Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1117-1123. doi:10.7150/jca.29212 This issue Cite
Review
Current Status of Research on the Period Family of Clock Genes in the Occurrence and Development of Cancer
Department of Oral and Maxillofacial Surgery, The First Affliated Hospital of Chongqing Medical University, 1 Youyi Road, Yuzhong District, Chongqing 400016, People's Republic of China
Received 2018-8-14; Accepted 2019-1-3; Published 2019-1-29
Abstract
Several physiological activities of organisms are coordinated based on periodic variations of ~24 h, which is called a circadian rhythm. Circadian rhythms, driven by circadian clock genes, play an important role in the regulation of various complex life activities of organisms, in an orderly and coordinated manner. Period (Per)1/2/3 genes are important core clock genes and part of the Per gene family. Current research has demonstrated that the abnormal expression of Per genes and disruption of circadian rhythms can lead to the occurrence and development of cancer; however, the exact mechanism has not yet been elucidated. Further study on this mechanism may lead to the discovery of new, effective therapies for the prevention and treatment of cancer. The present review summarizes the status of current research with regards to the association between the abnormal expression and rhythmic variation of the Per gene family, and carcinogenesis and progression of cancer.
Keywords: clock genes, Period gene, circadian rhythm, cancer, carcinogenesis
1. Introduction
The Earth's rotation results in a 24-h cycle of light and darkness, and of temperature. In order to adapt to this kind of periodic environmental change, several physiological processes in organisms also exhibit a periodic fluctuation of ~24 h, known as a circadian rhythm [1]. Circadian rhythms are the result of long-term evolution of organisms that has helped them adapt to environmental changes and to regulate physiological activities [2]. Circadian rhythms are driven by endogenous timing systems, known as circadian clocks, which can be found in almost every living organism on earth, from simple single-cellular cyanobacteria to mammals and humans [3]. Circadian clocks consist of the central and peripheral clocks [4]. The central clock is located in the hypothalamic suprachiasmatic nucleus (SCN). As a central pacemaker, SCN receives, converts and integrates the light-dark cycle signal from the retina-hypothalamus pathway and issues synchronization instructions that, in addition to regulating its own tissue rhythm, synchronize peripheral clocks to regulate the rhythm of peripheral effectors via endocrine hormones and autonomic nervous systems pathways [5]. The peripheral clock regulated by the rhythmic oscillations of the central clock is present in almost the entirety of peripheral tissues and organs, including the liver, heart, lung and kidney. As a result, many important physiological factors, such as blood pressure, body temperature, metabolism and immunity, exhibit robust circadian oscillations and rhythmic variations [3]. Mammalian peripheral clocks with autonomic rhythms can be independent of the central clock [6,7]. Circadian rhythms in organisms enable various complex life activities to be carried out in an orderly and synergistic manner, and play a crucial role in their regulation [2,3]. The disruption of circadian rhythms can lead to a number of diseases, including cancer. Women with long-term night shifts reportedly exhibit a significantly higher incidence of breast cancer than those with normal working habits [8], while men on night shifts have been associated with a significantly higher incidence of prostate cancer [9]. Of note, Junghynes et al found that resynchronizing a dysregulated circadian rhythm using melatonin can reduce the incidence of tumors [10]. Recently, Mocellin S and Benna C et al reported that genetic variation of clock genes was significantly associated with the predisposition to various types of cancer [11,12]. These results indicate that further study on the regulatory mechanism of circadian rhythm disruption in tumorigenesis may lead to new ideas and methods for the prevention and treatment of cancer.
2. Molecular mechanisms of circadian rhythms
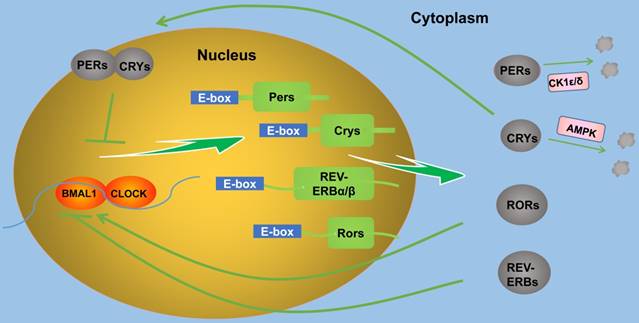
At the molecular level, the mechanism of circadian rhythms produced by the central and peripheral circadian clocks are practically the same: A series of circadian clock genes and their protein products form several positive and negative feedback loops at the transcription and translation levels and generate circadian oscillations [3,7]. Core clock genes reported to date include Period (Per)1/2/3, circadian locomotor output cycles kaput (Clock), Brain and muscle Arnt-like protein 1 (Bmal1), cryptochrome (Cry) 1/2, timeless (Tim), casein kinase 1 (CK1) ε/δ, neuronal PAS domain protein 2 (NPAS2), nuclear receptor subfamilies (REV-ERBs), retinoid-related orphan receptors (RORs), and differentiated embryo-chondrocyte expressed gene (Dec) 1/2 [1,13]. These clock genes are located in the SCN and almost the entirety of peripheral tissue cells [1,4,14]. In mammals, rhythm generation is mainly regulated by two interactive transcription-translation feedback loops (Figure 1) [2]. In the first major loop, Bmal1 and Clock (which can be replaced by NPAS2) in the cytoplasm form Bmal1/Clock (NPAS2) heterodimers through their common PAS domain, and then enter the nucleus, bind and activate the transcription of Per1/2/3 and Cry1/2 genes via E-boxes in gene promoters, thus forming a positive feedback loop. The translated Per and Cry proteins in the cytoplasm form hetero-multimeric complexes and then transfer to the nucleus and inactivate Bmal1/Clock (NPAS2) heterodimers, thus inhibiting the transcription of Per and Cry genes and forming a negative feedback loop [15]. In the above process, CK1ε/δ and adenosine monophosphate-activated protein kinase (AMPK) also accelerate or slow down the degradation of Per and Cry proteins to regulate the circadian rhythm length through phosphorylation [15,16]. In the second major loop, Bmal1/Clock heterodimers enter the nucleus and activate the transcription of REV-ERBs and RORs gene via E-boxes in gene promoters. ROR proteins act as a transcription factor and activate the transcription of Bmal1, thus forming a positive feedback loop, whereas REV-ERB proteins inhibit the transcription of Bmal1, thus forming a negative feedback loop [17]. There are other ways to regulate circadian rhythms. Sirtuin1 can bind to Bmal1/Clock heterodimers and promote Per2 protein deacetylation and degradation [18,19]. Dec1/2 inhibit their own expression by competitively binding to Bmal1/Clock heterodimers [20].
The molecular network of circadian clock produced by the interactive transcription-translation feedback loops (TTFL).
Period Genes and Cancer
| Genetic changes | Possible mechanism | Cancer phenotype | Reference |
|---|---|---|---|
| Per1 overexpression | Up-regulated Wee1, CRE-BP1, CDK1 and GADD45A | Increased G0/G1 phase, inhibites cholangiocarcinoma cells growth | 42 |
| Interacts with androgen receptor | Growth inhibition and apoptosis in prostate cancer | 29 | |
| Dependence of p53, downregulation of cyclin B1 and Cdc2 | Growth inhibition of human cancer cells | 38 | |
| Per1 silence | Interacts with cell cycle checkpoint proteins ATM and Chk2 | Increased proliferation | 38 |
| Per1 knockdown | Disruptes Cyclin-CDK-CKI network, affects clock gene network, increased Ki-67,MDM2, Bcl-2, MMP2 and MMP9, decreased Bax | Increases proliferation and decreases apoptosis in oral cancer cells | 13,43, 46 |
| Per2 overexpression | Up-regulated p53, down-regulated c-Myc and CyclinB1 | Increases cell apoptosis in K562 leukemia cells | 44 |
| Increased G0/G1 phase | Promotes cells apoptosis in MG63 osteosarcoma cells | 45 | |
| Down-regulated PI3K and PKB | Inhibities ovarian cancer | 48 | |
| Inhibities OCT1-mediated EMT genes | Reduces the invasion ability of breast cancer cells | 35 | |
| Per2 knockdown | Increased β-catenin and CyclinD | Promotes the proliferation of colon cancer cells | 49 |
| Increased phosphorylation of AKT and mTOR | Decreases cell apoptosis in A549/cisplatin lung adenocarcinoma cells | 47 | |
| Activates MDM2-p53 pathway | Regulates apoptosis in glioma cells | 50 | |
| Per2S662G mutation | Affects cell cycle | Increased cancer risk | 63 |
| Per3 overexpression | Increased p53, CyclinB1, CDC2, Bid and cleaved-caspase3/8, decreased Bcl2 | Inhibition of cell proliferation and metastasis in colorectal cancer | 51 |
On the one hand, clock genes generate their own circadian rhythm, on the other hand, they could regulate the expression of several other important genes in the genomes. Current studies have shown that clock genes can regulate ~43% of protein-coding genes in the genomes of mammals, suggesting that multiple important and complex physiological functions in the body can be regulated in a highly orderly way through the rhythmic expression of clock genes [21]. The abnormal expression of clock genes and disruption of circadian rhythms can markedly affect the development of several diseases, including cancer, cardiovascular disease, diabetes and depression [8,9,22,23].
3. Per gene expression variations in tumorigenesis
The Per gene family, which consists of the Per1/2/3 genes, is the core member of the circadian clock genes. Alterations in Per gene expression have been shown to be associated with the occurrence, development and prognosis of cancer (Table 1) [13,14]. Per1/2 expression levels have been found significantly downregulated in gastric, colorectal, pancreatic, prostate and breast cancer, head and neck squamous cell carcinoma (HNSCC), non-small cell lung cancer (NSCLC), chronic lymphocytic leukemia, melanoma and hepatocellular carcinoma [24-33]. Per3 expression has been found to be significantly downregulated in colorectal cancer [25], HNSCC [26], NSCLC [27], pancreatic cancer [28] and hepatocellular carcinoma [32].
Zhao et al reported that Per1 expression was negatively correlated with clinical stage and pathological differentiation in gastric cancer; moreover, low Per2 expression was found to be significantly correlated with clinical stage and invasion depth; the survival time of patients exhibiting low Per1/2 expression was shorter than that of patients exhibiting high expression [24]. Xiong et al showed that low Per2 expression was closely associated with clinical stage and cervical lymph node metastasis in oral SCC. Patients exhibiting low Per2 expression had a shorter survival time than those exhibiting high expression [34]. Hus et al reported that the low Per3 expression in HNSCC was closely associated with tumor size and invasion depth. In addition, the low expression of Per1/3 was found to be associated with a shorter survival time [26]. Liu et al verified that the decrease in Per1/2/3 expression in NSCLC was associated with poor differentiation, tumor size, high TNM stage and lymph node metastasis. In addition, patients exhibiting low Per1/2/3 expression had a shorter survival time than those exhibiting high Per1/2/3 expression [27]. Hwang-verslues et al reported that Per2 expression was negatively correlated with tumor size, clinical stage and histological grade in breast cancer. Patients exhibiting low Per2 expression had a worse survival rate [35]. Cadenas et al found that high Per1/2/3 expression in breast cancer was associated with histological grade and metastasis, and high Per2/3 expression with improved metastasis-free survival [36]. Tavano et al reported that patients with pancreatic cancer exhibiting high Per2 expression had a longer survival time than those exhibiting low expression [28]. Wang et al found that patients with colorectal cancer exhibiting high Per1/3 expression had a longer survival time than those exhibiting low expression [25]. However, Oshima et al demonstrated that high Per2 expression was associated with improved survival rates in colorectal cancer [37]. Recently, based on genome-wide association studies (GWASs), Mocellin S et al showed that genetic variation of Per1 and Per2 were significantly associated with the susceptibility to breast and lung cancer, respectively, while Per3 genetic variation was significantly associated with the susceptibility to prostate and lung cancer [11].
3.1. Role of Per genes in DNA damage response and cell cycle
Altered Per expression may be responsible for the development of tumors through the regulation of DNA damage response and cell cycle. Gery et al performed coimmunoprecipitation experiments and proved that Per1 could bind to phosphorylated checkpoint kinase 2 (Chk2). However, the phosphorylation of the Chk2 Thr-68 site was blocked following ionizing radiation in the Per1 siRNA-transfected HCT116 cells, suggesting that Per1 can regulate the critical DNA damage activation control point and thus participate in DNA damage response by binding to Chk2 [38]. Yang et al reported that the downregulation of Per2 in HCT116 colon cancer cells resulted in a higher level of phosphorylated Akt at the T308 site. Following the induction of DNA double-strand breaks by a low dose of doxorubicin in Per2 knockdown cells, the phosphorylation of Chk2 T68 was delayed, with the delay being dependent on Akt activity [39]. Per2 can also regulate the p53 signaling pathway in response to DNA damage [40]. Im et al showed that the DNA damage-induced Chk2 activation in Per3 knockout HeLa cells was abolished. Of note, the overexpression of Per3 stimulated the phosphorylation of Chk2 and activated the ATM-Chk2 pathway without DNA damage [41]. Therefore, Per expression can markedly influence DNA damage repair.
Per genes plays an important role in cell cycle regulation. Han et al reported that the overexpression of Per1 in cholangiocarcinoma cells significantly upregulated the expression of the cell cycle-associated molecules Wee1, CRE-BP1, CDK1 and GADD45A. In addition, the number of cells in the G0/G1 phase was shown to increase, whereas that in the G2/M phase to decrease [42]. Fu et al found that the expression of cyclin D1 and E were significantly increased following the silencing of the Per1 gene in SCC15 cells. Therefore, the ability to initiate transition through G1 phase was accelerated, and the transition from the G1 to the S phase was enhanced, resulting in the shortening of the cell cycle and the increase of cell proliferation [43]. Sun et al showed that the overexpression of Per2 in leukemia cells upregulated p53 and downregulated c-Myc and cyclin B1, resulting in cell cycle arrest in the G2/M phase, which inhibited cell proliferation and increased cell apoptosis [44]. Cheng et al found that the overexpression of Per2 in MG63 osteosarcoma cells increased the cell population in G0/G1 phase and promoted cell apoptosis [45]. The above results suggested that Per1/2 expression may be involved in tumor progression through the regulation of cell cycle-related genes.
3.2. Role of Per genes in cell proliferation, apoptosis and metastasis
Per genes have a significant effect on cell proliferation, apoptosis and metastasis-related genes, which can lead to the promotion of cancer occurrence and development. Li et al found the expression levels of Ki-67, MDM2, Bcl-2 and MMP2/9 increased and those of Bax decreased following Per1 knockdown in SCC15 cells, which led to a decreased apoptosis and increased cell proliferation [46]. The overexpression of Per1 has been shown to inhibit proliferation, epithelial-mesenchymal transition (EMT), angiogenesis and metastasis in cholangiocarcinoma cells [42]. Gery et al suggested that c-Myc and p53 downregulation following Per1 silencing results in cell proliferation [38]. Chen et al showed that Per2 knockdown increased the phosphorylation of Akt protein and mTOR and decreased cell apoptosis in A549/cisplatin lung adenocarcinoma cells [47]. Wang et al reported that Per2 overexpression downregulated PI3K and PKB, inhibiting tumor growth in nude mice xenografts of ovarian cancer [48]. Per2 acted as a transcriptional corepressor, which could recruit EZH2, SUZ12 and HDAC2 to the OCT1 binding sites of EMT gene (including TWIST1 and SLUG) promoters in order to inhibit the expression of these genes, thus reducing the invasion ability of tumor cells [35]. Wood et al have demonstrated that the downregulation of Per2 increases β-catenin and cyclin D, thus promoting the proliferation of colon cancer cells [49]. Per2 downregulation decreases the activity of Bmal1/Clock heterodimers through a negative feedback loop, induces the expression of oncogene c-Myc and deregulates cyclin D1, thus promoting cancer cell proliferation [14]. A recent study showed that Per2 knockdown in glioma cells led to a decrease in ATM and p53 and an increase in c-Myc and MDM2, suggesting that Per2 regulates apoptosis through the MDM2-p53 pathway [50]. Of note, Hong et al found that Per3 was downregulated in human colorectal cancer. The overexpression of Per3 increased the expression of p53, cyclin B1, CDC2, Bid and cleaved-caspases 3/8, and decreased the expression of Bcl-2, inhibiting cell proliferation and metastasis [51].
3.3. Role of Per genes in tumor cells metabolism
Per gene involvement in tumor development may be achieved through its altering of the metabolism of tumor cells [52]. Papagiannakopoulos et al showed that, compared with Per2+/+ mouse lung cancer cells, Per2 mutant cells significantly increased the glucose and glutamine consumption and lactate excretion level, indicating that Per2 mutations may lead to changes in cancer cell glucose metabolism [53]. Per genes may affect tumor progression through hormone level regulation. In breast cancer cells, Per2 inhibited the transcriptional activation of estrogen receptor α (ERα). Furthermore, Per2 overexpression inhibited cell growth through the degradation of ERα [54]. Kuo et al proved that Per1 promoter methylation was negatively correlated with estrogen receptor positivity in breast cancer, suggesting that the methylation of the Per1 gene may affect estrogen receptor expression [55]. Cao et al showed that Per1 inhibited the transcriptional activation of androgen receptor (AR), and the activated AR promoted the transcription of Per1 through binding to androgen response elements of the Per1 promoter, thus forming a feedback loop. This feedback loop may play an important role in the development of prostate cancer [29].
3.4. Role of Per genes in tumor immune and inflammatory responses
Immune function and inflammatory reaction balance in tumor cells may be disrupted by altered Per gene expression. Arjona et al reported that the RNAi-mediated Per2 knockdown in the natural killer (NK) cell line RNK16 decreased the expression of granzyme B and perforin, impairing the function of NK cells [56]. Miyazaki et al showed that the cytotoxicity of CLT/NK cells to sarcoma cells was enhanced following the overexpression of Per2 that caused tumor growth inhibition [57]. Of note, Mteyrek et al demonstrated that the inflammatory mediators IL-6 and TNF-α in the Per2m/m group were significantly higher than those in the wild type (WT) group in the liver cancer initiation phase, which further accelerated the process of liver carcinogenesis [58].
4. Per gene rhythm variations in tumorigenesis
Per gene expression exhibits a periodic rhythm of ~24 h in mammalian cells. Changes in the Per expression and rhythm are closely associated with the occurrence and progression of cancer. Lee et al showed that mice displayed early senescence and increased neoplastic and hyperplastic phenotypes following the loss of the Per1/2 circadian rhythm [59]. Zhanfeng et al showed that the expression of Per1/2 in gliomas still exhibited a periodic rhythm, but the period was shortened to ~12 h [60]. Ye H and Tan XM et al reported that the acrophase of Per1/2 mRNA in the precancerous lesion stage occurred 9.66 and 2.94 h earlier than that of normal buccal mucosa in golden hamsters, respectively. The mesor and amplitude of Per1/2 mRNA in the cancer stage were significantly lower than those in the precancerous lesion stage [61,62]. In cholangiocarcinoma cells, the Per1 expression level was markedly decreased and the daily rhythmicity in the Per1 expression was lost. Further studies showed that miR-34a binding to Per1 3'UTR led to a decrease in the Per1 expression and a circadian rhythm disorder in cholangiocarcinoma cells; however, the inhibition of miR-34a can restore the Per1 24-h circadian rhythm, thus reducing cell proliferation and infiltration [42]. Compared to the WT in mouse livers treated with diethylnitrosamine, the circadian rhythm phase of Bmal1, Clock, Cry1/2 and REV-ERBα in Per2m/m was advanced, and that the circadian amplitudes of these Clock genes were decreased, apart from that of Cry2. In addition, the rhythmic expression of ATM, Wee1 and Ccnb1 was lost, the 24-h mean mRNA expression of c-Myc was increased and the p53 rhythmic expression was deregulated in Per2m/m. These events further promoted the initiation and progression of liver tumors [58]. Gu et al reported that the acrophase and valley phase of p21 and cyclin D in Per2 mutant mice changed compared with the WT, which may be the potential carcinogenic mechanism of mice [63]. The above studies suggested that the rhythmic variations in Per expression are associated with tumor progression, but the specific mechanism needs to be explored in depth.
5. Conclusion and perspective
The circadian clock exists in the majority of living organisms, including human beings, and helps them adapt to the circadian variations of the earth's environment. The circadian clock is a result of the long-term evolution of these organisms and it plays a key role in regulating life activities. The Per gene family is a key clock gene family, which has a major involvement in circadian rhythm maintenance and regulation, cell proliferation, apoptosis, migration, metabolism, cell cycle, and immune and inflammatory responses in cancer. The abnormal expression and rhythm of Per genes is closely associated with the occurrence and development of cancer, but the underlying mechanism remains unclear. The main issues that need to be explored in depth include: i) The specific pathways through which the Per gene family regulates cell proliferation, apoptosis, metabolism and cell cycle in cancer; ii) The specific mechanisms that cause changes in Per gene expression and rhythm in cancer; iii) The way in which the disruption of the circadian rhythm of Per gene expression can lead to tumorigenesis. In conclusion, a further and in-depth exploration of the association between Per genes and tumorigenesis may provide new strategies and effective targets for cancer prevention and treatment.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;8:3
2. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935-41
3. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517-49
4. Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919-22
5. Buijs RM, Hermes MH, Kalsbeek A. The suprachiasmatic nucleus-paraventricular nucleus interactions: a bridge to the neuroendocrine and autonomic nervous system. Prog Brain Res. 1998;119:365-82
6. Canaple L, Kakizawa T, Laudet V. The days and nights of cancer cells. Cancer Res. 2003;63:7545-52
7. Xiang S, Mao L, Duplessis T. et al. Oscillation of clock and clock controlled genes induced by serum shock in human breast epithelial and breast cancer cells: regulation by melatonin. Breast Cancer (Auckl). 2012;6:137-50
8. Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74-7
9. Kubo T, Ozasa K, Mikami K. et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549-55
10. Jung-Hynes B, Huang W, Reiter RJ. et al. Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J Pineal Res. 2010;49:60-8
11. Mocellin S, Tropea S, Benna C. et al. Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies. BMC Med. 2018;16:20
12. Benna C, Helfrich-Förster C, Rajendran S. et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget. 2017;8:23978-23995
13. Zhao Q, Zheng G, Yang K. et al. The clock gene PER1 plays an important role in regulating the clock gene network in human oral squamous cell carcinoma cells. Oncotarget. 2016;7:70290-70302
14. Fu L, Pelicano H, Liu J. et al. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41-50
15. Hurley JM, Loros JJ, Dunlap JC. Circadian Oscillators: Around the Transcription-Translation Feedback Loop and on to Output. Trends Biochem Sci. 2016;41:834-846
16. Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164-179
17. Preitner N, Damiola F, Lopez-Molina L. et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251-60
18. Nakahata Y, Kaluzova M, Grimaldi B. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329-40
19. Asher G, Gatfield D, Stratmann M. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317-28
20. Nakashima A, Kawamoto T, Honda KK. et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol. 2008;28:4080-92
21. Zhang R, Lahens NF, Ballance HI. et al. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111:16219-24
22. Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728-42
23. Pinho M, Sehmbi M, Cudney LE. et al. The association between biological rhythms, depression, and functioning in bipolar disorder: a large multi-center study. Acta Psychiatr Scand. 2016;133:102-108
24. Zhao H, Zeng ZL, Yang J. et al. Prognostic relevance of Period1 (Per1) and Period2 (Per2) expression in human gastric cancer. Int J Clin Exp Pathol. 2014;7:619-30
25. Wang Y, Cheng Y, Yu G. et al. Expression of PER, CRY, and TIM genes for the pathological features of colorectal cancer patients. Onco Targets Ther. 2016;9:1997-2005
26. Hsu CM, Lin SF, Lu CT. et al. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149-55
27. Liu B, Xu K, Jiang Y. et al. Aberrant expression of Per1, Per2 and Per3 and their prognostic relevance in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:7863-71
28. Tavano F, Pazienza V, Fontana A. et al. SIRT1 and circadian gene expression in pancreatic ductal adenocarcinoma: Effect of starvation. Chronobiol Int. 2015;32:497-512
29. Cao Q, Gery S, Dashti A. et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69:7619-25
30. Rana S, Munawar M, Shahid A. et al. Deregulated expression of circadian clock and clock-controlled cell cycle genes in chronic lymphocytic leukemia. Mol Biol Rep. 2014;41:95-103
31. Lengyel Z, Lovig Cs, Kommedal S. et al. Altered expression patterns of clock gene mRNAs and clock proteins in human skin tumors. Tumour Biol. 2013;34:811-9
32. Lin YM, Chang JH, Yeh KT. et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925-33
33. Winter SL, Bosnoyan-Collins L, Pinnaduwage D. et al. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797-800
34. Xiong H, Yang Y, Yang K. et al. Loss of the clock gene PER2 is associated with cancer development and altered expression of important tumor-related genes in oral cancer. Int J Oncol. 2018;52:279-287
35. Hwang-Verslues WW, Chang PH, Jeng YM. et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci U S A. 2013;110:12331-6
36. Cadenas C, van de Sandt L, Edlund K. et al. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle. 2014;13:3282-91
37. Oshima T, Takenoshita S, Akaike M. et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25:1439-46
38. Gery S, Komatsu N, Baldjyan L. et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375-82
39. Yang X, He X, Yang Z. et al. Mammalian PER2 regulates AKT activation and DNA damage response. Biochem Cell Biol. 2012;90:675-82
40. Gotoh T, Vila-Caballer M, Liu J. et al. Association of the circadian factor Period 2 to p53 influences p53's function in DNA-damage signaling. Mol Biol Cell. 2015;26:359-72
41. Im JS, Jung BH, Kim SE. et al. Per3, a circadian gene, is required for Chk2 activation in human cells. FEBS Lett. 2010;584:4731-4
42. Han Y, Meng F, Venter J. et al. MiR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J Hepatol. 2016;64:1295-304
43. Fu XJ, Li HX, Yang K. et al. The important tumor suppressor role of PER1 in regulating the cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma cells. Onco Targets Ther. 2016;9:2237-45
44. Sun CM, Huang SF, Zeng JM. et al. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res. 2010;16:403-11
45. Cheng AY, Zhang Y, Mei HJ. et al. Construction of a plasmid for overexpression of human circadian gene period2 and its biological activity in osteosarcoma cells. Tumour Biol. 2015;36:3735-43
46. Li HX, Fu XJ, Yang K. et al. The clock gene PER1 suppresses expression of tumor-related genes in human oral squamous cell carcinoma. Oncotarget. 2016;7:20574-83
47. Chen B, Tan Y, Liang Y. et al. Per2 participates in AKT-mediated drug resistance in A549/DDP lung adenocarcinoma cells. Oncol Lett. 2017;13:423-428
48. Wang Z, Li L, Wang Y. Effects of Per2 overexpression on growth inhibition and metastasis, and on MTA1, nm23-H1 and the autophagy-associated PI3K/PKB signaling pathway in nude mice xenograft models of ovarian cancer. Mol Med Rep. 2016;13:4561-8
49. Wood PA, Yang X, Taber A. et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786-93
50. Zhanfeng N, Chengquan W, Hechun X. et al. Period2 downregulation inhibits glioma cell apoptosis by activating the MDM2-TP53 pathway. Oncotarget. 2016;7:27350-62
51. Hong Z, Feng Z, Sai Z. et al. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014;47:500-5
52. Altman BJ. Cancer Clocks Out for Lunch: Disruption of Circadian Rhythm and Metabolic Oscillation in Cancer. Front Cell Dev Biol. 2016;4:62
53. Papagiannakopoulos T, Bauer MR, Davidson SM. et al. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016;24:324-31
54. Gery S, Virk RK, Chumakov K. et al. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916-20
55. Kuo SJ, Chen ST, Yeh KT. et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454:467-74
56. Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469-76
57. Miyazaki K, Wakabayashi M, Hara Y. et al. Tumor growth suppression in vivo by overexpression of the circadian component, PER2. Genes to Cells. 2010;15:351-8
58. Mteyrek A, Filipski E, Guettier C. et al. Clock gene Per2 as a controller of liver carcinogenesis. Oncotarget. 2016;7:85832-85847
59. Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525-30
60. Zhanfeng N, Yanhui L, Zhou F. et al. Circadian genes Per1 and Per2 increase radiosensitivity of glioma in vivo. Oncotarget. 2015;6:9951-8
61. Ye H, Yang K, Tan XM. et al. Daily rhythm variations of the clock gene PER1 and cancer-related genes during various stages of carcinogenesis in a golden hamster model of buccal mucosa carcinoma. Onco Targets Ther. 2015;8:1419-26
62. Tan XM, Ye H, Yang K. et al. Circadian variations of clock gene Per2 and cell cycle genes in different stages of carcinogenesis in golden hamster buccal mucosa. Sci Rep. 2015;5:9997
63. Gu X, Xing L, Shi G. et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012;19:397-405
Author contact
![]() Corresponding author: Kai Yang. Tel/Fax: +86 23 89012569, E-mail: cqfyykcom
Corresponding author: Kai Yang. Tel/Fax: +86 23 89012569, E-mail: cqfyykcom

 Global reach, higher impact
Global reach, higher impact