3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(4):949-956. doi:10.7150/jca.27224 This issue Cite
Review
CHFR Promoter Hypermethylation Is Associated with Gastric Cancer and Plays a Protective Role in Gastric Cancer Process
1. Department of Medical Oncology, Sir Run Run Shaw Hospital, Medical School of Zhejiang University, Hangzhou, China
2. Laboratory of Cancer Biology, Key Lab of Biotherapy, Sir Run Run Shaw Hospital, Medical School of Zhejiang University, Hangzhou, China
# These authors contribute to this work equally.
Received 2018-5-12; Accepted 2018-12-5; Published 2019-1-29
Abstract
Background: Chromosomally unstable tumors account for 50% of gastric cancer. CHFR plays a role in controlling chromosomal instability and its inactivation will eventually lead to tumorigenesis. In addition to genetic deletion, DNA methylation could silence the expression of many cancer-related genes including CHFR. Its methylation was found to be associated with the initiation and progression of gastric cancer.
Methods: We performed a meta-analysis involving methylation analyses of CHFR promoter in gastric cancer. Nineteen studies with 1,249 tumor tissues and 745 normal tissues had been included in current study.
Results: We found that CHFR methylation was significantly higher in gastric cancer (studies numbers = 15, cases/controls = 862/745, odds ratio (OR) = 7.46, 95% confidence index (95% CI) = 4.99-11.14). Methylation array data was also obtained from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas network (TCGA). There were 7 out of 13 CHFR methylation probes target to the same CpG island region (hg19, 131973620-131975130) showed the CHFR methylation was higher in gastric cancers than normal controls. Eight probes showed CHFR promoter hypermethylation was associated with longer overall survival of gastric cancer patients (Hazard Ratio < 1).
Conclusions: The CHFR promoter hypermethylation was associated with gastric cancer and played a protective role in gastric cancer process. Its methylation could be a potential biomarker for the diagnosis and prognosis of gastric cancer.
Keywords: CHFR, gastric cancer, methylation, diagnosis biomarker
Introduction
Gastric cancer is the fourth most common cancer and the second leading cause of cancer death worldwide [1]. Chromosomally unstable tumors account for 50% of gastric cancer based on The Cancer Genome Atlas network (TCGA) molecular classification [2]. Chromosomal instability refers to the rate that chromosomes or large portions of chromosomes are changed [3].
CHFR (checkpoint with forkhead associated and ring finger domains) gene locates at chromosome 12q24.33. It encodes an E3 ubiquitin ligase to promote the ubiquitination-dependent degradation of mitosis-related oncoproteins such as Kiff22 [4], PLK1 [5] and Aurora A [6]. By doing so, it functions as an early mitotic checkpoint that delays the entry from prophase into metaphase in response to mitotic stresses. Cancer cell lines with function loss of CHFR could bypass mitotic arrest induced by mitotic stresses [7]. CHFR expression is reduced in 20-50% of primary tumors and tumor cell lines [6].
Several studies suggested that CHFR gene expression is frequently silenced by promoter hypermethylation in gastric cancer [8-12]. Additionally, CHFR promoter methylation improved the gastric cancer sensitivity to microtubule inhibitors such as docetaxel and paclitaxel [13, 14]. However, the samples of these studies were too small to confirm solid results. Meta-analysis is a classical statistical method that can pool data from different studies and enhance the power to draw a more comprehensive conclusion than a single study [15]. Therefore, we performed a meta-analysis to explore the relevance of CHFR promoter methylation to gastric cancer. Moreover, public data from TCGA and Gene Expression Omnibus (GEO) database were also collected to further validate the influence of CHFR methylation on the prognosis of gastric cancer patients.
Methods
Data collection
We collected studies from online index database PubMed, PubMed Centra (PMC), Embase, and Chinese database (WeiPu, WanFang and Chinese National Knowledge Infrastructure) without time and language limitation. The search keywords for PubMed were “((((((((((((checkpoint with forkhead and ring finger domains)) OR CHFR) OR RNF116) OR RNF196)) AND ((Gastric Cancers OR (Cancer of the Stomach) OR (Gastric Cancer) OR (Stomach Cancers) OR (Cancer of Stomach) OR (Gastric Neoplasm) OR (Gastric Neoplasms) OR (Stomach Neoplasm))))) AND methylation [tiab])) NOT (((animals [mh] NOT humans [mh]))))) NOT ("review" [Publication Type])”. Similar search keywords were applied for other database. The search was updated before July, 2018. We would include studies met the follow items: (1) it is an original human clinical trial on association between CHFR promoter methylation and gastric cancer; (2) the study should involve sufficient data to calculate the odds ratios (OR) and 95%confidence index (95% CI) between CHFR methylation and gastric cancer, and if not, it should have data of CHFR methylation of relevant clinical characteristics; (3) the controls should be adjacent tissue from same patient, or benign lesion/normal gastric tissues from health population. Two reviewers have independently processed the study selection (DD and BZ), and disagreements of decisions were solved by discussion and subsequent consensus. If the same population was used in different studies, the most recent, largest, or best-quality one would be included.
Data extraction
The data extraction from articles included the name of first author, year of publication, the region where the samples were from, numbers of cases and controls, methods of detecting methylation status, sample types, tumor stages, control type and study type. The extraction was performed by two independent reviewers (DD and BZ).
Data analyses of meta-analysis
All the statistical analyses were performed by Stata software 11.0[16]. The OR and 95% CI were calculated. Cochran's Q statistic and I² test [17] were used to calculate statistical heterogeneity. Since random and fix model present similar results when heterogeneity is low, the random effect model was used in current meta-analyses [18]. Funnel plots were drawn to observe the potential publication bias. Begg and Egger analyses were also analyzed [19, 20]. A p value lower than 0.05 was considered significant.
Validation analysis by TCGA and GEO database
We further collected data from public functional genomics data repository to validate the association between CHFR methylation and gastric cancer of current meta-analysis. A case-control designed and Infinium HumanMethylation27 BeadChip (HM27K) technical based study was selected from GEO database (GSE30601) [21], which involved 203 gastric tumors and 94 matched non-malignant gastric samples from Singapore. Moreover, prognostic analysis of CHFR was also performed in MethSurv [22], which contained TCGA stomach adenocarcinoma cohort that involved 395 patients whose cancer methylome status was tested by Infinium HumanMethylation450 BeadChip (HM450K). The follow-up time of this cohort was 449 days and 155 deaths were found after follow-up. The beta value of every CpG site was calculated as M/(M + U + 100), where M and U were methylated and unmethylated intensities, respectively. Different methylation status was defined by certain beta-value level (fully unmethylated, 0 < beta-value <= 0.2; partially methylated, 0.2 < beta-value < 0.6; fully methylated, 0.6 <= beta-value <= 1).
Results
Data collection
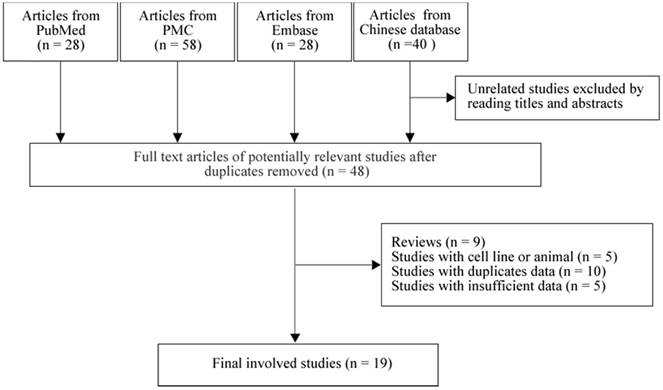
As in Figure 1, we found 28 studies from PubMed, 58 studies from PMC, 28 studies from Embase and 40 studies from Chinese database. By reading titles and abstracts, and removing the duplicate studies, 48 full-text articles were selected for further screening. We next excluded 9 reviews, 5 cell line and animal based studies, 10 studies with duplicate population and 5 studies with no sufficient data for calculating. Finally, 19 studies [8-12, 14, 23-35] with 1,249 tumor tissues and 745 normal tissues were involved in our meta-analysis. Among them, there were 10 studies from Japan [11, 12, 25, 28-30, 32, 33, 36, 37], 7 studies from China [8-10, 14, 23, 24, 26] and 2 studies from Korea [27, 31]. There were 3 prospective studies [14, 23, 28] and 16 retrospective [8-12, 24-27, 29-33, 36, 37] studies.
The association between CHFR methylation and gastric cancer
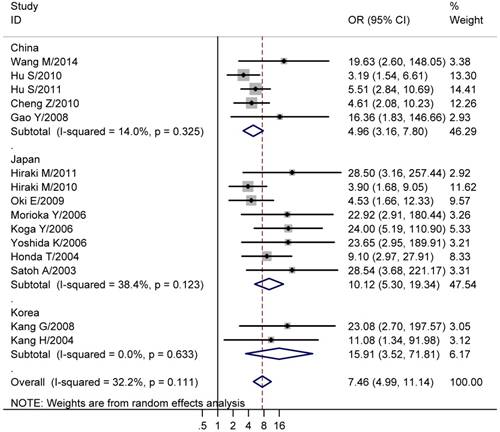
We next explored the association between CHFR methylation and gastric cancer. The results showed that the CHFR methylation was significantly higher in gastric cancer than normal tissues (studies numbers = 15, cases/controls = 862/745, OR = 7.46, 95% CI = 4.99-11.14, I² = 32.20%, Table 2, Figure 2). Since the involved studies were mainly performed in China, Japan and Korea, a further subgroup population based analysis was conducted. The results showed CHFR methylation was significantly higher in gastric cancers in all of the three regions (China: studies numbers = 5, cases/controls = 394/323, OR = 4.96, 95% CI = 3.16-7.80, I² = 14.00%; Japan: studies numbers = 8, cases/controls = 400/383, OR = 10.12, 95% CI = 5.30-19.34, I² = 38.40%; Korea: Studies numbers = 2, cases/controls = 68/39, OR = 15.91, 95% CI = 3.52-71.81, I² = 0.00%, Table 2, Figure 2). Moreover, we analyzed the association between CHFR methylation and different clinical features of gastric cancer patients that comprised gender (males vs. females), age (>= 60 vs. < 60 years), tumor histology (intestinal type vs. diffused and mixed type), differentiation level (high vs. low/intermediate), tumor size (>= 5cm vs. < 5cm), degree of T stage (T3-4 vs. T1-2), tumor stage (III+IV vs. I+II), lymph node metastasis (positive vs. negative), distant metastasis (positive vs. negative) and P53 statue (mutation vs. wild). We found the CHFR methylation level was significant associated with the tumor size of gastric cancer (studies numbers = 6, cases/controls = 236/186, OR = 1.71, 95% CI = 1.11-2.65, I² = 16.00%, Table 2). No significant results were found in other analyses (Table 2).
Publications bias
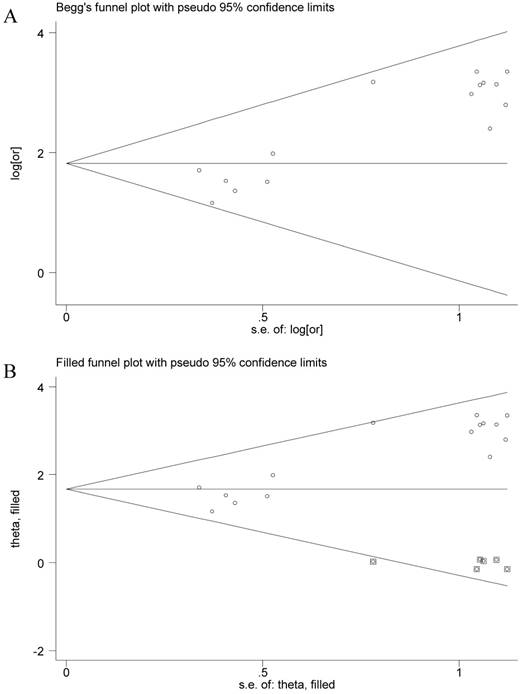
The funnel plot presented asymmetry shape of all involved studies, and the Egger test showed a significant result for funnel plot asymmetry, indicating potential publication bias in current meta-analysis. Therefore, a nonparametric trim was performed to assess the influence of these asymmetry studies. Six missing studies were filled in new pooled funnel plots, and the new plots presented symmetrical shape (Figure 3) meanwhile the CHFR methylation still significantly associated with gastric cancer (OR = 5.31, 95% CI = 3.45-8.14), suggesting that our results were solid.
Flowchart of selection process in the meta-analyses
Characteristics of the involved in current meta-analyses
| Author/Year | Regions | Cases/Controls | Methods | Samples | Tumor stages | Control type | Study type | Reference |
|---|---|---|---|---|---|---|---|---|
| Li Y/2015 | China | 102/0 | MSP | Tissues | I-IV | NA | Prospective | [14] |
| Guo He/2014 | China | 70/0 | MSP | Tissues | I-IV | NA | Retrospective | [8] |
| Wang M/2014 | China | 117/46 | MethyLight | Tissues | NA | Adjacent tissue | Prospective | [23] |
| Hiraki M/2011 | Japan | 127/20 | q-MSP | Tissues | NA | Adjacent tissue | Retrospective | [36] |
| Hu S/2010 | China | 70/70 | MSP | Tissues | I-IV | Adjacent tissue | Retrospective | [24] |
| Hu S/2011 | China | 123/123 | MSP | Tissues | I-IV | Adjacent tissue | Retrospective | [9] |
| Cheng Z/2010 | China | 64/64 | MSP and COBRA | Tissues | I-IV | Adjacent tissue | Retrospective | [10] |
| Hiraki M/2010 | Japan | 49/49 | q-MSP | Tissues | I-IV | Adjacent gastric mucosa and adjacent regional LN | Retrospective | [25] |
| Oki E/2009 | Japan | 59/59 | MSP | Tissues | I-IV | Adjacent tissue | Retrospective | [11] |
| Gao Y/2008 | China | 20/20 | MSP | Tissues | I-IV | Adjacent tissue | Retrospective | [26] |
| Kang G/2008 | Korea | 25/25 | MSP | Tissues | NA | chronic gastritis tissues from other patients | Retrospective | [27] |
| Mitsuno M/2007 | Japan | 56/0 | MSP | Tissues | I-IV | NA | Prospective | [28] |
| Morioka Y/2006 | Japan | 53/53 | MSP | Tissues | I-IV | Adjacent tissue | Retrospective | [12] |
| Koga Y/2006 | Japan | 46/46 | MSP | Tissues | NA | Adjacent tissue | Retrospective | [29] |
| Yoshida K/2006 | Japan | 41/41 | COBRA | Tissues | NA | Adjacent tissue | Retrospective | [37] |
| Homma N/2004 | Japan | 52/0 | MSP | Tissues | NA | Adjacent tissue | Retrospective | [30] |
| Honda T/2004 | Japan | 71/71 | MSP | Tissues | NA | Adjacent tissue | Retrospective | [32] |
| Kang H/2004 | Korea | 43/14 | COBRA | Tissues | I-IV | Adjacent tissue | Retrospective | [31] |
| Satoh A/2003 | Japan | 61/44 | COBRA | Tissues | NA | Adjacent tissue | Retrospective | [33] |
MSP: methylation-specific polymerase chain reaction; COBRA: Combined Bisulfite Restriction Analysis; NA: not available.
Meta-analyses of CHFR methylation with gastric cancer
| Group | Studies number | Cases/Controls | OR (95% CI) | I² (%) | p value of I² | Begg | Egger |
|---|---|---|---|---|---|---|---|
| Studies with all races (Cancer tissues vs. Normal tissues) | 15 | 862/745 | 7.46 (4.99, 11.14) | 32.20 | 0.111 | 0.023 | < 0.001 |
| China group (Cancer tissues vs. Normal tissues) | 5 | 394/323 | 4.96 (3.16, 7.80) | 14.00 | 0.325 | 0.462 | 0.099 |
| Japan group (Cancer tissues vs. Normal tissues) | 8 | 400/383 | 10.12 (5.30, 19.34) | 38.40 | 0.123 | 0.108 | 0.001 |
| Korea group (Cancer tissues vs. Normal tissues) | 2 | 68/39 | 15.91 (3.52, 71.81) | 0.00 | 0.633 | NA | NA |
| Gender (Males vs. Females) | 13 | 518/307 | 0.81 (0.58, 1.12) | 0.00 | 0.565 | 0.200 | 0.257 |
| Age (>=60 years vs. <60 years) | 5 | 196/233 | 0.73 (0.49, 1.09) | 0.00 | 0.512 | 0.462 | 0.492 |
| Tumor histology (Intestinal type vs. Diffused and mixed type) | 5 | 166/198 | 1.26 (0.79, 2.03) | 9.20 | 0.354 | 0.462 | 0.554 |
| Differentiation (Well and moderately differentiated vs. Poorly differentiated ) | 6 | 233/214 | 0.99 (0.47, 2.11) | 73.00 | 0.002 | 0.707 | 0.744 |
| Tumor size (>=5cm vs. <5cm) | 6 | 236/186 | 1.71 (1.11, 2.65) | 16.00 | 0.311 | 0.260 | 0.141 |
| Tumor invasion (T3-4 vs. T1-2) | 8 | 315/158 | 0.84 (0.55, 1.27) | 0.00 | 0.658 | 0.536 | 0.979 |
| Tumor stage (III/IV vs. I/II) | 11 | 492/235 | 1.03 (0.74, 1.43) | 0.00 | 0.674 | 0.350 | 0.548 |
| Lymph node metastasis (N2/N3/N4 vs. N0/N1) | 8 | 319/190 | 1.08 (0.73, 1.61) | 0.00 | 0.601 | 0.902 | 0.617 |
| Distant metastasis (M1 vs. MO) | 4 | 44/236 | 1.32 (0.64, 2.73) | 0.00 | 0.510 | 0.734 | 0.847 |
| P53 (Mutant vs. Wild) | 2 | 21/76 | 0.30 (0.07, 1.22) | 28.70 | 0.236 | 1.000 | NA |
OR: odds ratio; 95% CI, 95%confidence index; NA: not available.
Forest plots of CHFR methylation with gastric cancer. The large diamond at the bottle of the table represents the pooled odds ratio of all studies. The width of the diamond represents with 95% CI.
Funnel plots of CHFR methylation with gastric cancer. A, Funnel plots of association between CHFR promoter methylation and gastric cancer in tissues before nonparametric trim; B, Funnel plots of association between CHFR promoter methylation and gastric cancer in tissues after nonparametric trim or, odds ratio; SE, standard error; One cycle represents one individual study.
Data analyses from GEO and TCGA database
As shown in Table 3, after performing differential analyses between gastric cancers and matched gastric non-malignant samples from GSE30601, thirteen statistically significant probes were selected. Among them, seven strongly associated probes (cg00338702, cg04044684, cg17387870, cg19027571, cg20535781, cg23653008, cg27040423) (p < 0.001) showed the CHFR methylation was higher in cancers than normal tissue. These 7 probes were from the same CpG island region (hg19, 131973620-131975130). The mean beta-value difference between groups was all over 0.1 in these 7 probes (Table 3, Figure S1). On the other side, other probes presented limited mean beta-value difference (one was 0.1036 while the others were less than 0.034, Table 3, Figure S2). Moreover, the mean beta-value was extremely high and presented as fully methylated status in both groups when used the other 6 probes (Table 3, Figure S2), which might not reflect the promoter methylation status of CHFR. Hence, the overall methylation statue of the 7 sites in CpG island region 131973620-131975130 might present the association between CHFR promoter methylation and gastric cancer. Interestingly, prognostic analysis showed that CHFR promoter hypermethylation was associated with longer overall survival in gastric cancer patients. Eight probes were found to be significantly predicting a better prognosis of gastric cancer patients. Previous study found CHFR promoter methylation could predict the sensitivity of gastric cancers to microtubule inhibitors [33], which might explain why CHFR promoter hypermethylation played a protective role in gastric cancer.
Significantly differential CHFR methylation probes from GEO database
| ID | adj.P.Val | Mean ± SEM of cancers | Mean ± SEM of normal tissues | Difference between means | RANGE_START | RANGE_END | GB_ACC |
|---|---|---|---|---|---|---|---|
| cg00338702 | <0.001 | 0.2998 ± 0.01393 | 0.1611 ± 0.008613 | -0.1387 ± 0.02131 | 131973620 | 131975130 | NM_018223.1 |
| cg04044684 | <0.001 | 0.3134 ± 0.01461 | 0.1214 ± 0.008635 | -0.1919 ± 0.02228 | 131973620 | 131975130 | NT_024477.13 |
| cg17387870 | <0.001 | 0.4747 ± 0.01233 | 0.3712 ± 0.009551 | -0.1036 ± 0.01926 | 131973620 | 131975130 | NM_018223.1 |
| cg19027571 | <0.001 | 0.2856 ± 0.01635 | 0.1291 ± 0.008534 | -0.1565 ± 0.02474 | 131973620 | 131975130 | NT_024477.13 |
| cg20535781 | <0.001 | 0.4656 ± 0.008622 | 0.3156 ± 0.007398 | -0.1501 ± 0.01364 | 131973620 | 131975130 | NT_024477.13 |
| cg23653008 | <0.001 | 0.1908 ± 0.01391 | 0.04342 ± 0.006138 | -0.1474 ± 0.02088 | 131973620 | 131975130 | NT_024477.13 |
| cg27040423 | <0.001 | 0.2112 ± 0.01223 | 0.09632 ± 0.005376 | -0.1148 ± 0.01835 | 131973620 | 131975130 | NM_018223.1 |
| cg17003970 | <0.001 | 0.7878 ± 0.01172 | 0.8941 ± 0.002406 | 0.1063 ± 0.01731 | 131945626 | 131946355 | NT_024477.13 |
| cg17524886 | <0.001 | 0.8925 ± 0.004785 | 0.9249 ± 0.002214 | 0.03244 ± 0.007198 | 131934293 | 131934917 | NT_024477.13 |
| cg17904068 | 0.012 | 0.9083 ± 0.003359 | 0.8936 ± 0.003251 | -0.01467 ± 0.005411 | 131930864 | 131931625 | NT_024477.13 |
| cg20066677 | 0.037 | 0.8355 ± 0.004471 | 0.8513 ± 0.003603 | 0.01576 ± 0.007017 | 131934293 | 131934917 | NT_024477.13 |
| cg21432513 | 0.005 | 0.8854 ± 0.007080 | 0.9169 ± 0.001714 | 0.03145 ± 0.01048 | 131945626 | 131946355 | NT_024477.13 |
| cg22381955 | <0.001 | 0.9643 ± 0.002186 | 0.9766 ± 0.0008457 | 0.01227 ± 0.003267 | 131939857 | 131940762 | NT_024477.13 |
SEM, standard error of the mean; RANGE_START POSITION. Chromosomal start position of oligo relative to chromosome accession version; RANGE_END Chromosomal end position of oligo relative to chromosome accession version; GB_ACC, GenBank accession; the probes from the CpG island region 131973620-131975130 were bolded.
CHFR played a protective roles in gastric cancer prognosis
| Name | HR (95% CI) | p value | CHR | MAPINFO | UCSC_RefGene_Group | Relation_to_UCSC_CpG_Island |
|---|---|---|---|---|---|---|
| cg00470794 | 0.631 (0.416, 0.959) | 0.031 | 12 | 1.33E+08 | TSS1500 | Island |
| cg02667335 | 0.660 (0.437, 0.997) | 0.048 | 12 | 1.33E+08 | TSS1500 | Island |
| cg04044684 | 0.644 (0.466, 0.891) | 0.0079 | 12 | 1.33E+08 | TSS1500 | Island |
| cg07951978 | 0.607 (0.397, 0.927) | 0.021 | 12 | 1.33E+08 | TSS1500 | Island |
| cg15063355 | 0.622 (0.407, 0.950) | 0.028 | 12 | 1.33E+08 | TSS1500 | Island |
| cg18533833 | 0.657 (0.433, 0.998) | 0.049 | 12 | 1.33E+08 | TSS1500 | Island |
| cg26832509 | 0.644 (0.424, 0.978) | 0.039 | 12 | 1.33E+08 | TSS1500 | Island |
| cg27382164 | 0.587 (0.384, 0.897) | 0.014 | 12 | 1.33E+08 | TSS1500 | Island |
HR, hazard ratio; 95% CI, 95%confidence index; CICHR, chromosome number of the site; MAPINFO, position of the site in the genome.
Discussion
CHFR is a key player in controlling chromosomal integrity. Compared with other mitotic checkpoint genes, CHFR is more frequently inactivated in cancers [7]. CHFR inactivation in cancers might be a result of promoter CpG island methylation meanwhile CHFR hypermethylation was found in a series of cancers [38]. Expression of CHFR mRNA was downregulated in 53% gastric cancers, and 16% of this 53% were found to be caused by DNA methylation [39]. Studies found that CHFR could play multiple roles in regulating growth rate, cellular invasion, motility, and genomic stability in cancer cells [40, 41]. The current study found CHFR promoter hypermethylation was associated with gastric cancer in eastern Asian countries that included China, Japan, and Korea. We also found CHFR promoter hypermethylation was associated with larger tumor size. To our knowledge, this is the most comprehensive meta-analysis of CHFR methylation and gastric cancer.
The association between CHFR inactivation and cancer prognosis were widely studied. The immunohistochemistry showed that diminished expression of CHFR protein was associated with poor prognosis of smoking-related squamous cell carcinoma of the lung [42]. Hypermethylation of CHFR was found to be a predictor for worse outcomes in lung cancer [43], colorectal cancer [44]. Interestingly, on the other side, CHFR inactivation caused by methylation could also predict the sensitivity to microtubule inhibitors (such as docetaxel and paclitaxel) in various cancers [29, 33, 45-48] including gastric cancer [29, 33]. CHFR inactivation that caused the fast entry from prophase into metaphase might be a reason for the sensitivity to microtubule inhibitors [38]. Docetaxel and paclitaxel are included in first and second line treatment of gastric cancer according to National Comprehensive Cancer Network (NCCN). Our results showed that CHFR methylation played a protective role in gastric cancer. A study [14] found that when treated with docetaxel, overall survival of gastric cancer patients was longer in the CHFR methylated group than in the CHFR unmethylated group, while in the groups treated with oxaliplatin, there was no significant difference in overall survival between the CHFR methylated group and the unmethylated group. We hypothesized that the protective role of CHFR methylation in gastric cancer might be a result of alternation of microtubule inhibitors sensitivity. The 8 methylation probes we found in current studies might be good markers for the prognosis of gastric cancer.
There are some limitations in current studies. First, the studies involved in current meta-analysis were mostly from eastern Asia, and the GEO data was also from Singapore, whose population are mostly Chinese, we expect to see more results from other regions. Second, the samples we collected were tissues, which were hard to obtain and might limit the clinical use. Studies from serum or gastric juice are needed for further study. Third, different methylation test methods were involved in current meta-analysis, which might cause systemic bias, however, no significant heterogeneity was found in current meta-analysis. Finally, more prognostic studies are needed for further analysis.
In conclusion, we found CHFR promoter hypermethylation was associated with gastric cancer and played a protective role in gastric cancer process.
Abbreviations
CHFR: checkpoint with forkhead associated and ring finger domains; TCGA: Cancer Genome Atlas network; GEO: Gene Expression Omnibus; PMC: PubMed Centra; OR: odds ratio; 95% CI: 95% confidence index; HR: hazard ratio; HM27K: Infinium HumanMethylation27 BeadChip; HM450K: Infinium HumanMethylation450 BeadChip; NCCN: National Comprehensive Cancer Network; MSP: methylation-specific polymerase chain reaction; COBRA: Combined Bisulfite Restriction Analysis; NA: not available; SEM: standard error of the mean; GB_ACC: GenBank accession; CICHR: chromosome number of the site; MAPINFO: position of the site in the genome.
Supplementary Material
Supplementary figures.
Acknowledgements
This grant was supported by the National Natural Science Foundation of China (81372178, 81502386, 81772944) and Zhejiang Provincial Program for High-level Innovative Healthcare talents.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-64
2. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-9
3. Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining 'chromosomal instability'. Trends Genet. 2008;24:64-9
4. Maddika S, Sy SM, Chen J. Functional interaction between Chfr and Kif22 controls genomic stability. J Biol Chem. 2009;284:12998-3003
5. Shtivelman E. Promotion of mitosis by activated protein kinase B after DNA damage involves polo-like kinase 1 and checkpoint protein CHFR. Mol Cancer Res. 2003;1:959-69
6. Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ. et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401-6
7. Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430-5
8. Guo H, Yan W, Yang Y, Guo M. [Promoter region methylation of DNA damage repair genes in human gastric cancer]. Zhonghua Yi Xue Za Zhi. 2014;94:2193-6
9. Hu SL, Huang DB, Sun YB, Wu L, Xu WP, Yin S. et al. Pathobiologic implications of methylation and expression status of Runx3 and CHFR genes in gastric cancer. Med Oncol. 2011;28:447-54
10. Cheng ZD, Hu SL, Sun YB, Xu WP, Shen G, Kong XY. Promoter methylation of CHFR gene in gastric carcinoma tissues detected using two methods. Chin J Cancer. 2010;29:163-6
11. Oki E, Zhao Y, Yoshida R, Masuda T, Ando K, Sugiyama M. et al. Checkpoint with forkhead-associated and ring finger promoter hypermethylation correlates with microsatellite instability in gastric cancer. World J Gastroenterol. 2009;15:2520-5
12. Morioka Y, Hibi K, Sakai M, Koike M, Fujiwara M, Kodera Y. et al. Aberrant methylation of the CHFR gene in digestive tract cancer. Anticancer Res. 2006;26:1791-5
13. Carrera C, Chinchilla N, Fronczek FR, Galindo JC, Macias FA. Structure-activity relationship studies of the phytotoxic properties of the diterpenic moiety of breviones. Pest Manag Sci. 2015;71:701-11
14. Li Y, Yang Y, Lu Y, Herman JG, Brock MV, Zhao P. et al. Predictive value of CHFR and MLH1 methylation in human gastric cancer. Gastric Cancer. 2015;18:280-7
15. Gopalakrishnan S, Ganeshkumar P. Systematic Reviews and Meta-analysis: Understanding the Best Evidence in Primary Healthcare. J Family Med Prim Care. 2013;2:9-14
16. Jamshidi Ardeshiri M, Moosazadeh M, Feizi Masouleh M, Kiani A, Fakhri M. Prevalence of smoking in 15-64 years old population of north of Iran: meta-analysis of the results of non-communicable diseases risk factors surveillance system. Acta Med Iran. 2013;51:494-500
17. Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010;39:932 author reply 3
18. Jackson D, White IR, Riley RD. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med. 2012;31:3805-20
19. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-34
20. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
21. Zouridis H, Deng N, Ivanova T, Zhu Y, Wong B, Huang D. et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4:156ra40
22. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277-88
23. Wang M, Shen L, Deng D. Association between CHFR methylation and chemosensitivity of paclitaxel in advanced gastric cancer. Med Oncol. 2014;31:907
24. Hu SL, Kong XY, Cheng ZD, Sun YB, Shen G, Xu WP. et al. Promoter methylation of p16, Runx3, DAPK and CHFR genes is frequent in gastric carcinoma. Tumori. 2010;96:726-33
25. Hiraki M, Kitajima Y, Sato S, Mitsuno M, Koga Y, Nakamura J. et al. Aberrant gene methylation in the lymph nodes provides a possible marker for diagnosing micrometastasis in gastric cancer. Ann Surg Oncol. 2010;17:1177-86
26. Gao YJ, Xin Y, Zhang JJ, Zhou J. Mechanism and pathobiologic implications of CHFR promoter methylation in gastric carcinoma. World J Gastroenterol. 2008;14:5000-7
27. Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ. et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161-70
28. Mitsuno M, Kitajima Y, Ide T, Ohtaka K, Tanaka M, Satoh S. et al. Aberrant methylation of p16 predicts candidates for 5-fluorouracil-based adjuvant therapy in gastric cancer patients. J Gastroenterol. 2007;42:866-73
29. Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41:133-9
30. Homma N, Tamura G, Honda T, Jin Z, Ohmura K, Kawata S. et al. Hypermethylation of Chfr and hMLH1 in gastric noninvasive and early invasive neoplasias. Virchows Arch. 2005;446:120-6
31. Kang HC, Kim IJ, Park JH, Shin Y, Park HW, Ku JL. et al. Promoter hypermethylation and silencing of CHFR mitotic stress checkpoint gene in human gastric cancers. Oncol Rep. 2004;12:129-33
32. Honda T, Tamura G, Waki T, Kawata S, Nishizuka S, Motoyama T. Promoter hypermethylation of the Chfr gene in neoplastic and non-neoplastic gastric epithelia. Br J Cancer. 2004;90:2013-6
33. Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K. et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606-13
34. Jensen J, Counte MA, Glandon GL. Elderly health beliefs, attitudes, and maintenance. Prev Med. 1992;21:483-97
35. Sato Y, Yamashita M, Hayashi K, Tokoro T. [Abnormality of visual field in pathological myopia]. Nippon Ganka Gakkai Zasshi. 1984;88:977-82
36. Hiraki M, Kitajima Y, Koga Y, Tanaka T, Nakamura J, Hashiguchi K. et al. Aberrant gene methylation is a biomarker for the detection of cancer cells in peritoneal wash samples from advanced gastric cancer patients. Ann Surg Oncol. 2011;18:3013-9
37. Yoshida K, Hamai Y, Suzuki T, Sanada Y, Oue N, Yasui W. DNA methylation of CHFR is not a predictor of the response to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Anticancer Res. 2006;26:49-54
38. Derks S, Cleven AH, Melotte V, Smits KM, Brandes JC, Azad N. et al. Emerging evidence for CHFR as a cancer biomarker: from tumor biology to precision medicine. Cancer Metastasis Rev. 2014;33:161-71
39. Privette LM, Petty EM. CHFR: A Novel Mitotic Checkpoint Protein and Regulator of Tumorigenesis. Transl Oncol. 2008;1:57-64
40. Privette LM, Weier JF, Nguyen HN, Yu X, Petty EM. Loss of CHFR in human mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia. 2008;10:643-52
41. Privette LM, Gonzalez ME, Ding L, Kleer CG, Petty EM. Altered expression of the early mitotic checkpoint protein, CHFR, in breast cancers: implications for tumor suppression. Cancer Res. 2007;67:6064-74
42. Takeshita M, Koga T, Takayama K, Kouso H, Nishimura-Ikeda Y, Yoshino I. et al. CHFR expression is preferentially impaired in smoking-related squamous cell carcinoma of the lung, and the diminished expression significantly harms outcomes. Int J Cancer. 2008;123:1623-30
43. Koga T, Takeshita M, Yano T, Maehara Y, Sueishi K. CHFR hypermethylation and EGFR mutation are mutually exclusive and exhibit contrastive clinical backgrounds and outcomes in non-small cell lung cancer. Int J Cancer. 2011;128:1009-17
44. Tanaka M, Chang P, Li Y, Li D, Overman M, Maru DM. et al. Association of CHFR promoter methylation with disease recurrence in locally advanced colon cancer. Clin Cancer Res. 2011;17:4531-40
45. Takeshita M, Koga T, Takayama K, Yano T, Maehara Y, Nakanishi Y. et al. Alternative efficacy-predicting markers for paclitaxel instead of CHFR in non-small-cell lung cancer. Cancer Biol Ther. 2010;10:933-41
46. Banno K, Yanokura M, Kawaguchi M, Kuwabara Y, Akiyoshi J, Kobayashi Y. et al. Epigenetic inactivation of the CHFR gene in cervical cancer contributes to sensitivity to taxanes. Int J Oncol. 2007;31:713-20
47. Yanokura M, Banno K, Kawaguchi M, Hirao N, Hirasawa A, Susumu N. et al. Relationship of aberrant DNA hypermethylation of CHFR with sensitivity to taxanes in endometrial cancer. Oncol Rep. 2007;17:41-8
48. Ogi K, Toyota M, Mita H, Satoh A, Kashima L, Sasaki Y. et al. Small interfering RNA-induced CHFR silencing sensitizes oral squamous cell cancer cells to microtubule inhibitors. Cancer Biol Ther. 2005;4:773-80
Author contact
![]() Corresponding author: Dr. Xian Wang, email: wangx118edu.cn, Tel/Fax: +86 571 8600 6929
Corresponding author: Dr. Xian Wang, email: wangx118edu.cn, Tel/Fax: +86 571 8600 6929

 Global reach, higher impact
Global reach, higher impact