Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1086-1096. doi:10.7150/jca.29390 This issue Cite
Research Paper
Association between TNF-ɑ-308G/A polymorphism and esophageal cancer risk: An updated meta-analysis and trial sequential analysis
1. Department of Oncology, The Affiliated Sir Run Run Hospital, Nanjing Medical University, Nanjing, Jiangsu Province, China
2. Department of Oncology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu Province,China
3. Department of Thoracic surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu Province, China
4. Department of Urology, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu Province, China
† These three authors contribute equally to this article
Received 2018-8-21; Accepted 2018-12-14; Published 2019-1-29
Abstract
Background: TNF-α-308G/A (rs1800629) polymorphism has been previously implicated in the susceptibility to esophageal cancer, but results of these studies remained controversial or ambiguous. A meta-analysis was conducted to provide a more reliable conclusion about the association between TNF-ɑ-308G/A polymorphism and risk of esophageal cancer.
Methods: Databases such as PubMed, EMBASE, Web of Science and CNKI were searched for relevant articles published till June 1, 2018. We used the pooled odds ratios (ORs) with 95% confidence intervals (CIs) to evaluate the strength of such associations. Subgroup analysis was carried out according to ethnicity, source of controls and genotyping method. A trial sequential analysis (TSA) was performed to reduce the risk of type I error and evaluate whether the results of our meta-analysis were credible.
Results: A total of 9 published case-control studies with 1,435 esophageal cancer patients and 3,762 healthy controls were identified. Overall, our results indicated no significant correlation between TNF-α-308G/A polymorphism and increased risk of esophageal cancer in the fixed-effects model (allele model: pooled OR=1.11, 95% CI: 0.96-1.27, homozygote model: pooled OR=1.23, 95% CI: 0.77-1.95, heterozygote model: pooled OR=1.14, 95% CI: 0.97-1.35, dominant model: pooled OR=1.14, 95% CI: 0.97-1.34 and recessive model: pooled OR=1.00, 95% CI: 0.64-1.56). Subgroup analysis by ethnicity, source of controls and genotyping method showed no significant increase in the risk of esophageal cancer. TSA results need further investigation with a large sample size to certify such association.
Conclusions: This meta-analysis study suggested no significant association between TNF-ɑ-308G/A polymorphism and the risk of esophageal cancer.
Keywords: TNF-ɑ-308G/A, Polymorphism, Esophageal cancer, Risk, Meta-analysis.
Introduction
Esophageal cancer is considered as the eighth most common cancer and the sixth leading cause of cancer-related deaths in the world1. Its overall 5-year survival was less than 20% due to delayed diagnosis, even in the United States2. Esophageal cancer is a multifactorial disease involving intricate interactions between numerous genetic as well as various environmental factors, such as alcohol, smoking, poor diet, poor oral health, chemical carcinogens or occupational exposure3, 4. However, several genetic factors responsible for the esophageal cancer have not been clarified yet. Recent studies have shown that several single-nucleotide genetic polymorphisms (SNPs) were associated with the susceptibility to esophageal cancer5, 6. Among these, polymorphism of tumor necrosis factor alpha (TNF-α) is one of the most widely studied genes, possibly predicting the genetic risk of of esophageal cancer.
TNF-α is a pro-inflammatory cytokine, and is mainly secreted by monocytes and macrophages7. It plays a key role in host defense and inflammatory responses, but in some cases also triggers cell death and tissue degradation8, 9. Dysregulated expression of TNF-α was reported to be associated with various disorders, including inflammatory diseases (such as Rheumatoid arthritis, Crohn's disease), central nervous system diseases (Alzheimer's disease) and a variety of other tumors10-13. TNF-ɑ gene is located on human chromosome 6q21 within class III region of the major histocompatibility complex (MHC)14. A number of SNPs of TNF-α gene have been found, which include TNF-α-238 G/A (rs361525), TNF-α-308G/A (rs1800629), TNF-α-857C/T (rs179972), TNF-α-863C/A (rs1800630), TNF-α-509C/T (rs1800469) and TNF-α-1031T/C (rs1799964)15, 16. Among these, the most common TNF-α polymorphisms is present in the promoter region at position -308 and it has been studied most extensively 17-19.
Up to now, several studies have been performed to clarify the association between TNF-ɑ-308G/A genetic polymorphism and susceptibility to esophageal cancer. However, the results were still inconsistent. Therefore, we carried out this meta-analysis with all accessible case-control studies and trial sequential analysis (TSA), which showed that the present research was not enough to get such a conclusion, which also required other studies to confirm this conclusion. Therefore, the results of this meta-analysis demonstrated that no evidence supporting the relationship between TNF-ɑ-308G/A polymorphism and esophageal cancer risk was detected. More importantly, further studies were needed to give more comprehensive understanding of such association in the future.
Materials and Methods
Literature search
A total of nine published case-control studies were identified by searching PubMed, EMBASE, Web of Science and CNKI databases till June 1, 2018. The following index terms and Mesh terms were used for the search: “tumor necrosis factor alpha” or “TNF-ɑ”, “polymorphism” or “variants” and “esophageal cancer” or “esophageal tumor” or “ECa”. Moreover, we scanned the references of the original articles, and performed a manual search for additional literatures that might be identified. To avoid overlapping of the data, we checked carefully and selected the latest and more credible studies.
Inclusion and exclusion criteria
Inclusion criteria were as follows: (1) An independent case-control study; (2) Association between TNF-α-308G/A gene polymorphism and susceptibility to esophageal cancer; (3) The study should also contain abundant data of regarding the genotype frequency to evaluate whether such association was available.
Exclusion criteria were as follows: (1) Not case-control studies; (2) Studies not providing sufficient data to calculate the genotypic distributions of cases and controls; (3) Reviews or meta-analyses studies; (4) Previous duplicated publications.
Data extraction
The following information was extracted independently by two reviewers (FMYang and ZQQin) from each article: first author's name, year of publication, the number of esophageal cancer cases and controls, and genotypes or alleles of the TNF-α-308 G/A polymorphism. Any controversial issues were resolved through discussion with the third author until a consensus was reached.
Quality assessment
The quality of eligible articles was assessed using the Newcastle-Ottawa Quality Assessment Scale (NOS) for cohort and case-control studies. Quality assessment included the selection, comparability, exposure of a case-control study, and the outcome of a cohort study. Based on the scoring system, studies with scores >7 were considered to be of high quality.
Statistical analysis
The strength of association between TNF-α-308 G/A mutations and esophageal cancer risk was evaluated by the pooled odds ratios (ORs) with 95% confidence intervals (CIs). Five genetic comparison models for the meta-analysis used were as follows: (1) dominant model: (GA+AA) vsGG; (2) recessive model: AA vs (GA+GG); (3) homozygous model: AA vs GG; (4) heterozygous model: GA vs GG; and (5) allele model: A vs G. The chi-square (χ2) goodness of fit was adopted to evaluate Hardy-Weinberg equilibrium (HWE) in controls and P<0.05 was considered as statistically significant difference.
Pooled OR was calculated by using fixed-effects model (the Mantel-Haenszel method) or random-effects model (the DerSimonian and Laird method) according to the P values of study heterogeneities. If the P value was <0.05, the pooled OR was then calculated by the fixed-effects model, otherwise random-effects model was used. To verify the potential sources of heterogeneity, subgroup analyses were performed by ethnicity, source of controls and genotyping method. Furthermore, sensitivity analysis was conducted by sequentially excluding each individual study to examine the stability and reliability of the results. Publication bias was checked by Begg's funnel plots and Egger's linear regression test. All statistical analyses were performed using STATA software (version 12.0; StataCorp LP, College Station, TX).
Trial Sequential Analysis (TSA)
Conventional meta-analyses might obtain false positive results (type I errors) and false negative results (type II errors) due to systematic errors (bias) and random errors caused by sparse data and repetitive testing20-22. Therefore, we conducted TSA to reduce the risk of type I error by maintaining the overall 5% risk of a type I error and 20% risk of a type II error (power of 80%) to estimate the required information size23. In TSA, we constructed the cumulative Z-curve of each study and assessed its crossing of Z=1.96 (P=0.05) and the trial sequential monitoring boundaries24. When the cumulative Z-curve crosses the trial sequential monitoring boundary or the required information size has been reached, firm evidence was shown for the present mata-analysis study and further studies are not required. On the contrary, if the Z curve did not cross any of the boundaries, it is necessary to carry out an additional clinical trial to reach a consistent conclusion25. These analyses were done using TSA 0.9 (Copenhagen Trial Unit, Copenhagen, Denmark).
Results
Characteristics of the studies
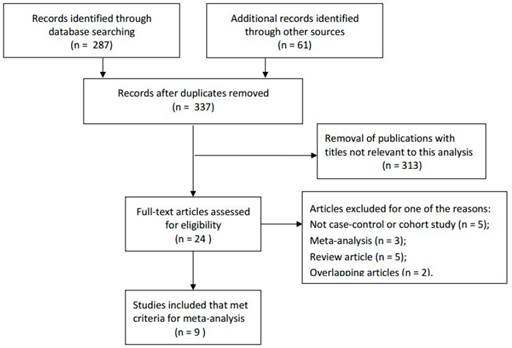
According to the inclusion and exclusion criteria, a total of 287 articles were initially identified through primary search of the relevant databases and reference lists. After reading the titles and abstracts, 9 full-text studies with a total of 1,435 esophageal cancer patients and 3,762 controls met the inclusion criteria and were involved in the present meta-analysis for further evaluation, which had been accrued between May 2003 and May 201526-34. In addition, all studies suggested that the genotypic distributions in the controls were consistent with Hardy-Weinberg equilibrium (HWE), except the study by Guo et al.34 For the source of samples, Among the 9 enrolled studies, DNA was extracted from whole blood in 8 studies26-30, 32-34, while only 1 study used Frozen tissue to extract DNA31. So we decided not to carry out the subgroup analysis by source of samples. The flowchart of literature search and selection procedure was shown in Figure 1. In this meta-analysis, the baseline characteristics of the studies associated with the risk of esophageal cancer were comprehensively listed in Table 1. Among the 9 enrolled studies, 6 studies were based on Asian population, 1 study was based on Caucasian population and the remaining 2 studies included mixed population. Furthermore, we included 6 population-based studies, including 1 hospital-based study and the remaining 2 unknown-control of source studies, to distinguish between different sources of control group. Different genotyping methods applied were as follows: TaqManSNP (TaqMan), polymerase chain reaction (PCR), SNPlex and Sequenom.
Flow diagram of literature search and selection process.
Characteristics of studies that investigated the association between TNF-α-308G/A polymorphism and esophageal cancer risk.
| TNF-α-308G/A | Case (n) | Control(n) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Author | Country | Ethnicity | SOC | Genotyping | Case | Control | GG | GA | AA | GG | GA | AA | HWE |
| 2015 | Cui | China | Asian | NR | PCR | 212 | 200 | 150 | 57 | 5 | 140 | 58 | 2 | Y |
| 2014 | Wang | China | Asian | PB | PCR-RFLP | 33 | 50 | 3 | 26 | 4 | 11 | 25 | 14 | Y |
| 2013 | Umar | India | Asian | NR | PCR-RFLP | 290 | 311 | 227 | 62 | 1 | 268 | 42 | 1 | Y |
| 2011 | Zhang | China | Asian | HB | PCR-SSP | 120 | 95 | 99 | 19 | 2 | 82 | 12 | 1 | Y |
| 2010 | David | Australia | Caucasian | PB | Sequenom | 207 | 1293 | 128 | 71 | 8 | 842 | 403 | 48 | Y |
| 2010 | Zhao | China | Asian | PB | PCR | 202 | 317 | 141 | 56 | 5 | 228 | 83 | 6 | Y |
| 2010 | Oh | USA | Mix | PB | SNPlex | 27 | 849 | 19 | 8 | 0 | 641 | 195 | 13 | Y |
| 2005 | Guo | China | Asian | PB | PCR-RFLP | 291 | 437 | 266 | 21 | 4 | 391 | 40 | 6 | N |
Meta-analysis of association between TNF-α-308G/A polymorphism and esophageal cancer risk after the elimination of Hamasaki et al study.
| Variables | No. of studies | Dominant model | Recessive model | Homozygous model | Heterozygous model | Allele model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P values | I-squared (%) | OR (95% CI) | P values | I-squared (%) | OR (95% CI) | P values | I-squared (%) | OR (95% CI) | P values | I-squared (%) | OR (95% CI) | P values | I-squared (%) | ||
| rs1800629 -308G/A | (GA + AA) vs. GG | AA vs. (GA + GG) | AA vs. GG | GA vs. GG | A vs. G | |||||||||||
| All | 9 | 1.14(0.97-1.34) | 0.316 | 14.1 | 1.00(0.64-1.56) | 0.824 | 0 | 1.23(0.77-1.95) | 0.999 | 0 | 1.14(0.97-1.35) | 0.18 | 29.8 | 1.11(0.96-1.27) | 0.596 | 0 |
| Ethnicity | ||||||||||||||||
| Asian | 6 | 1.17(0.96-1.44) | 0.158 | 37.3 | 0.94(0.52-1.69) | 0.52 | 0 | 1.32(0.69-2.53) | 0.978 | 0 | 1.17(0.95-1.44) | 0.084 | 48.4 | 1.13(0.94-1.34) | 0.336 | 12.3 |
| Caucasian | 1 | 1.15(0.85-1.56) | NA | NA | 1.04(0.49-2.24) | NA | NA | 1.10(0.51-2.37) | NA | NA | 1.16(0.85-1.59) | NA | NA | 1.11(0.86-1.44) | NA | NA |
| Mix | 2 | 0.94(0.55-1.62) | 0.349 | 0 | 1.28(0.31-5.23) | 0.92 | 0 | 1.23(0.30-5.10) | 0.993 | 0 | 0.94(0.54-1.65) | 0.25 | 24.3 | 0.95((0.59-1.55) | 0.528 | 0 |
| Source of control | ||||||||||||||||
| NR | 2 | 1.29(0.96-1.74) | 0.057 | 72.5 | 1.96(0.49-7.94) | 0.62 | 0 | 1.97(0.49-8.04) | 0.679 | 0 | 1.27(0.94-1.72) | 0.039 | 76.5 | 1.27(0.97-1.67) | 0.097 | 63.6 |
| PB | 6 | 1.07(0.88-1.31) | 0.479 | 0 | 0.90(0.55-1.47) | 0.691 | 0 | 1.13(0.68-1.88) | 1 | 0 | 1.08(0.88-1.32) | 0.28 | 20.3 | 1.04(0.88-1.23) | 0.862 | 0 |
| HB | 1 | 1.34(0.63-2.84) | NA | NA | 1.59(0.14-17.84) | NA | NA | 1.66(0.15-18.60) | NA | NA | 1.31(0.60-2.86) | NA | NA | 1.33(0.67-2.67) | NA | NA |
| Genotyping | ||||||||||||||||
| PCR | 6 | 1.17(0.96-1.44) | 0.158 | 37.3 | 0.94(0.52-1.69) | 0.52 | 0 | 1.32(0.69-2.53) | 0.978 | 0 | 1.17(0.95-1.44) | 0.084 | 48.4 | 1.13(0.94-1.34) | 0.336 | 12.3 |
| Sequenom | 1 | 1.15(0.85-1.56) | NA | NA | 1.04(0.49-2.24) | NA | NA | 1.10(0.51-2.37) | NA | NA | 1.16(0.85-1.59) | NA | NA | 1.11(0.86-1.44) | NA | NA |
| SNPlex | 1 | 1.30(0.56-3.01) | NA | NA | 1.13(0.07-19.44) | NA | NA | 1.22(0.07-21.24) | NA | NA | 1.38(0.60-3.21) | NA | NA | 1.16(0.54-2.50) | NA | NA |
| Taqman | 1 | 0.77(0.38-1.56) | NA | NA | 1.33(0.26-6.80) | NA | NA | 1.24(0.24-6.35) | NA | NA | 0.71(0.33-1.52) | NA | NA | 0.85(0.45-1.58) | NA | NA |
NA: Not Applicable.
Quantitative synthesis results
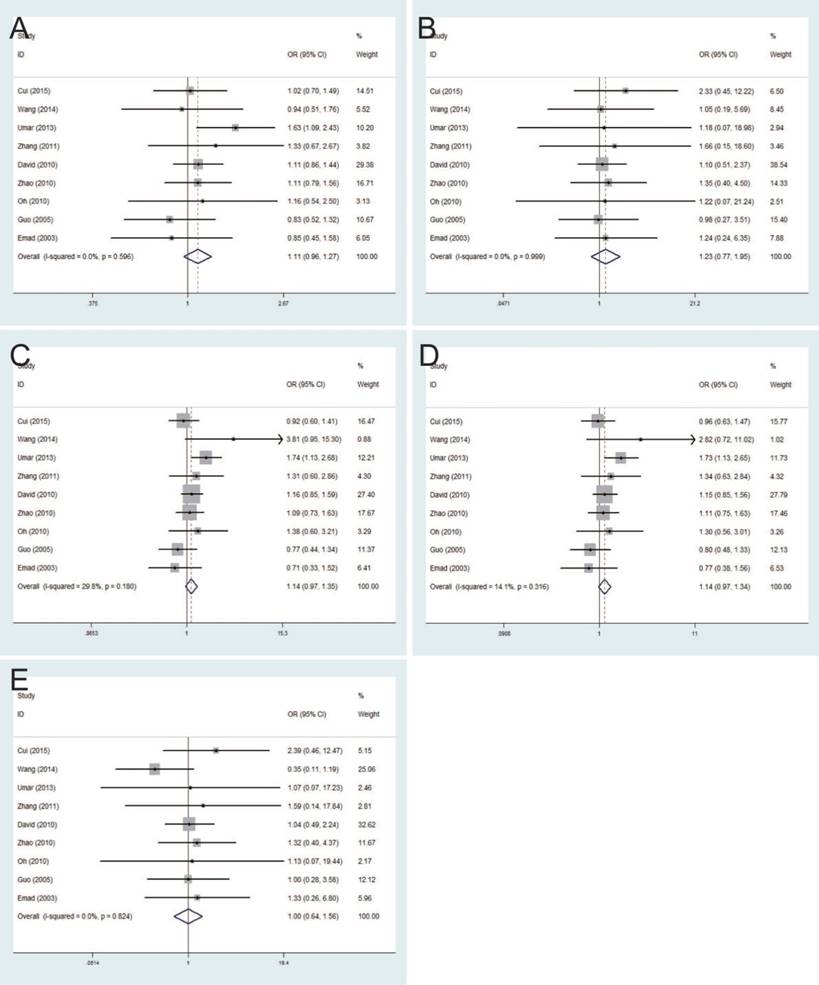
The strength of association between TNF-ɑ-308G/A polymorphism and esophageal cancer risk was evaluated by the pooled ORs with 95% CIs based on five genetic comparison models. Summary of all results regarding the relationship between TNF-ɑ-308G/A polymorphisms and esophageal cancer risk in the 9 studies was provided in Table 2. Results of this meta-analysis demonstrated no significant relationship between TNF-ɑ-308G/A polymorphism and esophageal cancer risk with the fixed-effects model, with the pooled ORs and 95% CIs in allele model (pooled OR=1.11, 95% CI: 0.96-1.27), homozygote model (pooled OR=1.23, 95% CI: 0.77-1.95), heterozygote model (pooled OR=1.14, 95% CI: 0.97-1.35), dominant model (pooled OR=1.14, 95% CI: 0.97-1.34) and recessive model (pooled OR=1.00, 95% CI: 0.64-1.56) (Figure 2).
In the subgroup analysis by ethnicity, results showed no statistical significance in the Asian, Caucasian, and Mixed populations. Moreover, subgroup analysis by control source groups were also performed, and no statistically significant results were detected in the population-based control group and hospital-based control group. In addition, in the subgroup analysis by different genotyping methods, no significant results of such association were found using TaqMan, PCR, Sequenom and SNPlex, respectively (Table 2). In general, there was no association between TNF-ɑ-308G/Apolymorphism and esophageal cancer risk in these five genetic comparison models.
Test of heterogeneity
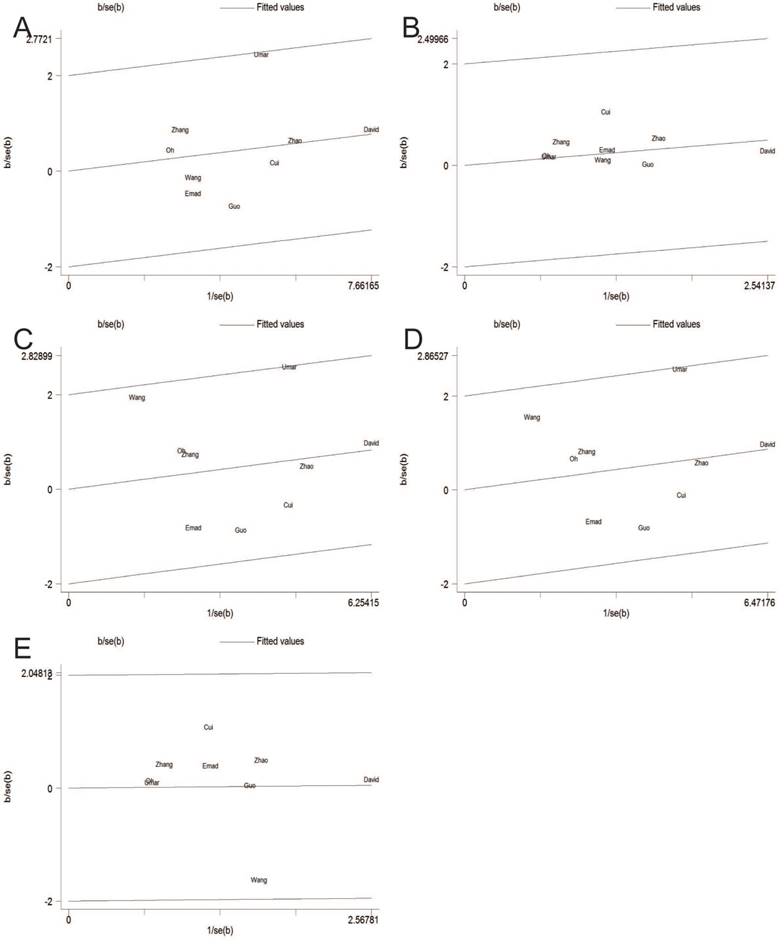
Heterogeneity was observed in the overall genetic models, but it was interesting that subgroup analyses could decrease the heterogeneity. Thus, neither ethnicity nor source of controls was performed for substantial heterogeneity. Figure 3 showed analysis of a Galbraith radial plot in dominant model, suggesting no significant heterogeneity between the studies.
Forest plots of the association between TNF-α-308G/A polymorphism and esophageal cancer susceptibility in fixed-effects model. A: allele model; B: homozygote model; C: heterozygote model; D: dominant model; E: recessive model.
Galbraith plot of the association between TNF-ɑ-308G/A polymorphism and esophageal cancer susceptibility in fixed-effects model. A: allele model; B: homozygote model; C: heterozygote model; D: dominant model; E: recessive model.
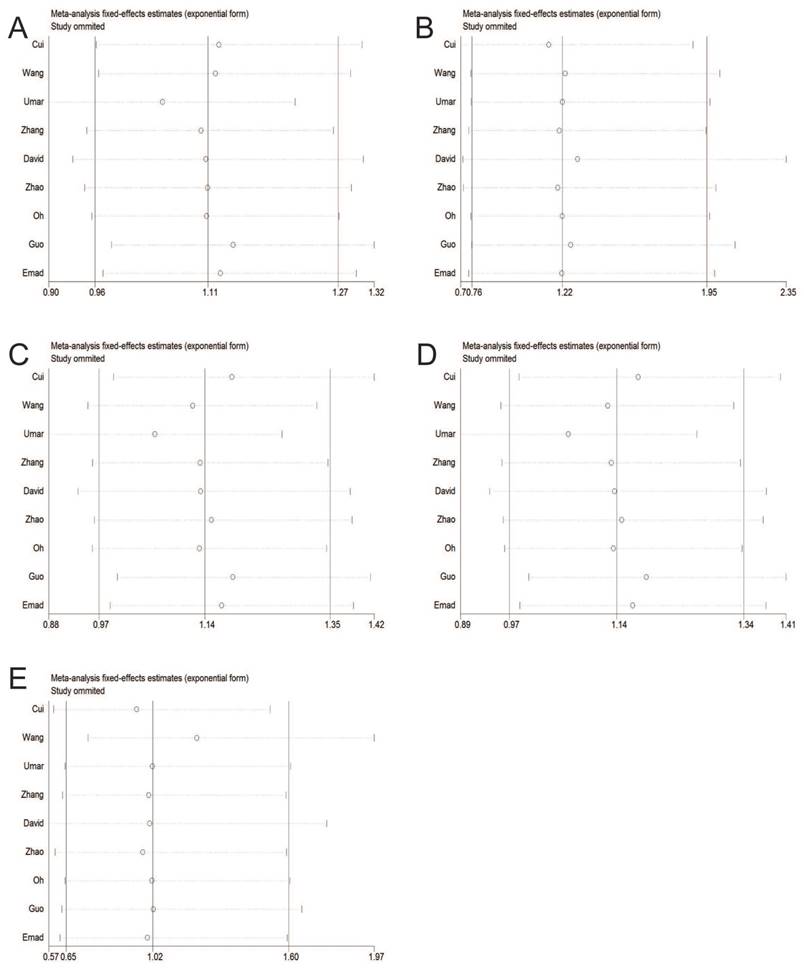
Sensitivity analysis
Sensitivity analysis was performed to explore the influence of each study on the pooled ORs. Figure 4 showed that the pooled ORs were not substantially altered, which resulted in the reliable and comprehensive meta-analysis study.
Publication bias
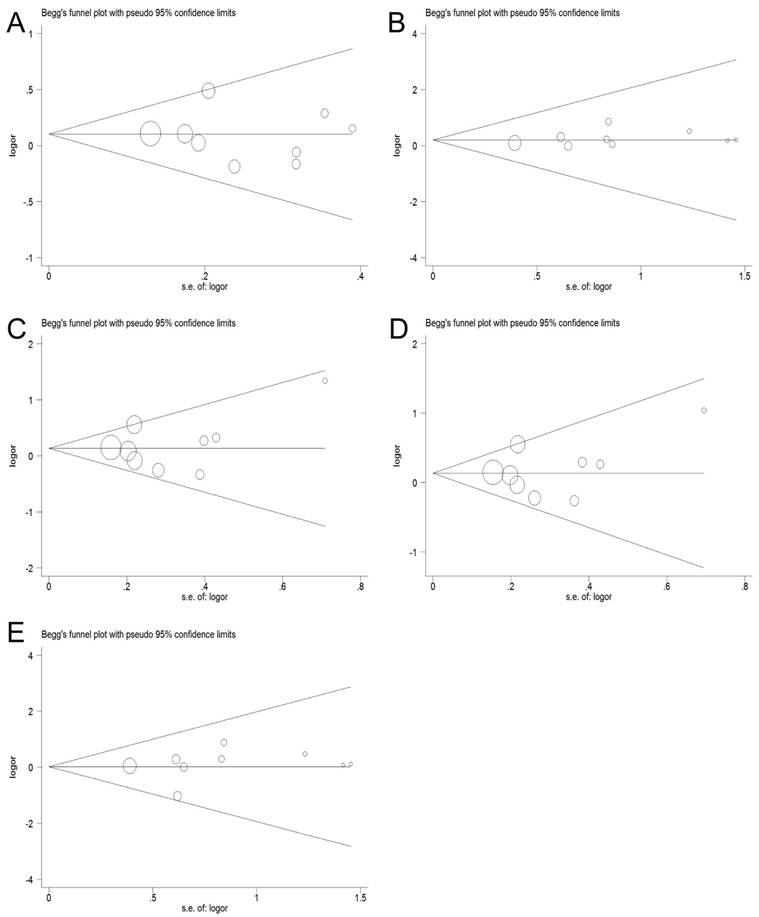
Publication bias of the included studies was assessed by Begg's funnel plot and Egger's test. The funnel plot of the TNF-α-308G/A polymorphism did not reveal any evidence of clear asymmetry, indicating that there was no significant publication bias in all the studies, as evidenced by the Egger's test (allele model: P=0.717, homozygous model: P=0.336, heterozygous model: P=0.636, dominant model: P=0.680 and recessive model: P=0.560), (Figure 5).
Sensitivity analysis in fixed-effects model. A: allele model; B: homozygote model; C: heterozygote model; D: dominant model; E: recessive model.
Begg's funnel plot of publication bias test. A: allele model; B: homozygote model; C: heterozygote model; D: dominant model; E: recessive model.
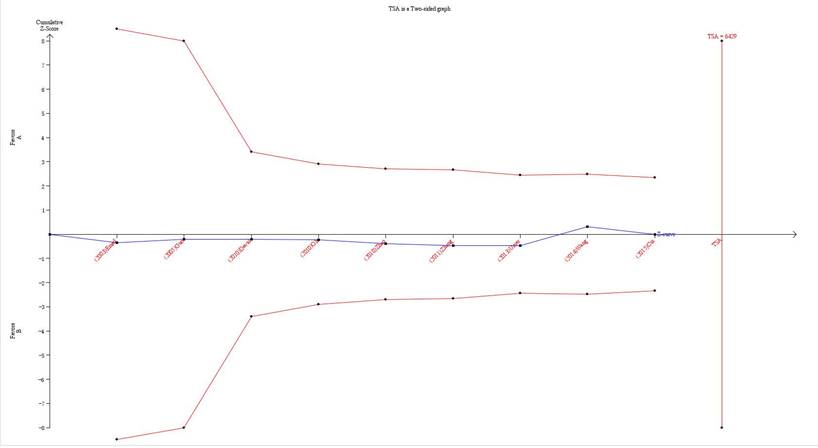
Trial Sequential Analysis results
In our current study, the cumulative Z-curve (the blue line) did not exceed the information size (vertical red line), and the total number of cases and controls were less than the required information size (Figure 6). Therefore, our results require further investigation in a sufficiently large number of participants to certify the associations in well-designed studies.
Discussion
TNF-α gene is encoded in class III major histocompatibility complex (6p21.3). As a potent pro-inflammatory cytokine, TNF-α plays an important role in the inflammatory and immune responses35. However, the effect of TNF-α on tumors remained unclear. Previous studies have suggested that dysregulated expression of TNF-α might promote the occurrence and development of tumors11, 36, 37. Notably, TNF-α production is regulated by SNP in the promoter region. At least 12 SNPs have been identified in the TNF-α gene, and the most studied SNP is TNF-α-308G/A (rs1800629) 38. Both in vivo and in vitro studies have demonstrated that TNF-α-308G/A was involved in the occurrence and development of tumors by regulating the production of TNF-α39, 40.
Trial sequential analysis of the association between TNF-α-308G/A polymorphism and the risk of esophageal cancer. The required information size was calculated based on a two side α= 5%, β= 15% (power 85%), and a relative risk reduction of 20%.
To date, some studies have investigated whether TNF-α-308G/A polymorphism was associated with the risk of esophageal cancer. The studies due to limited sample size and other reasons ultimately led to conflicting results. In a word, there is no definitive conclusion about the role of rs1800629 in esophageal cancer risk. Findings by Umar et al. study suggested that TNF-α-308 G>A polymorphism enhanced the risk of esophageal cancer, especially in females and in patients with regional lymph node involvement28. On the contrary, results of Cui et al. study showed lack of association of TNF-α-308G/A polymorphism with ECa risk26. What's more, another study by Guo et al. found no significant difference in the overall genetypic distribution of TNF-α-308G/A polymorphism among ECa patients and controls 34. Hence, there were no consistent conclusions about the role of TNF-α-308G/A gene polymorphism in esophageal cancer risk. Hence, we aimed to elucidate whether TNF-α-308G/A gene polymorphism was associated with the susceptibility to esophageal cancer in our meta-analysis. In addition, TSA was applied to effectively reduce the risk of type I errors and assess whether the required information size has been reached.
Our present meta-analysis study collected 1,435 esophageal cancer patients and 3,762 healthy controls from 9 case-controlled studies to investigate the association between -308G/A polymorphism in the TNF-α gene and esophageal cancer risk. As a powerful tool, our meta-analysis made the conclusion more credible compared with a single study, especially in analyzing the unexplained associations41. With the development of the current meta-analysis study, a more comprehensive understanding of the relationship between rs1800629 and the risk of esophageal cancer by different subgroup analysis was performed. As a consequence, we took advantage of the meta-analysis to explain this possible association. Our study results revealed no significant relationship between TNF-α-308G/A polymorphism and increased risk of esophageal cancer. This contradiction could be caused by several factors, including the differences in sample size, genotyping methods, study design, statistical methods and so on.
Three subgroup meta-analyses were conducted by ethnicity, source of controls and genotyping method. In the ethnic subgroup, the TNF-α-308G/A allele was not responsible for the increased risk of esophageal cancer in Caucasians, Africans, and Asians. However, the results might not be conclusive due to relatively small number of Caucasians used in the meta-analysis. Besides, as Caucasians include mixed populations from different geographic regions and other ethnic groups, there was a significant inter-study heterogeneity among Caucasians, leading to the negative results of our analysis. Meanwhile, in the subgroup analysis by source of controls, no significant results were found in both population-based control group and hospital-based control group. The possible reason was that people in the control group might be exposed to other risks of esophageal cancer, thus affecting the results. After stratification according to different genotyping methods, no statistically significant difference about such association in TaqMan, PCR, PCR-RFLP and so on were found. Different genotyping methods might also deviate the results because of their own strengths and weaknesses in various aspects. Therefore, adopting the same appropriate genotyping method might make meta-analysis results more impersonal and reliable. More importantly, it was necessary to have a unified inclusion criteria and a larger sample size of relevant studies.
TSA is a powerful and useful approach in summarizing the evidence and providing the required information size in meta-analyses 42. In order to reduce the risk of type I error and estimate whether further trials are needed, TSA was implied to calculate the required information size for the meta-analysis with the adaptation of monitoring boundaries43. If the cumulative Z-curve crosses the trial sequential monitoring boundary or the required information size, it shows firm evidence for such study. If not, it is necessary to perform an additional clinical trial to reach for a consistent conclusion44. As shown in our study, the cumulative Z-curve did not reach the perpendicular line (required information size), which meant that our results needed further firm evidence regarding the effect.
Furthermore, our meta-analysis has few limitations that need to be emphasized: (1) Most of the populations involved in these case-control studies were Caucasians and Asians, and hence the results might be applicable only to the two races. Further studies with more data are required to investigate the association in other populations. (2) The sample size of each study included in this analysis was relatively small, resulting in the lack of strong statistical persuasion to reveal the real relationship. Hence, further studies with abundant and comprehensive data were required to verify the association. (3) Since our meta-analysis only selected previously published studies, unpublished studies can be omitted and the results are negative, which may bias the results. (4) As a multifactorial disease, the risk of developing esophageal cancer was closely related to the environment, diet, occupational exposure and the interaction of various genetic factors, but not by any single factor. Therefore, we need further studies with more raw data controlling the variable factors to achieve more accurate results about the association. Additionally, the incidence of esophageal cancer was different among different races. Majority of the studies included were investigated in Asian population in this meta-analysis. Therefore, the outcome of this ethnic sub-group analysis might be affected.
Conclusion
In conclusion, our meta-analysis study demonstrated no evidence supporting the relationship between TNF-α-308G/A polymorphism and esophageal cancer risk. More importantly, further studies were needed to give more comprehensive understanding regarding such association in the future.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China (no. 81874230), Jiangsu Social Development Project (BE2018726), the Natural Science Foundation of Jiangsu Province (Grants no. BK20171484), the Project of Invigorating Health Care through Science, Technology, and Education (Jiangsu Provincial Medical Youth Talent QNRC2016856), the Summit of the Six Top Talents Program of Jiangsu Province (2017-WSN-179), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1483), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Domper AM, Ferrandez AA, Lanas AA. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943
2. Palladino-Davis AG, Mendez BM, Fisichella PM, Davis CS. Dietary habits and esophageal cancer. Dis Esophagus. 2015;28:59-67
3. Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598-5606
4. Lin Y, Totsuka Y, He Y. et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233-242
5. Sadot E, Kraus S, Stein M. et al. CD24 gene polymorphism-a novel prognostic factor in esophageal cancer. Int J Biol Markers. 2014;29:e49-e54
6. Yun YX, Wang YP, Wang P. et al. CYP1A1 genetic polymorphisms and risk for esophageal cancer: a case-control study in central China. Asian Pac J Cancer Prev. 2014;14:6507-6512
7. Wallach D. The TNF cytokine family: one track in a road paved by many. Cytokine. 2013;63:225-229
8. Watanabe M, Tanaka K, Takizawa T. et al. Characterization of a canine tetranucleotide microsatellite marker located in the first intron of the tumor necrosis factor alpha gene. J Vet Med Sci. 2014;76:119-122
9. Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149-160
10. Jia T, Pan Y, Li J, Wang L. Strategies for active TNF-alpha vaccination in rheumatoid arthritis treatment. Vaccine. 2013;31:4063-4068
11. Zhu G, Du Q, Wang X, Tang N, She F, Chen Y. TNF-alpha promotes gallbladder cancer cell growth and invasion through autocrine mechanisms. Int J Mol Med. 2014;33:1431-1440
12. Chu CQ. Molecular probing of TNF: From identification of therapeutic target to guidance of therapy in inflammatory diseases. Cytokine. 2018;101:64-9
13. Uslu S, Akarkarasu ZE, Ozbabalik D. et al. Levels of amyloid beta-42, interleukin-6 and tumor necrosis factor-alpha in Alzheimer's disease and vascular dementia. Neurochem Res. 2012;37:1554-1559
14. Carroll MC, Katzman P, Alicot EM. et al. Linkage map of the human major histocompatibility complex including the tumor necrosis factor genes. Proc Natl Acad Sci U S A. 1987;84:8535-8539
15. Olaniyan SA, Amodu OK, Bakare AA, Troye-Blomberg M, Omotade OO, Rockett KA. Tumour necrosis factor alpha promoter polymorphism, TNF-238 is associated with severe clinical outcome of falciparum malaria in Ibadan southwest Nigeria. Acta Trop. 2016;161:62-67
16. Nguyen TN, Baaklini S, Koukouikila-Koussounda F. et al. Association of a functional TNF variant with Plasmodium falciparum parasitaemia in a congolese population. Genes Immun. 2017;18:152-157
17. Piotrowski P, Wudarski M, Sowinska A, Olesinska M, Jagodzinski PP. TNF-308 G/A polymorphism and risk of systemic lupus erythematosus in the Polish population. Mod Rheumatol. 2015;25:719-723
18. Roszak A, Misztal M, Sowinska A, Jagodzinski PP. TNF-alpha -308 G/A as a risk marker of cervical cancer progression in the Polish population. Mol Diagn Ther. 2015;19:53-57
19. Salama H, Hammad E. Risk Association between TNF-alpha-308 G>A and IL-6-174 G/C Polymorphisms and Recurrent Transient Ischemic Attacks. Egypt J Immunol. 2015;22:49-56
20. Wetterslev J, Thorlund K, Brok J, Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J Clin Epidemiol. 2008;61:64-75
21. Kulinskaya E, Wood J. Trial sequential methods for meta-analysis. Res Synth Methods. 2014;5:212-220
22. Brok J, Thorlund K, Gluud C, Wetterslev J. Trial sequential analysis reveals insufficient information size and potentially false positive results in many meta-analyses. J Clin Epidemiol. 2008;61:763-769
23. Keus F, Wetterslev J, Gluud C, Gooszen HG, van Laarhoven CJ. Trial sequential analyses of meta-analyses of complications in laparoscopic vs. small-incision cholecystectomy: more randomized patients are needed. J Clin Epidemiol. 2010;63:246-256
24. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539-1558
25. Wetterslev J, Thorlund K, Brok J, Gluud C. Estimating required information size by quantifying diversity in random-effects model meta-analyses. Bmc Med Res Methodol. 2009;9:86
26. Cui XB, Wang DD, Zhang HY. et al. Tumor necrosis factor-alpha gene 308G/A polymorphism is not associated with esophageal squamous cell carcinoma risk in Kazakh patients. Int J Clin Exp Pathol. 2015;8:9293-9299
27. Wang H, Liu H, Guli A. et al. p21 and TNF-α gene polymorphisms and susceptibility of Xinjiang Kazakh esophageal family. Chinese Journal of Cancer Prevention and Treatment. 2014;21:329-332
28. Umar M, Upadhyay R, Kumar S, Ghoshal UC, Mittal B. Association of common polymorphisms in TNFA, NFkB1 and NFKBIA with risk and prognosis of esophageal squamous cell carcinoma. Plos One. 2013;8:e81999
29. Whiteman DC, Parmar P, Fahey P. et al. Association of Helicobacter pylori infection with reduced risk for esophageal cancer is independent of environmental and genetic modifiers. Gastroenterology. 2010;139:73-83 e11-e12
30. El-Omar EM, Rabkin CS, Gammon MD. et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193-1201
31. Zhao W, Miao Z, Lu P, Xu F. TNF-α Polymorphism and Esophageal Squamous Cell Carcinoma. China Healthcare Innovation. 2010:6-8
32. Oh SS, Chang SC, Cai L. et al. Single nucleotide polymorphisms of 8 inflammation-related genes and their associations with smoking-related cancers. Int J Cancer. 2010;127:2169-2182
33. Ling-ling Z, Yue-ping L, DU Kun, Heng W, Shi-zheng L, Xiao-ling W. Association between 308 G/A tumor necrosis factor alpha gene polymorphism and prognosis of esophageal squamous cell carcinoma. Chinese Journal of Pathology. 2011;40:44-45
34. Guo W, Wang N, Li Y, Zhang JH. Polymorphisms in tumor necrosis factor genes and susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a population of high incidence region of North China. Chin Med J (Engl). 2005;118:1870-1878
35. Tahghighi F, Ziaee V, Moradinejad MH. et al. Tumor necrosis factor-alpha single nucleotide polymorphisms in juvenile systemic lupus erythematosus. Hum Immunol. 2015;76:533-536
36. Roszak A, Misztal M, Sowinska A, Jagodzinski PP. TNF-alpha -308 G/A as a risk marker of cervical cancer progression in the Polish population. Mol Diagn Ther. 2015;19:53-57
37. Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218-e228
38. Radouane A, Oudghiri M, Chakib A, Bennani S, Touitou I, Barat-Houari M. SNPs in the TNF-alpha gene promoter associated with Behcet's disease in Moroccan patients. Rheumatology (Oxford). 2012;51:1595-1599
39. Brinkman BM, Zuijdeest D, Kaijzel EL, Breedveld FC, Verweij CL. Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation. J Inflamm. 1995;46:32-41
40. Louis E, Franchimont D, Piron A. et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998;113:401-406
41. Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res. 2004;129:39-44
42. Miladinovic B, Kumar A, Hozo I, Mahony H, Djulbegovic B. Trial sequential analysis may be insufficient to draw firm conclusions regarding statistically significant treatment differences using observed intervention effects: a case study of meta-analyses of multiple myeloma trials. Contemp Clin Trials. 2013;34:257-261
43. Miladinovic B, Mhaskar R, Hozo I, Kumar A, Mahony H, Djulbegovic B. Optimal information size in trial sequential analysis of time-to-event outcomes reveals potentially inconclusive results because of the risk of random error. J Clin Epidemiol. 2013;66:654-659
44. Wetterslev J, Jakobsen JC, Gluud C. Trial Sequential Analysis in systematic reviews with meta-analysis. Bmc Med Res Methodol. 2017;17:39
Author contact
![]() Corresponding authors: Yongqian Shu. Address: Department of Oncology, the First Affiliated Hospital of Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People's Republic of China. E-mail: shuyongqian1998com; Hua Shen. Address: Department of Oncology, the First Affiliated Hospital of Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People's Republic of China. E-mail: medshenhuacom
Corresponding authors: Yongqian Shu. Address: Department of Oncology, the First Affiliated Hospital of Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People's Republic of China. E-mail: shuyongqian1998com; Hua Shen. Address: Department of Oncology, the First Affiliated Hospital of Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People's Republic of China. E-mail: medshenhuacom

 Global reach, higher impact
Global reach, higher impact