3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(4):903-910. doi:10.7150/jca.28169 This issue Cite
Research Paper
Regulatory sequence analysis of semaphorin 4D 5' non-coding region
1. Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College, Kunming 650118, PR China
2. Kunming Children's Hospital (The Affiliated Children's Hospital of Kunming Medical University), Institute of Yunnan Pediatric Disease Research, Kunming 650228, PR China
3. Yunnan Key Laboratory of Vaccine Research & Development on Severe Infectious Diseases, Kunming 650118, PR China
4. Kunming Medical University Haiyuan College, Kunming 650106, PR China
*These authors contributed equally to this work.
Received 2018-6-28; Accepted 2018-12-20; Published 2019-1-29
Abstract
Semaphorin 4D (Sema4D) has been proven to be one of the hypoxia effectors regulated by hypoxia inducible factor (HIF-1) in multiple cells, and play a role in angiogenesis like VEGF. However, the regulatory sequence characteristics of the Sema4D are not clarified. The possible hypoxia response element (HRE) sequences in 5' non-coding Region before ATG start codon of Sema4D were screened, followed by point mutagenesis and luciferase assay analysis. Sequencing and alignment of this region in 11 cancer cell lines and 4 normal cell lines were also performed, followed by cloning, mutation and luciferase assay analysis. The results showed that there were four possible HREs (HRE1-4) sequences in 1275bp range before ATG start codon. Among HRE1-4, HRE2 and HRE4 were functional HIF-1α binding sites. In addition, these two binding sites play different roles in the regulation of Sema4D expression in HUVEC and Caco-2 cells. There were three nucleotide variants (T471C/A600G/C862T) frequently detected in cancer cell lines. The site variation rates of T471C/A600G/C862T were 72.7%, 18.2%, and 72.7% in cancer cells respectively. Luciferase assays showed that T471C and C862T could significantly increase the expression efficiency of downstream target genes. Furthermore, secondary structure prediction showed that mutations at T471C and C862T apparently lead to change of the gene structure. Our study describes the sequence characteristics of 5' non-coding region of Sema4D, enhances our understanding of the regulatory mechanism of Sema4D and benefits the development of a possible anti-angiogenesis therapeutic strategy for malignancies.
Keywords: Hypoxia Inducible Factor 1α, Semaphorin 4D, 5' non-coding Region
Introduction
Semaphorin 4D (Sema4D) belongs to the semaphorin family, which binds to receptors such as Pleixn-B1 (transmembrane structure receptor), CD72 (cluster of differentiation antigen 72, mainly expresses on lymphocyte cells), then activates downstream signaling molecules. A previous study showed that Sema4D could be a pro-vascular element in cancer (1-3). In PAE cells, soluble Sema4D promoted cell migration and developed a “tubulogenic” phenotype (4). As a proangiogenic response effecter, the interaction of Sema4D with plexin-B1 is dependent on the COOH-terminal PDZ-binding motif of plexin-B1, which binds two guanine nucleotide exchange factors (GEFs) for Rho and is mediated by the activation of Rho-initiated (Ras homolog gene family) pathways (4-8). Sema4D is regulated by HIF-1 and plays a role in tumor-associated macrophage-induced metastatic behavior and is clinically significant in colon cancer (9). Furthermore, studies have shown HIF-mediated Sema4D induction in the generation of the pro-angiogenic phenotype in oral squamous cell carcinoma (OSCC) and have determined its biological significance for tumor growth and vascularity (10).
Hypoxia is the most common characteristic in the microenvironment of solid tumors because tumor progression and metastasis depend on the ability of cancer cells to initiate angiogenesis and ensure the delivery of oxygen and nutrients (11, 12). HIF-1 works in multiple cell-autonomous and non-cell-autonomous processes such as metabolism, angiogenesis, matrix invasion, and cancer metastasis by binding with target genes (13-15). HIF-1 consists of two parts, HIF-1α and HIF-1β. HIF-1α contacts DNA directly and binds to DNA much more strongly than HIF-1β (16). In hypoxia, the HIF-1 transcriptional complex drives the induction or repression of a myriad of genes through binding to hypoxia response elements (HREs, 5'-RCGTG-3') within the untranslated region of target genes (11). In hypoxic tissues, hypoxia inducible factor HIF-1α is accumulated and stimulates the transcription of genes that have HREs in their promoters, including Sema4D (17). Previous research showed that Sema4D is induced by hypoxia in a HIF-1-dependent manner and influences endothelial cell migration and tumor vascularity; however, the specific regulatory mechanism was not yet classified (10, 18).Sema4D has been proven to be a hypoxia effector and regulated by HIF-1 in cancer cells (10). However, the actual binding sequence of the HIF-1 complex is yet to be elucidated. The regulatory sequence characteristics of the Sema4D 5' non-coding region remain unclear in normal and cancer cells. It has been predicted that the Sema4D promoter region is likely regulated by a variety of factors. The sequence characteristics of the Semaphorin 4D 5' non-coding region require further elucidation. In this study, we attempted to determine the HRE binding sites, the important variants sequences before ATG start codon of SEMA 4D and possible biological effects.
Materials and Methods
Cell culture
Cancer cells, including A549 (lung cancer cell), Caco-2 (colorectal cancer cell), CNE (nasopharyngeal carcinoma cell), RD (rhabdomyoma cell), Tca8113 (tongue cancer cell), SK-OV-3 (ovarian cancer cell), Jurkat (acute T cell leukemia cell), HepG-2 (liver cancer cell), SK-N-SH (neuroblastoma cell), HL-60 (anterior myeloid cells), U937 (histiocytic lymphoma cell), and normal cells, including BEAS-2B (bronchial epithelial cell), HUVEC (human umbilical vein endothelial cell), MRC-5 (embryonic lung fibroblasts cell), 293T (renal epithelial cell), were kept by the Institute of Medical Biology, Chinese Academy of Medical Science & Peking Union Medical College. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM, 06-1055-57-1ACS, BI, USA) or Roswell Park Memorial Institute 1640 medium (RPMI 1640, 01-100-1ACS, BI, USA) with 10% fetal bovine serum, and incubated at 37 °C and 5% CO2. Cells were transfected with plasmid-lipofectamine 2000TM (11668019, Thermo Fisher Scientific, USA) mixture.
Immunoblot analysis
Cells were lysed in RIPA buffer (R0010, Solarbio, Beijing, China) with protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, Sigma, USA) for 20 min at 4 °C and collected in 1.5 mL Eppendorf tubes, followed by centrifugation (14000 g, 10 min). The precipitate was discarded and protein concentrations were measured using the BCA Protein Assay Kit (P0010S, Beyotime biotechnology, Beijing, China). Forty micrograms of protein from each sample were subjected to SDS-PAGE followed by transfer onto a PVDF membrane (IPVH00010, Immobilon P, Millipore, Bedford, MA). The membrane was then probed for HIF-1α, Sema4D, using β-actin as a loading control. The primary antibodies used were as follows: rabbit anti-Sema4D (610670, BD Biosciences, 150 kDa); mouse anti-HIF-1α (D43B5, Cell Signaling Technology, 120 kDa), rabbit anti-β-actin (P30002, Abmart, 42 kDa). The secondary antibodies used were as follows: goat anti-mouse IgG (H+L) peroxidase labeled (074-1806, KPL, MD, USA), goat anti-rabbit IgG (H+L) peroxidase labeled (074-1506, KPL, MD, USA). Antigen-antibody binding was detected using Luminata Crescendo Western HRP substrate (WBLUR0100, Millipore, MA, USA).
Hypoxia assays
Hypoxic environments were created by chemical methods. Caco-2 cells and HUVECs were cultured in DMEM with 0.2 mM CoCl2 at 37 °C and 5% CO2. For detecting whether cells become “hypoxic”, HIF-1α was detected by western blot since HIF-1α would accumulate in cells in hypoxic environment.
Chromatin Immunoprecipitation (ChIP)
1 x 106 cells on a 10 cm dish as appropriate were harvested after being incubated under hypoxia condition 12h. Then CHIP assay was performed with CHIP kit (17-295, Affiliate of Merck KGaA Darmstadt, Germany) according to the manufacturer's instructions. 4 HREs were detected respectively. IgG was set as negative control. Precipitated DNA amplified by PCR using the following primers. HRE1, 2: forward 5'-AAAGGTACACCAGGACAG-3', reverse 5'-ATGTACACCTGTAGAAACAC-3'; HRE3: forward 5'-GCGAAGCCAGGGTCTTCA-3', reverse 5'-GAGGCCTCACGTGCGTGT-3'; HRE4: forward 5'-AGCAGGTCATCACTCACCAT-3', reverse 5'-GGGCTTCAGCAGCAAAGG-3'. The VEGF promoter was used as positive control (HIF-1α), primer forward: 5'-ACAGACGTTCCTTAGTGCTGG-3', reverse 5'-AGCTGAGAACGGGAAGCTGTG-3'.
Nucleotide secondary structure prediction
The Sema4D 5' non-coding region sequence was obtained from NCBI (LOC105584284). HIF-1α-HREs within the Sema4D 5'non-coding region were scanned on the Promoter 2.0 Prediction Server of the JASPAR database. The JASPAR matrix model is ARNT-HIF1A (MA0259.1), an Arnt (10009) class of basic helix-loop-helix factors in the PAS domain factor family, and the relative profile score threshold was set at 85% (Table 1). DNA secondary structure analysis was performed in RNA structure 5.3.
Cloning of the regulatory sequence within Sema4D 5' non-coding region and luciferase assays
Genomic DNA of the 16 cell lines included in the study was isolated by the Wizard Genomic DNA Purification Kit (A1120, Promega, USA). The regulatory sequence was cloned by PCR. Primers for amplifying the regulatory sequence were designed by Premier Primer 5.0 (sense: 5'-TGAGGTTCTGTTTCGGCTGT-3', antisense: 5'-AATGCCATCGCTGTCCCAAA-3'). Plasmids (pRL-CMV vector, pGL3 luciferase reporter vector) for luciferase assays were purchased from Promega. The 5' untranslated region of Sema4D was cloned into the pGL3 luciferase reporter vector, while pRL-CMV acted as an internal reference. Detecting primers of pGL3 luciferase reporter vector are sense: 5'-CTAGCAAAATAGGCTGTCC-3', antisense: 5'-CTTTATGTTTTTGGCGTCTTCCA-3'. Both of these plasmids were transfected into Caco-2 and HUVEC lines. Cells were harvested in 40h after transfection, and luminescence was detected followed by Dual-Glo Luciferase assay system kit (E1910, promega, USA) introduction using Synergy 4 (BioTek). The parameters used were detection by luminescence detection method, endpoint read type, standard dynamic range, and the sensitivity was set to 200.
Statistical analysis
Fisher's Exact Test was used to determine the differences between different groups. P values were calculated to determine whether the two groups differed significantly from each other (* p<0.05; ** p<0.01).
Results
Accession Sequence ID
The regulatory sequences within Sema4D 5' non-coding region of all 15 cell lines included in the study were upload to GenBank, and the sequence IDs are shown below. A549, KX018325; Caco-2, KX018326; CNE, KX018327; RD, KX018328; 293T, KX018329; BEAS-2B, KX018330; HUVEC, KX018331; MRC-5, KX018332; Tca8113, KX789515; SK-OV-3, KX789516; Jurkat, KX789517; HepG-2, KX789518; SK-N-SH, KX789519; HL-60, KX789520; and U937, KX789522.
Sema4D's expression was correlated with HIF-1α
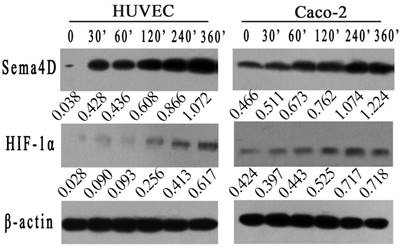
To further verified the correlation between HIF-1 and Sema4D in HUVECs or Caco-2 cells, both cells were cultured in a hypoxic incubator or in medium with 0.2 mM CoCl2. The expression levels of Sema4D and HIF-1α were detected by western blotting at different times (0', 30', 60', 120', 240', and 360'). Expression levels of HIF-1α in both Caco-2 cells and HUVECs were increased in both the hypoxic culture and CoCl2 (0.2 mM) treatment. Meanwhile, the expression level of Sema4D increased and correlated with HIF-1 in both cells (Fig. 1).
Expression of Sema4D correlated with expression of HIF-1α in both HUVECs and Caco-2 cells. HUVECs and Caco-2 cells were cultured in medium containing CoCl2 in incubator (5%CO2, 1%O2, 94%N2) . The expression of HIF-1α increased in both HUVECs and Caco-2 cells after 360 minutes in a hypoxic environment. In addition, the expression level of Sema4D correlated with the expression of HIF-1α in both HUVECs and Caco-2 cells. β-actin was used as the loading control. Expression of Sema4D and HIF-1α was analyzed by NIH image software (pixel intensity of scanning) relative to β-actin controls.
HRE4 and HRE2 are HIF-1α binding sites in HUVEC or Caco-2 cells
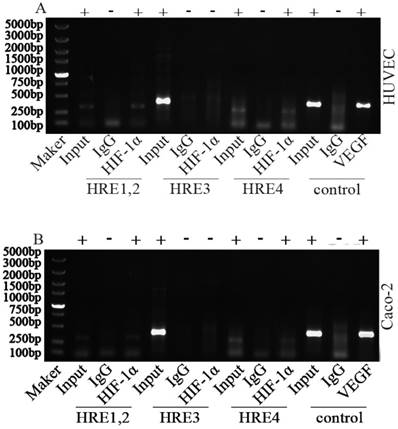
To investigate HIF-1 binding region, a CHIP assay have been performed. Fragments about 200bp bands were observed in the immunoprecipitated fragments from HRE1/2 and HRE4 in either HUVECs or Caco2 cells exposed to hypoxic conditions. The HRE sequence of VEGF promoter was used as the positive control (Fig. 1). Results show that HIF-1α could bind with HREs within Sema4D 5' non-coding region, and Sema4D is regulated by hypoxia in both HUVEC and Caco2 cells. Meanwhile, this regulation occurs at the level of transcription (Fig. 2).
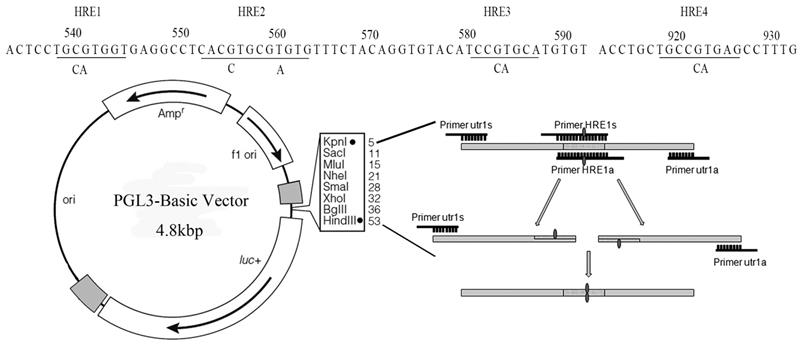
The promoter of Sema4D was predicted in the Promoter 2.0 Prediction Server. Then, the 1275 bp 5' non-coding region upstream of Sema4D was screened for candidate HIF-1α binding sites. After scanning the HIF-1α binding sites within the Sema4D 5' non-coding region in the JASPAR database, 4 promising HREs were obtained, which were corresponding to other 3 HRE sites showed in figure 3: 537-CTGCGTGG-544 (HRE1), 552-TCACGTGC-559 (HRE2), 553-CACGTG-558 (HRE2), and 556-GTGCGTGT-563 (HRE3) (Table 1, Table 2, Figure 3).
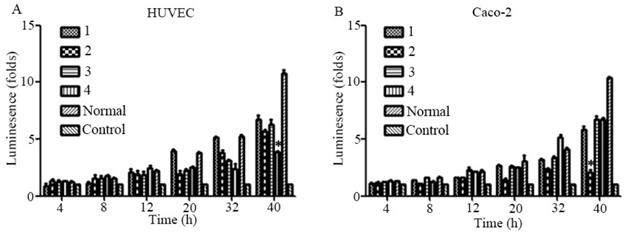
The Sema4D gene contains four HRE sequences that were cloned, and site-directed mutagenesis within the HRE core sequences was conducted by overlap PCR (Fig. 3). Regulatory sequences containing Mutated HREs were cloned into luciferase reporter vectors. In HUVECs, the mutation of HRE4 results in an enormous reduction of luciferase compared to the normal group (positive control). However, in Caco-2 cells, the mutation of HRE2 caused a tremendous reduction of luciferase compared to the normal Sema4D 5' non-coding region sequence (Fig. 4).
Distinct site mutations (T471C/A600G/C862T) within Sema4D 5' non-coding region in cancer cells
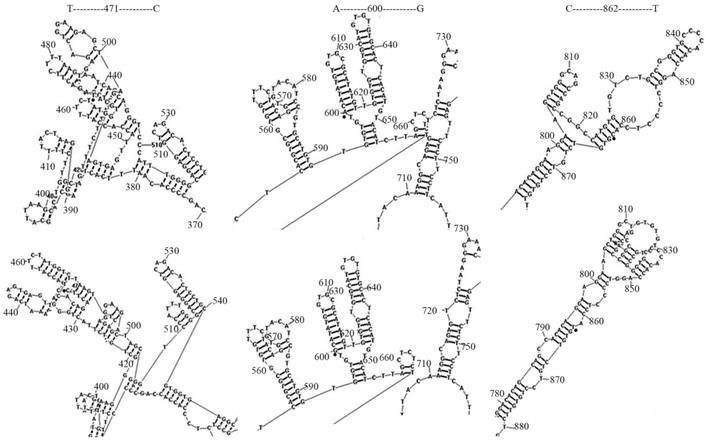
The regulatory sequences within Sema4D 5' non-coding regions of 11 cancer cell lines, including for A549, Caco-2, CNE, RD, Tca-8113, SK-OV-3, Jurkat, HepG-2, HL-60, SK-N-SH and U937, and 4 normal cell lines, namely, 293T, BEAS-2B, HUVEC, MRC-5, were sequenced and aligned. There were three distinct mutation sites (T471C/A600G/C862T) were detected (Table 3). The mutation rates of T471C/A600G/C862T were 72.7%, 18.2%, and 72.7% in cancer cells, respectively. In normal cells, these 3 mutation rates were not dectected. The secondary structure of the regulatory sequences containing these distinct mutation sites was predicted using RNA structure 5.3 (Function-fold DNA single strand). The point mutations T471C and C862T led to fewer DNA ring structures, while C862T increased the distance between 665T-720T. As a result, point mutations on T471C and C862T led to change of the gene structure (Fig. 5).
Chromatin immunoprecipitation assay of Sema4D 5' non-coding region HREs. HUVECs and Caco-2 cells were cultured under hypoxia condition (medium containing CoCl2 or cultured in an anoxic box containing GENbag). Chromatin lysates from these cells were immunoprecipitated with anti-HIF-1 antibodies according to the manufacturer's instructions. Then purified DNA was subjected to PCR with primers pairs HRE1/2 (due to their close proximity), HRE 3 and HRE 4. The VEGF promoter was used as positive control (HIF-1α). HIF-1α could bind with HRE1, HRE2, HRE4 (+) but did not bind with HRE 3 (-) in both HUVEC (A) and Caco-2 cells (B). IgG was set as negative control. Size markers are shown on the left.
HIF-1α binding site model in JASPAR database
| JASPAR ID | Name | Species | Class | Family | Sequence |
|---|---|---|---|---|---|
| MA0004.1 | Arnt | 10090 | Basic helix-loop-helix factors (bHLH) | PAS domain factors |  |
| MA0259.1 | ARNT::HIF1A | 10090;10117;9606;9986 |  |
Two matrix model of HIF-1α binding site were search in JASPAR.
Cloning of the Sema4D 5' non-coding region which containing mutant sites. All four specific HREs (RCGTG) were mutated (RCCAG) by overlap PCR. The Sema4D 5' UTR with site mutations were cloned into the pGL3-Basic vector.
Luciferase assays of the Sema4D 5' non-coding region containing mutant HREs. HUVECs and Caco-2 cells were cultured in medium containing 0.2 mM CoCl2 and infected with luciferase reporter vectors containing five mutation combinations. A, The Sema4D 5' non-coding region containing mutated HRE4 resulted in a significant decrease of luciferase in HUVECs. Mutation of HRE1, HRE2, HRE3 resulted in a decrease ranging from 10- 40% (A). B, Sema4D 5' untranslated region containing mutated HRE2 resulted in a significant decrease of luciferase in Caco-2 cells (B). Sample scores were analyzed by the Kruskal-Wallis Test. (* P<0.05, ** P<0.01, compared to the normal group, bars indicate mean of three individual experiments ± standard error).
Four predicted HRE site for HIF-1 by JASPAR
| Model ID | Model name | score | Relative score | Strand | Start | End | Site sequence |
|---|---|---|---|---|---|---|---|
| MA0259.1 | ARNT::HIF1A | 7.416 | 0.887142780249 | 1 | 537 | 544 | CTGCGTGG |
| MA0259.1 | ARNT::HIF1A | 6.649 | 0.864261295358 | 1 | 552 | 559 | TCACGTGC |
| MA0004.1 | Arnt | 10.351 | 0.999997274644 | 1 | 553 | 558 | CACGTG |
| MA0259.1 | ARNT::HIF1A | 7.809 | 0.898866930917 | 1 | 556 | 563 | GTGCGTGT |
Putative sites were predicted in JASPAR database within model Arnt\ARNT::HIFA, and relative profile score threshold was set on 85%.
Three important mutation sites among 15 cells
| Mutation Sites | 471 | P value | 600 | P value | 862 | P value | |
|---|---|---|---|---|---|---|---|
| Rf sequence | AL590233.23 | T | A | C | |||
| Cancer cell | A549 | C | A | T | |||
| Caco-2 | C | A | C | ||||
| CNE | C | G | T | ||||
| RD | C | A | T | ||||
| Tca8113 | C | A | T | ||||
| SK-OV-3 | C | G | T | ||||
| Jurkat | C | A | T | ||||
| SK-N-SH | T | A | T | ||||
| HepG-2 | C | A | T | ||||
| U937 | T | A | C | ||||
| HL-60 | T | A | C | ||||
| Normal cell | 293T | T | A | C | |||
| BEAS-2B | T | A | C | ||||
| MRC-5 | T | A | C | ||||
| HUVEC | T | A | C | ||||
| Total | Mutate ratio | 53.3% | 0.015 | 13.3% | 0.352 | 53.3% | 0.015 |
The reference sequence (Rf sequence) AL590233.23 is human genomic DNA from clone RP13-93L13 on chromosome 9 (95588~96862) contains Semaphorin4D cds. There was significant difference among cell lines including 11 tumor cell lines and 4 normal cell lines on site mutations T471C and C862T within Sema4D 5' Non-coding Conserved Region (P=0.015).
The biological effects of T471C/A600A/C862C nucleotide variants within the Sema4D 5' non-coding region in cancer cells
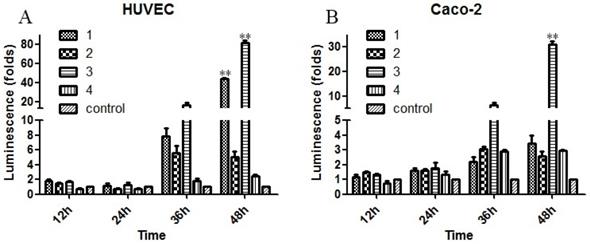
To evaluate the biological effects of the previously mentioned mutations, the Sema4D regulatory sequences with combinations of the three typical mutation sites in cancer cells or normal cells were cloned into luciferase report vectors, including group1 (T471C/A600A/C862C), group2 (C471C/A600A/C862T), group3 (T471C/A600A/C862T), group4 (T471T/A600G/C862C), and group5 (T471T/A600G/C862C, acted as a control). The results showed that simultaneous mutation of T471C and C862T caused obviously higher gene expression efficiency. The single point mutation of A600G did not cause a significant change in the expression efficiency of luciferase (Fig. 6).
Discussion
Sema4D was up-regulated expression in many tumor tissues and play a critical endothelial chemoattractant role in adenocarcinomas of the prostate, colon, breast and squamous cell carcinomas of the head, neck or lung under hypoxia condition (2, 19). In hypoxic tissues, hypoxia inducible factor HIF-1α is accumulated and stimulates the transcription of genes that have HREs in their promoters by binding HRE, for example VEGF (10, 17). Liao's and Zhou's research showed that Sema4D is induced by hypoxia in a HIF-1-dependent manner and influences endothelial cell migration and tumor vascularity; (10, 18). However, the specific regulatory mechanism was not yet classified.
Mutation on Sema4D 5' non-coding region led to DNA secondary structure change. The Sema4D 5'non-coding region of ten cancer cell lines A549, Caco-2, CNE, RD, Tca-8113, SK-OV-3, Jurkat, HepG-2, SK-N-SH, U937, HL-60 were sequenced and aligned. Two distinct site mutations (T471C/C862T) of the Sema4D 5' non-coding region were detected in 7 cancer cell lines. The A600G mutation was detected in 2 cancer cell lines. These three mutations caused significant changes in the DNA secondary structure. The T471C point mutation caused fewer DNA rings between 390G-400G and loosened the tight structure between 450T-510C. The mutation at C862T exposed the retraction structure between 810C-830T, while C862T increased the distance between 665T-720T.
Mutations of the Sema4D promoter affect the promoting efficiency. Four kinds of luciferase reporter plasmids containing four combinations of site mutations were infected into Caco-2 cells and HUVECs, including 1(T471C/A600A/C862C), 2 (T471T/A600A/C862T), 3 (T471C/A600A/C862T) and 4 (T471T/A600G/C862C). The normal Sema4D 5' un-coding region (T471T/A600A/C862C) acted as the control. Cells were cultured in either normal medium or medium containing 0.2 mM CoCl2. Luciferase reporter plasmids containing a combination of 1 and 3 promoted the expression of luciferase when compared to the control in both HUVECs (A) and Caco-2 cells (B). The sample score was analyzed by the Kruskal-Wallis Test. (* P<0.05, ** P<0.01, compared to the normal group, bars indicate mean of three individual experiments ± standard error).
In this study, our result demonstrated that HIF could regulate Sema4D transcript by binding HRE within Sema4D 5'uncoding region in both HUVECs and Caco-2 cells, then play a role in vascular induce under hypoxia condition which was similar to VEGF. In addition, the result showed that key HREs varied between tumor cells and normal cells, few study report similar result before and it interested us a lot. 3 distinct mutation sites were frequently detected within 5' un-coding region among 11 tumor cell strains (distinguish from 4 normal cells) and DNA secondary structure were predicted online. The result exhibit diversity in the predict DNA secondary structure among different Sema4D 5' uncoding region nucleotide variants. In addition, expression efficiency differs among cancer cells and normal cells with different nucleotide variants. It indicated that 5'uncoding region contained diversity nucleotide variants of cancer cells contributed to the difference of Sema4D expression. These data enhance our understanding of the regulatory mechanism of Sema4D expression. Considering Sema4D play a role in inducing vascular, the study will benefit the development of a possible therapeutic strategy for malignancies.
Four possible HREs in the Sema4D 5' non-coding region have been screened. The possible HRE sequences were then cloned in vectors, and site-directed mutation of the HRE core sequences was conducted by site-directed mutagenesis. The luciferase assay results showed that the key HIF-1α binding HRE sequence was HRE4 in HUVECs. While, the key HIF-1α binding HRE sequence was HRE2 in Caco-2 cells. Therefore, HIF-1 mainly combines with one of the four core HREs, and the key HRE is distinct in different cells.
Genomic DNA of 11 cancer cells (A549, Caco-2, CNE, RD, Tca8113, SK-OV-3, Jurkat, HepG-2, HL-60, U937, and SK-N-SH) and normal cells (293T, BEAS-2B, HUVEC, MRC-5,) were extracted, followed by sequencing of the Sema4D 5'non-coding region. Three distinctive point mutations (T471C/A600G/C862T) were detected in the Sema4D 5' non-coding region in cancer cells. Except for SK-N-SH, HL-60, U937, the T471C mutation was detected in the other 8 cancer cell lines. Meanwhile, the C862T mutation was detected in 8 of the 11 cancer cell lines except for Caco-2 cell line. A600G is detected in several types of cancer cell lines, including CNE and Tca-8113, while this mutation was not detected in 4 of the normal cell lines analyzed. Meanwhile, T471C/C862T mutations were also rarely found in these normal cell lines. Our result reveals that the nucleotide variant T471C and C862T are quite important in cancer cells, which benefit from the expression of Sema4D. The T471C point mutation caused fewer DNA rings between 390G-400G and loosened the tight structure between 450T-510C. The mutation at C862T exposed the retraction structure between 810C-830T, and increased the distance between 665T-720T (Fig. 5). These mutations which were close to the HIF-1α binding site may affect the HIF-1α or other factors combination due to the nucleotide secondary structure change.
There is a great need to enhance our understanding of the regulatory mechanism of the Sema4D 5' non-coding region. In this study, we showed that HRE4 and HRE2 play distinct regulatory roles in HIF-1 induced Sema4D unregulated expression in different cells. Additionally, the mutation sites of the Sema4D 5' non-coding region are distinctly different between normal and cancer cells, which affect the expression of Sema4D and lead to different biological effects.
Acknowledgements
Our research was supported by the Foundation of the CAMS Initiative for Innovative Medicine (CAMS-I2M) (grant no. 2016-12M-1-19), the Project of Yunnan Province Medical Science and Technology (grant no. 2017NS113), the Joint Fund of the Yunnan Science and Technology Department and Kunming Medical University (grant no. 2018FE001(-270) and grant no.2014FZ058).
Author contributions
Lijuan Qiu, Jia Luo, Yujiao Zhao: Acquisition and analysis of data; drafting of the manuscript. Lijuan Qiu, Yue Pan, Junying Chen, Hongchao Jiang, Juemin Xi, Xiaodan Wang: technical or material support. Qiangming Sun: Study concept and design; critical revision of the manuscript; obtained funding; study supervision. The authors declare that there is no conflict of interest regarding the publication of this paper. I confirm that appropriate statistical analyses have been performed, and all sequence information included in this study can be downloaded from GenBank.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proceedings of the National Academy of Sciences of the United States of America 103: 9017-9022. 2006
2. Wu M, Li J, Gao Q, Ye F. The role of Sema4D/CD100 as a therapeutic target for tumor microenvironments and for autoimmune, neuroimmune and bone diseases. Expert opinion on therapeutic targets: 1-17. 2016
3. Swiercz JM, Worzfeld T, Offermanns S. ErbB-2 and met reciprocally regulate cellular signaling via plexin-B1. The Journal of biological chemistry 283: 1893-1901. 2008
4. Basile JR, Barac A, Zhu T, Guan KL, Gutkind JS. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer research 64: 5212-5224. 2004
5. Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proceedings of the National Academy of Sciences of the United States of America 99: 12085-12090. 2002
6. Hirotani M, Ohoka Y, Yamamoto T. et al. Interaction of plexin-B1 with PDZ domain-containing Rho guanine nucleotide exchange factors. Biochemical and biophysical research communications 297: 32-37. 2002
7. Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. The Journal of biological chemistry 277: 43115-43120. 2002
8. Driessens MH, Olivo C, Nagata K, Inagaki M, Collard JG. B plexins activate Rho through PDZ-RhoGEF. FEBS letters 529: 168-172. 2002
9. Mu L, Wang J, Chen Y. et al. Hypoxia-inducible factor-1alpha and semaphorin4D genes involved with tumor-associated macrophage-induced metastatic behavior and clinical significance in colon cancer. Chinese medical journal 127: 3568-3575. 2014
10. Zhou H, Yang YH, Binmadi NO, Proia P, Basile JR. The hypoxia-inducible factor-responsive proteins semaphorin 4D and vascular endothelial growth factor promote tumor growth and angiogenesis in oral squamous cell carcinoma. Experimental cell research 318: 1685-1698. 2012
11. Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. The EMBO journal 19: 4298-4309. 2000
12. Sotoodehnejadnematalahi F, Burke B. Human activated macrophages and hypoxia: a comprehensive review of the literature. Iran J Basic Med Sci 17: 820-830. 2014
13. Shah YM. The role of hypoxia in intestinal inflammation. Molecular and cellular pediatrics 3: 1. 2016
14. Cummins EP, Keogh CE, Crean D, Taylor CT. The role of HIF in immunity and inflammation. Molecular aspects of medicine. 2016;47-48:24-34
15. Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta pharmaceutica Sinica. B 5: 378-389. 2015
16. Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. The Journal of biological chemistry 270: 1230-1237. 1995
17. Lee M. Hypoxia targeting gene expression for breast cancer gene therapy. Adv Drug Deliv Rev 61: 842-849. 2009
18. Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev 26: 281-290. 2007
19. Jiang H, Chen C, Sun Q. et al. The role of semaphorin 4D in tumor development and angiogenesis in human breast cancer. OncoTargets and therapy 9: 5737-5750. 2016
Author contact
![]() Corresponding author: Qiangming Sun, E-mail: msun08com. Current postal address: Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS & PUMC), 935 Jiao Ling Road, Kunming, Yunnan Province 650118, P.R. China. Telephone number: 86-871-68335165; Fax number: 86-871-68334483
Corresponding author: Qiangming Sun, E-mail: msun08com. Current postal address: Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS & PUMC), 935 Jiao Ling Road, Kunming, Yunnan Province 650118, P.R. China. Telephone number: 86-871-68335165; Fax number: 86-871-68334483

 Global reach, higher impact
Global reach, higher impact