarding LUAD and LUSC. No significant differences in TRIM15 expression concerning the different stages were seen, but in patients with stageⅠ-Ⅱ LUSC and Ⅲ-Ⅳ LUAD, low TRIM15 expression had a significantly better prognosis (Supplementary Fig. 4). Also, the univariable Cox proportion hazards model analyzed the prognostic value of all TRIM gene family members (Supplementary Table 2) using the clinical data from the TCGA.
Heatmaps showed the expression of the tripartite motif-containing (TRIM) family members in lung squamous cell carcinoma (LUSC) (A) and lung adenocarcinoma (LUAD) tissues, (B) the degree of color differences indicated the level of gene expression. Expression of TRIM members was ranked from the highest, to the most similar, and then to the lowest compared with normal expression. The top and bottom of the heatmaps contained the least similar expression ranking compared with normal tissue expression.
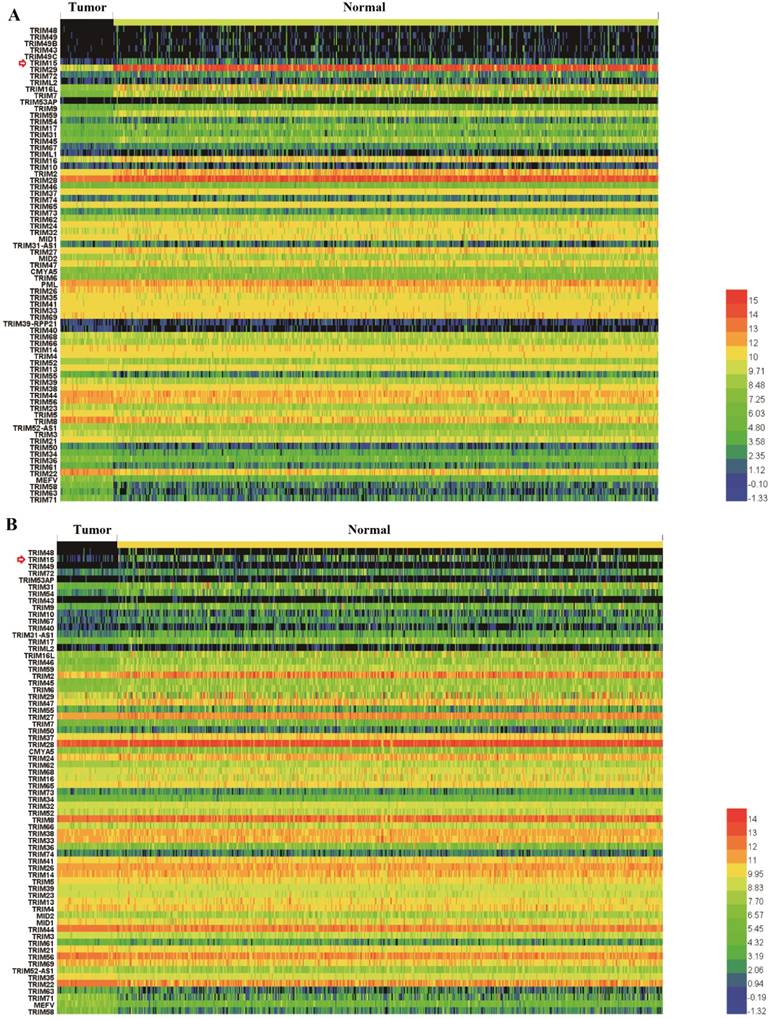
The Kaplan-Meyer survival curves of tripartite motif-containing 15 (TRIM15) (A), TRIM2 (B), and TRIM58(C) in LUSC and LUAD.
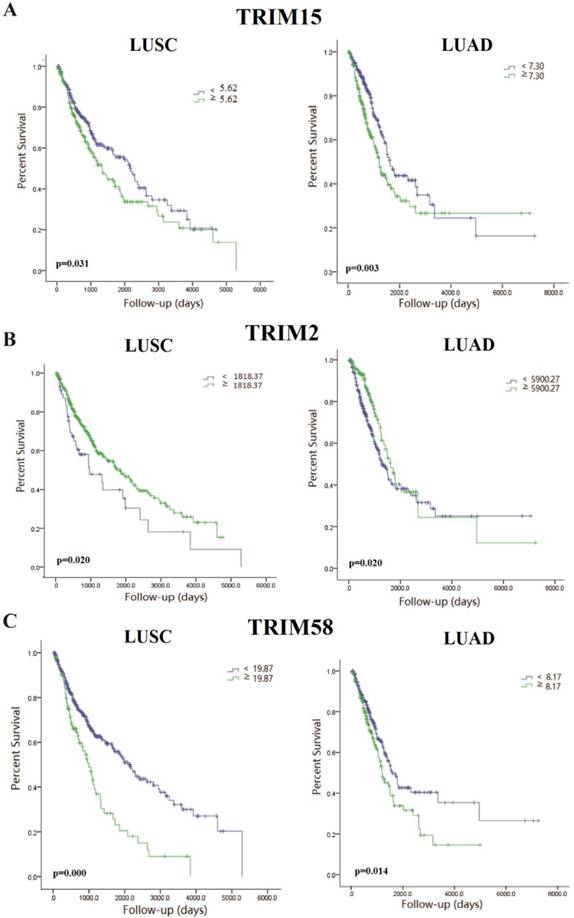
To further clarify the relationship between TRIM15 expression and prognoses, we analyzed the relationship between TRIM15 and several important mutations of lung cancer. We performed a relationship between TRIM15 expression and prognoses in EGFR, LKB1, and KRAS wild-type and mutant LUAD patients and p53 wild-type and mutant LUAD and LUSC patients. We also saw differences in TRIM15 expression between mutant and wild-type patient tissues concerning all of the above four genes. We found that patients with EGFR mutations had significantly higher TRIM15 expression compared with that of wild-type patients (Supplementary Fig. 5C). In EGFR mutations LUAD patients, patients with low TRIM15 expression had a marginally different prognosis compared with those with high TRIM15 expression (Supplementary Fig. 5K). In KRAS mutations LUAD patients, patients with low TRIM15 expression had a better prognosis compared with those with high TRIM15 expression (Supplementary Fig. 5M).
Verification of the reliability of TRIM15 as a prognostic indicator
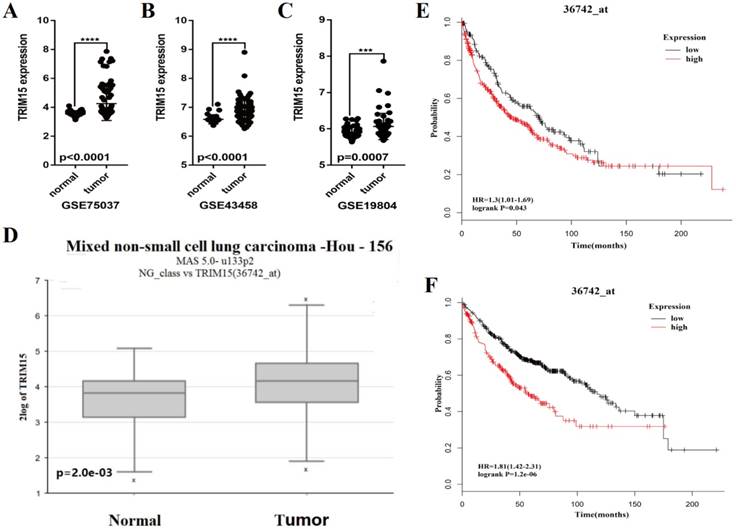
We analyzed the expression of TRIM15 using the Series Matrix File of GSE75037, GSE19804 and GSE43458 database. The expression of TRIM15 was significantly higher in tumor tissue than normal tissues (p<0.0001 in GSE75037 and GSE43458, p=.0007 in GSE19804), as show in Fig. 3A-C. The data was confirmed in R2 (36742_at). TRIM15 was up-regulated (p=2e-03, Fig. 3D) in LUAD. Meanwhile, in the dataset (36742_at) in Kaplan-Meier Plotter, the univariate Cox regression analysis showed significant in prognosis in both LUSC and LUAD (HR = 1.3, CI 1.01-1.68, p=0.043 in LUSC and HR=1.81, CI 1.42-2.31, p=1.2e-06 in LUAD) (Fig. 3E-F). We analyzed the prognosis of the whole family of TRIM family using Kaplan-Meier Plotter, and the detail data is shown in supplementary table 3.
TRIM15 was upregulated in tumor tissue compared with matched normal tissue
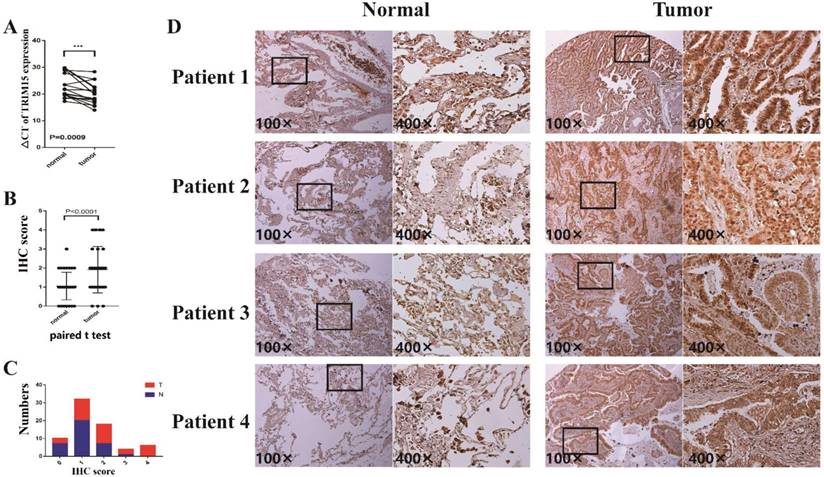
The clinicopathologic features of 17 patients were shown in supplementary table 4. In 17 pairs of tissues, TRIM15 was up-regulated in 16 patients (Fig. 4A). The TRIM15 expression was significantly higher in NSCLC tissues than in matched normal tissues (p =0.0009).
In tissue microarray analysis containing 35 paired tissues, TRIM15 expression was significant higher in tumor tissues compared with that of the matched normal tissues (p<0.0001), as shown in Fig. 4B. The proportions of tumor tissues and the normal tissues in each score and the expression levels of normal tissues and tumor tissues are shown in Fig. 4C-D.
Gene set enrichment analysis (GSEA)
High TRIM15 expression was closely associated with glycolysis, oxidative phosphorylation, the cell cycle, and processes associated with the proteasome and interferon. All p-values were and false discovery rates (FDRs) were <0.001 (Supplementary Fig. 6). Based on the results of the GSEA analysis, cell cycle analysis showed that the cell cycle changed in cells with TRIM15 knocked down (Supplementary Fig. 7).
The scatter diagram showed that the tripartite motif-containing 15 (TRIM15) was upregulated in tumor tissues compared with normal tissues in three GEO databases (p<0.0001 in GSE75037 (A), p<0.0001 in GSE43458 (B), and p=0.0007 in GSE19804 (C)); (D) TRIM15 upregulation was significant (p=2e-03) in the dataset 36742 _at from R2. (E) The univariate Cox regression analysis of TRIM15 in LUSC (HR = 1.3, CI 1.01-1.69, p=0.043) identified in the 36742_at dataset using the Kaplan-Meier Plotter. (F) The univariate Cox regression analysis of TRIM15 LUAD (HR=1.81, CI 1.42-2.31, p=1.2e-06) identified in the 36742 dataset using the Kaplan-Meier Plotter.
(A) The ΔCT of tripartite motif-containing 15 (TRIM15) expression was significantly lower (the lower ΔCT means higher expression of TRIM15) in non-small cell lung cancer (NSCLC) tissues than that in matched normal tissues (p =0.0009) in 17 patients analyzed by quantitative real-time PCR and normalized to GAPDH expression; (B) The paired t-test showed that TRIM15 expression was significantly higher in tumor tissues compared with matched normal tissues (p<0.0001); (C) The proportions of tumor tissues and the normal tissues in each score; (D) The expression level of tumor tissues compared with normal tissues.
Discussion
According to previous research, TRIM family proteins that account for a large proportion of E3s, play essential roles in many fundamental biological processes, especially in that of cancer development [27]. Researchers have paid more attention to certain TRIM family members and abundant research has been performed regarding TRIM protein associations with tumors. And although the current research about TRIM family members and various cancer types are abundant, systematic studies looking at TRIM family members in lung cancer has never been performed, and only a small number have been functionally validated. Therefore, we analyzed 78 TRIM members to screen for important biomarkers that could provide a diagnosis or prognosis in LUSC and LUAD.
During our analysis, three members (TRIM2, TRIM15, and TRIM58) had significant expression differences, and their K-M curves showed prognostic value in both LUSC and LUAD. We selected TRIM15 as an indicator of NSCLC diagnosis and prognosis. TRIM58 expression was lower in tumor tissues compared with normal tissues, but among tumor tissues, the lower expression was associated with better prognoses. In this case, the results of expression and prognosis were mutually exclusive. If we selected to use TRIM58 as the diagnosis and prognosis indicator, the results would be confusing because it would be difficult to identify if lower TRIM58 expression indicated a normal condition or a poor prognosis. It was the same for TRIM2. However, overexpression of TRIM15 was associated with poor prognoses and was overexpressed in tumors compared with normal tissues with no expression in normal tissues. Thus, it could be used as an important indicator not only for diagnoses but also for prognoses in future clinical studies. We assessed upregulated TRIM15 expression in 17 pairs of NSCLC tissues by RT-PCR (p=0.0009) and 35 pairs of tissues by immunohistochemistry (p<0.0001), and results showed that TRIM15 expression was significantly higher in NSCLC tissues than in matched normal tissues.
TRIM15 is also known as RNF93 or ZNFB7. There three common motifs in the N-terminal region that are common to all members of the TRIM family of proteins: a RING domain, one or two B-box domains, and a coiled-coil domain. In the C-terminal region, they contain a PRY/SPRY domain [28]. TRIM15 research involving the regulation of inflammatory and innate immune signaling in response to antiretroviral activity has been demonstrated [29, 30], and authors identified that type Ⅰ interferon secretion was strongly dependent on TRIM15, which was in agreement with our GSEA analysis. Some research on gastrointestinal tumors has shown that TRIM15 plays a vital role in tumor development, but no researches concerning lung cancer have been reported. Previous studies demonstrated TRIM15 to be a possible candidate gene for the diagnosis of gastric cancer due to changes in DNA methylation patterns [31]. A recent study reported that TRIM15 was downregulated and functioned as a tumor suppressor in colon cancer [32]. Authors stated that the mechanism might be related to the TRIM15 PRY/SPRY regions rather than RING domains and that the PRY/SPRY regions were associated with cell migration. In another study, TRIM15 was described as a focal adhesion protein, and cell migration was impaired in TRIM15-depleted cells. TRIM15-depleted cells had reduced disassembly rates, continual growth of focal adhesions, and cell sizes were increased with greater focal adhesion durations. In our study, TRIM15 was not expressed in normal lung tissues and was noticeably increased in NSCLC tissues. Moreover, overexpressed TRIM15 indicated poorer prognoses, which could be related to high TRIM15 expression promoting migration.
In addition, given the critical role of mutations such as p53, KRAS, LKB1 and p53 in lung adenocarcinoma and p53 in squamous cell carcinoma [33-35], we analyzed the association between TRIM15 and these mutations. We found that patients with EGFR mutations had significantly higher TRIM15 expression compared with wild-type patients. And in EGFR mutations LUAD patients, prognosis of patients with low TRIM15 expression were marginally different compared with those with high TRIM15 expression. Also, in KRAS mutations LUAD patients, patients with low TRIM15 expression had better prognoses compared with LUAD patients with high TRIM15 expression. The link with these important mutations also indirectly indicates that TRIM15 can be used as a prognostic indicator in lung cancer patients.
To investigate which biological processes TRIM15 participates in, we performed GSEA analysis. Results showed that many biological processes and signaling pathways were strongly impacted with TRIM15 upregulation, including glycolysis, oxidative phosphorylation, the cell cycle, and processes associated with proteasomes. Also, our cell cycle test results showed that knocking down TRIM15 significantly affected cell cycle progression, which indicates that TRIM15 might be necessary for tumor cell development.
To our knowledge, little is known about the biological properties of TRIM15 in lung cancer patients, and until now, no reports investigating the possible association between TRIM15 expression and lung cancer prognosis have been published. In this study, we explored the association between TRIM15 gene expression and the prognosis of LUAD and LUSC patients and identified, for the first time, that TRIM15 was a lung cancer oncogene. TRIM15 could be used to diagnose or predict prognoses of lung cancer patients. Moreover, as a lung cancer oncogene, it could become a therapeutic target in the treatment of lung cancer patients. Further research looking at how TRIM15 affects tumorigenesis and tumor progression mechanistically is needed, and could become a vitally important factor for future lung cancer research.
Abbreviations
TCGA: The Cancer Genome Atlas; GEO: Gene Expression Omnibus; IHC: Immunohistochemistry; GSEA: Gene set enrichment analysis; LUSC: Lung squamous cell carcinoma; LUAD: Lung adenocarcinoma; NSCLC: Non-small cell lung cancer; E1s: E1 ubiquitin-activating enzymes; E2s: E2 ubiquitin-conjugating enzymes; E3s: E3 ubiquitin ligases; Ub: ubiquitin; RING- finger-containing protein family: the really interesting new gene - finger-containing protein family; RBR family: RING-Between RING-RING family; TRIM: tripartite motif-containing; EDTA: Ethylenediaminetetraacetic acid; ROC: Receiver operating characteristic; FDR: False discovery rate.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The work was supported by Provincial Science Foundation of Shandong (ZR2014HM100), National Natural Science Foundation of China (81672288) and Key Research Development Plan of Shandong Province (2015GSF118109).
Compliance with Ethical Standards
This study was approved by the Shandong Provincial Hospital's Ethical Review Committee. Our research involved human participants. And, the authors declare no competing financial interests.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Lemjabbar-Alaoui H, Hassan OU, Yang Y-W, Buchanan P. Lung cancer: Biology and treatment options. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2015;1856:189-210
3. Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nature reviews Molecular cell biology. 2011;12:439
4. Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425-79
5. Pickart CM. Mechanisms underlying ubiquitination. Annual review of biochemistry. 2001;70:503-33
6. Ford JM, Baron EL, Hanawalt PC. Human fibroblasts expressing the human papillomavirus E6 gene are deficient in global genomic nucleotide excision repair and sensitive to ultraviolet irradiation. Cancer research. 1998;58:599-603
7. Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL. et al. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proceedings of the National Academy of Sciences. 1991;88:139-43
8. Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D. et al. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes & development. 1995;9:1586-97
9. Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitinproteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773-85
10. Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays in biochemistry. 2005;41:15-30
11. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399-434
12. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L. et al. The tripartite motif family identifies cell compartments. The EMBO journal. 2001;20:2140-51
13. Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D. et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer cell. 2016;29:846-58
14. Tsai W-W, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S. et al. TRIM24 links a noncanonical histone signature to breast cancer. Nature. 2010;468:927
15. Liu X, Huang Y, Yang D, Li X, Liang J, Lin L. et al. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PLoS One. 2014;9:e85462
16. Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K. et al. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Annals of surgical oncology. 2007;14:2543-9
17. Sho T, Tsukiyama T, Sato T, Kondo T, Cheng J, Saku T. et al. TRIM29 negatively regulates p53 via inhibition of Tip60. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2011;1813:1245-53
18. Wang L, Yang H, Palmbos PL, Ney G, Detzler TA, Coleman D. et al. ATDC/TRIM29 phosphorylation by ATM/MAPKAP kinase 2 mediates radioresistance in pancreatic cancer cells. Cancer research. 2014;74:1778-88
19. Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L. et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and β-catenin stabilization. Cancer cell. 2009;15:207-19
20. Palmbos PL, Wang L, Yang H, Wang Y, Leflein J, Ahmet ML. et al. ATDC/TRIM29 Drives Invasive Bladder Cancer Formation through miRNA-Mediated and Epigenetic Mechanisms. Cancer research. 2015;75:5155-66
21. Lai W, Zheng X, Huang Q, Wu X, Yang M. Down-regulating ATDC inhibits the proliferation of esophageal carcinoma cells. European review for medical and pharmacological sciences. 2014;18:3511-6
22. Ai L, Kim W-J, Alpay M, Tang M, Pardo CE, Hatakeyama S. et al. TRIM29 suppresses TWIST1 and invasive breast cancer behavior. Cancer research. 2014;74:4875-87
23. Zeng SX, Cai QC, Guo CH, Zhi LQ, Dai X, Zhang DF. et al. High expression of TRIM29 (ATDC) contributes to poor prognosis and tumor metastasis by inducing epithelialmesenchymal transition in osteosarcoma. Oncology reports. 2017;38:1645-54
24. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41:D991-D5
25. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241
26. Zhang S, Qiu C, Wang L, Liu Q, Du J. The elevated level of ERalpha36 is correlated with nodal metastasis and poor prognosis in lung adenocarcinoma. Steroids. 2014;87:39-45
27. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends in biochemical sciences. 2017;42:297-311
28. James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proceedings of the National Academy of Sciences. 2007;104:6200-5
29. Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J. et al. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. Journal of virology. 2013;87:257-72
30. Uchil PD, Quinlan BD, Chan W-T, Luna JM, Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS pathogens. 2008;4:e16
31. Cheng Y, Yan Z, Liu Y, Liang C, Xia H, Feng J. et al. Analysis of DNA methylation patterns associated with the gastric cancer genome. Oncology letters. 2014;7:1021-6
32. Lee O-H, Lee J, Lee KH, Woo YM, Kang J-H, Yoon H-G. et al. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2015;1853:409-21
33. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science (New York, NY). 2004;304:1497-500
34. Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD. et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:2890-6
35. Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science (New York, NY). 1991;253:49-53
Author contact
![]() Corresponding author: Jiajun Du, Department of Thoracic Surgery and Institute of Oncology, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Jinan, 250021 P.R. China. Tel: +86-0531-6877-7100; Fax: +86-0531-6877-7100; E-mail: dujiajunedu.cn
Corresponding author: Jiajun Du, Department of Thoracic Surgery and Institute of Oncology, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Jinan, 250021 P.R. China. Tel: +86-0531-6877-7100; Fax: +86-0531-6877-7100; E-mail: dujiajunedu.cn

 Global reach, higher impact
Global reach, higher impact