3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1138-1144. doi:10.7150/jca.29086 This issue Cite
Research Paper
Relationship of common variants in Interleukin 33 gene with susceptibility and prognosis of osteosarcoma in Han Chinese population
1. Department of Oncology, Shaanxi Traditional Chinese Medicine Hospital, Xi'an, China
2. Department of Hand & Foot Microsurgery, the Second Affiliated Hospital of Yanan University, Yulin, China
3. The First Department of Oncology, Affiliated Hospital of Shaanxi University of Traditional Chinese Medicine, Xianyang, China
4. Department of Joint Surgery, Honghui Hospital, Xi'an Jiaotong University, Xi'an, China.
Received 2018-8-8; Accepted 2019-1-4; Published 2019-1-29
Abstract
Osteosarcoma (OS) is one of the most common malignant bone tumors. Many previous studies have indicated that OS is a complex disease and that its development may be affected by multiple genetic factors, which may contribute to its carcinogenesis. The aim of the present study was to evaluate the relationship of IL-33 with susceptibility and prognosis of OS in Han Chinese individuals. A total of 1,605 study subjects including 507 OS patients and 1,098 controls were recruited. Eighteen SNPs mapped to IL-33 were selected for genotyping. Genetic associations between selected SNPs and OS disease status were evaluated. Survival analyses, including Kaplan-Meier analysis and Cox model fitting for significant SNPs, were performed. The functional consequences of significant SNPs were analyzed using a publicly available database. SNP rs1048274 was identified to be significantly associated with OS disease status (OR=0.75, P=1.53×10-4). Compared to the GA and GG groups, OS patients with the AA genotype of rs1048274 had better survival rate. The hazard ratio of SNP rs1048274 (AA group compared to GG+GA group) was 0.35 (95% confidence interval of 0.25-0.5) following adjustment for several clinical variables. In conclusion, our results suggested that IL-33 may play a key role in the etiology of OS, indicating IL-33 as a potential genetic risk factor of the development and prognosis of OS.
Keywords: Osteosarcoma susceptibility, IL-33, Common variants, Prognosis, Case-control studies
Introduction
Osteosarcoma (OS) is one of the most common malignant bone tumors and occurs predominantly in children and adolescents 1. Because of its highly malignant and invasive nature, the metastasis of OS often occurs early in patients and, thus, the prognosis is poor 2. Although some patients benefit from surgery and chemotherapy, the 5-year survival rate of OS is still lower than 50% 3. Therefore, it is an urgent problem to clarify the pathology and the molecular mechanism of OS. Previous research have indicated that the process of OS development is complicated and may be influenced by multiple factors 4-5. Only a portion of people who are exposed to the same environmental factors will develop OS, suggesting that genetic factors may contribute to the carcinogenesis of OS. Therefore, it is necessary to find the genes responsible for susceptibility to OS.
Interleukin-33 (IL-33) was found to be predominantly expressed in the nuclei of stromal cells and activated myeloid cells. Previous studies have confirmed that IL-33 plays an important regulatory role in many physiological and pathological conditions, such as tissue repair, autoimmune diseases and cancer, and it is also considered to be a damage-associated molecular pattern (DAMP) molecule 6. IL-33 is a cytokine member of the IL-1 family, which includes IL-18 and IL-1 7. Previous studies have shown that high levels of both IL-1α and IL-18 have a positive effect on suppressing the metastasis of OS 8-9. Additionally, it has been found that the expression of IL-33 in tumor cells can enhance immunogenicity and also promote type 1 antitumor immune responses via CD8+ T cells and NK cells 10-11. Studies have shown that many cancers can cause an increase in serum IL-33 levels, such as gastric cancer, hepatocellular carcinoma, and lung cancer 12-15. Moreover, IL-33 is an inhibitor of bone reabsorption that blocks osteoclastic activity in in vitro experiments 16; in vivo, researchers have also shown that IL-33 can have anti-osteoclastogenic effects, reducing osteoclast formation and activity by inducing their apoptosis in mice 17. Taken together, these studies suggest that IL-33 may be important to the occurrence and development of OS.
A recent genome-wide association study (GWAS) involving 632 OS patients, including 523 European patients and 109 from Brazilian patients, produced highly significant association of GLDC/IL33 locus at 9p24.1 with the risk of OS. Using publicly available data, Koster et al. found that the OS susceptible single nucleotide polymorphism (SNP) in GLDC/IL33 locus associated with low expression of IL-33, which was also significantly correlated with poor survival in OS patients 18. This result suggested that IL-33 might play some roles in the pathogenesis of OS. However, to date, only one report correlating IL-33 with OS is available. Importantly, the study only evaluated OS patients of European and Brazilian descent. Given that different ethnic populations may exhibit genetic heterogeneity of OS, there is a need to confirm these findings with additional samples from different populations. Therefore, in the present study, we aimed to determine whether IL-33 was associated with susceptibility to OS in Han Chinese population.
Methods
Study subjects
A total of 1,605 study subjects including 1,098 healthy controls and 507 OS patients were recruited from 3 clinical centers located in Xi'an (Shaanxi Traditional Chinese Medicine Hospital, Affiliated Hospital of Shaanxi University of Traditional Chinese Medicine and Honghui Hospital of Xi'an Jiaotong University). All patients were diagnosed with OS based on histological and pathological evidence by at least two experienced pathologists. All healthy controls were identified by health screening and recruited from the same hospitals mentioned above. All subjects were Han Chinese individuals from the same geographic region of Xi'an, China. This study has been approved by the Medical Ethics Committee of Xi'an Jiaotong University. Informed consent forms were obtained from all participants. The characteristic information for all study subjects were summarized in Table 1. No differences were identified for age, gender or family history between OS case and control groups.
Distributions of selected variables in all subjects
| Variables | Subjects (N=1605) | Statistics | P-value | |
|---|---|---|---|---|
| OS (N=507) | Control (N=1098) | |||
| Mean age, years (%) | 21.08±6.1 | 21.12±5.8 | T=-0.12 | 0.90 |
| ≤20 | 318 (62.7) | 700 (63.8) | ||
| >20 | 189 (37.3) | 398 (36.2) | χ2=0.12 | 0.73 |
| Gender (%) | ||||
| Male | 261 (51.5) | 563 (51.3) | ||
| Female | 246 (48.5) | 535 (48.7) | χ2= 0.0005 | 0.98 |
| Cancer family history (%) | ||||
| Yes | 158 (31.2) | 333 (30.3) | ||
| No | 349 (68.8) | 765 (69.7) | χ2= 0.08 | 0.78 |
| Tumor location (%) | ||||
| Long tubular bones | 408 (80.5) | - | ||
| Axial skeleton | 99 (19.5) | - | - | - |
| Pathological fracture (%) | ||||
| Yes | 87 (17.2) | - | ||
| No | 420 (82.8) | - | - | - |
| Metastasis (%) | ||||
| Yes | 112 (22.1) | - | ||
| No | 395 (77.9) | - | - | - |
| Enneking stages (%) | ||||
| I | 47 (9.3) | - | ||
| II | 391 (77.1) | - | ||
| III | 69 (13.6) | - | - | - |
SNP selection and Genotyping
SNPs in the gene region of the IL-33 with minor allele frequencies (MAF) ≥ 0.01 based on 1000 genome data were extracted. Then, MAF ≥ 0.01 by pair-wise tagging and r2 ≥ 0.8 were used as selection criteria for tag SNPs, and 18 tag SNPs were generated for genotyping. The information regarding the 18 tag SNPs is summarized in Supplemental Table S1. Genomic DNA was isolated from the peripheral blood using a Tiangen DNA extraction kit (Tiangen Biotech Co. Ltd, Beijing, China). Sequenom MassARRAY platform was utilized for SNP genotyping. Typer 4.0 was used for genotyping results processing. We randomly re-performed the analysis on 5% of the samples and found a concordance of 100%. The MAFs of these SNPs in our study subjects ranged from 0.039-0.484, and all of these SNPs were in Hardy-Weinberg equilibrium (HWE) in our controls (Supplemental Table S1). Power analyses were performed using GAS power calculator, and the results were showed in Ssupplemental Figure S1. As seen from this figure, 500 OS cases could achieve statistical power of 78.7% to detect a SNP with relative risk equal to 1.4 at α=0.05 level.
Statistics and bioinformatics analysis
HWE was tested for all SNPs by Haploview v4.2 19. Differences of multiple clinical and characteristic variables between cases and controls were examined by Student's t and χ2 tests. Genetic associations between selected SNPs and OS risk were tested using Plink 20. The linkage disequilibrium (LD) structure was plotted using Haploview v4.2 19. Survival analyses, including Kaplan-Meier analysis and Cox model fitting for significant SNPs, were performed using R project (using “survival” and “survminer” packages) 21. Bonferroni corrections were applied if necessary to address the problem of multiple comparisons. To examine the functional consequences of the significant SNP, we performed bioinformatics analyses using the GTEx 22 and the PolymiRTS Database 23. The eQTL effects of significant SNPs on gene IL-33 were investigated using gene expression data from multiple human tissues that were extracted from GTEx. The effects of an SNP on miRNA-mediated gene expression were evaluated using the PolymiRTS Database.
Results of single marker based association analyses
| CHR | SNP | POS | A1 | F_A | F_U | χ2 | P | OR |
|---|---|---|---|---|---|---|---|---|
| 9 | rs1157505 | 6216240 | G | 0.265 | 0.262 | 0.043 | 0.84 | 1.02 |
| 9 | rs11791561 | 6216516 | C | 0.449 | 0.453 | 0.054 | 0.82 | 0.98 |
| 9 | rs72614080 | 6218595 | T | 0.274 | 0.279 | 0.071 | 0.79 | 0.98 |
| 9 | rs1891385 | 6219845 | C | 0.223 | 0.226 | 0.035 | 0.85 | 0.98 |
| 9 | rs143589217 | 6220187 | A | 0.048 | 0.047 | 0.031 | 0.86 | 1.03 |
| 9 | rs16924144 | 6221246 | C | 0.314 | 0.317 | 0.036 | 0.85 | 0.98 |
| 9 | rs183791076 | 6222545 | G | 0.052 | 0.053 | 0.004 | 0.95 | 0.99 |
| 9 | rs10975495 | 6223819 | A | 0.057 | 0.055 | 0.086 | 0.77 | 1.05 |
| 9 | rs1418385 | 6226295 | T | 0.476 | 0.472 | 0.058 | 0.81 | 1.02 |
| 9 | rs16924159 | 6229417 | A | 0.298 | 0.293 | 0.084 | 0.77 | 1.02 |
| 9 | rs189217453 | 6230731 | T | 0.050 | 0.048 | 0.092 | 0.76 | 1.06 |
| 9 | rs182211245 | 6230736 | C | 0.050 | 0.051 | 0.019 | 0.89 | 0.98 |
| 9 | rs117414011 | 6242939 | C | 0.071 | 0.076 | 0.256 | 0.61 | 0.93 |
| 9 | rs4742170 | 6242950 | C | 0.487 | 0.482 | 0.068 | 0.79 | 1.02 |
| 9 | rs79981454 | 6247232 | C | 0.042 | 0.038 | 0.391 | 0.53 | 1.13 |
| 9 | rs76864631 | 6248408 | A | 0.248 | 0.242 | 0.124 | 0.73 | 1.03 |
| 9 | rs10758751 | 6252044 | C | 0.074 | 0.077 | 0.064 | 0.80 | 0.96 |
| 9 | rs1048274 | 6256292 | A | 0.414 | 0.486 | 14.340 | 1.53×10-4 | 0.75 |
POS: position; A1: tested allele; F_A: allele frequency in cases; F_U: allele frequency in controls. Threshold of P value was 0.0028 (0.05/18). Significant results were highlighted in bold.
Results
Single marker based association analyses
SNP rs1048274, which is located at 3' untranslated region of IL-33, was identified to be significantly associated with the disease status of OS (Table 2), and its significance was retained after Bonferroni correction (Table 2). Our results showed that the minor allele (A) of this SNP had significant protective effect against OS (OR=0.75, P=1.53×10-4). The LD structure of our 18 selected tag SNPs is shown in Supplemental Figure S2, but there was no LD block which included the associated SNP of rs1048274. Furthermore, we examined the distribution of multiple clinical variables in the OS groups with different genotypes of rs1048274 (Table 3). None of those variables were identified as significantly associated with any of the genotypes of rs1048274 within OS patients.
Association between genotypes of rs1048274 and several clinical variables within our OS subjects
| Genotypes of rs1048274 (N=507) | Statistics | P | |||
|---|---|---|---|---|---|
| AA (N=83) | AG (N=254) | GG (N=170) | |||
| Age in years, mean±sd | 20.6±5.8 | 21.4±6.2 | 20.8±6.0 | F=0.03 | 0.87 |
| Gender (%) | |||||
| Male (N=261) | 43 (16.5) | 130 (49.8) | 88 (33.7) | ||
| Female (N=246) | 40 (16.3) | 124 (50.4) | 82 (33.3) | χ2=0.02 | 0.99 |
| Tumor location (%) | |||||
| Long tubular bones (N=408) | 62 (15.2) | 209 (51.2) | 137 (33.6) | ||
| Axial skeleton (N=99) | 21 (21.2) | 45 (45.5) | 33 (33.3) | χ2=2.29 | 0.32 |
| Pathological fracture (%) | |||||
| Yes (N=87) | 13 (14.9) | 47 (54.0) | 27 (31.1) | ||
| No (N=420) | 70 (16.7) | 207 (49.3) | 143 (34.0) | χ2=0.65 | 0.72 |
| Metastasis (%) | |||||
| Yes (N=112) | 18 (16.1) | 55 (49.1) | 39 (34.8) | ||
| No (N=395) | 65 (16.5) | 199 (50.4) | 131 (33.1) | χ2=0.11 | 0.95 |
| Enneking stages (%) | |||||
| I (N=47) | 11 (23.4) | 23 (48.9) | 13 (27.7) | ||
| II (N=391) | 58 (14.8) | 199 (50.9) | 134 (34.3) | ||
| III (N=69) | 14 (20.3) | 32 (46.4) | 23 (33.3) | χ2=3.43 | 0.49 |
Survival analyses
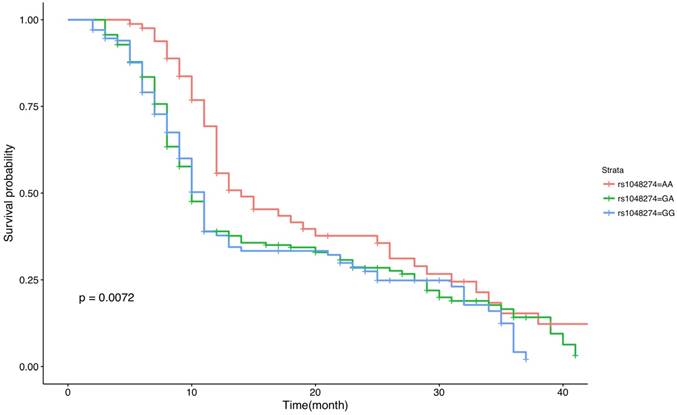
An overall survival plot for all of our OS patients stratified by genotypes of rs1048274 (AA, GA and GG groups) was made, and Kaplan-Meier analysis was conducted (Figure 1). Significant differences of survival curves were obtained (P=0.0072) between the three genotype groups. Compared to the GA and GG groups, OS patients with the AA genotype of rs1048274 had better survival rate. Next, we performed Kaplan-Meier analysis stratified by Enneking stages (Supplemental Figure S3-S5) and gender (Supplemental Figure S6 and S7). The patterns of the survival curves for each stratification variable were similar to the overall survival curves. For all survival curves, OS patients with AA genotype had better survival rate, although this statistical difference was not very significant when the curves were stratified by gender (P=0.025 for male and P=0.13 for female).
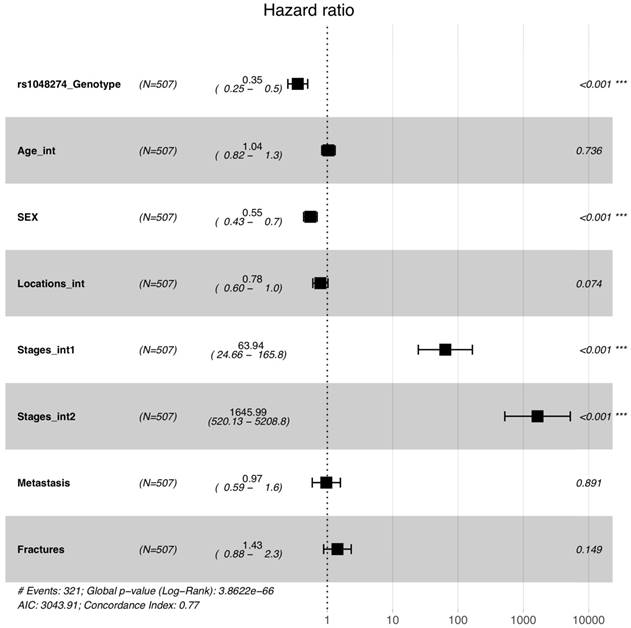
A Cox model adjusted with multiple clinical variables was fitted (Figure 2) to the curves. The coding scheme is shown in Supplemental Table S2. Since the survival curves of the GG and GA groups were very similar to each other (Figure 1), we combined the two groups in the Cox model. The adjusted hazard ratio of SNP rs1048274 (AA group compared to GG+GA group) was 0.35 (with 95% confidence interval of 0.25-0.5). The two other variables, gender and Enneking stages, were also significant in our Cox model.
Kaplan-Meier analysis and survival plot for the overall survival of OS patients with different genotypes of s1048274. P values for Kaplan-Meier analysis are indicated on the plot.
Forest plot for the survival analysis of rs1048274 using a Cox model after adjustment for multiple clinical variables.
Bioinformatics analyses
Gene expression data of IL-33 from 47 human tissues was extracted. The most significant eQTL finding for rs1048274 was identified in the esophagus (P=0.076 Supplemental Table S3). However, this hit failed to survive after Bonferroni correction. According to the genomic database from the University of California, Santa Cruz (http://genome.ucsc.edu/), rs1048274 acts as a 3' untranslated region (3' UTR) variant, which may affect microRNA binding. We used a free online tool (PolymiRTS Database) to examine the predicted target gain or loss in microRNA binding and found that the A allele of rs1048274 causes the loss of binding of microRNA/SNP target duplexes of two miRNAs (hsa-miR-543 and hsa-miR-570-3p) and IL-33. Comparatively, the G allele of rs1048274 maintains a stable microRNA-binding site at the 3' UTR region of IL-33.
Discussion
Previous GWAS have linked IL-33 to several disease states including asthma 24-25 and periodontal microbiota 26. GWAS using European and Brazilian samples conducted by Koster et al. have shown that IL-33 might contribute to susceptibility to the risk of OS. However, this evidence was weak because the significant SNP identified by this GWAS was located in the GLDC gene and was only linked with IL-33 based on publicly available data on gene expressions. In addition, study conducted by Koster et al. was based on European populations, and the genetic structure of OS related SNPs of IL33 was still unclear for Chinese population considering the differences in linkage disequilibrium structure between the two ethnic groups. To the best of our knowledge, the present study is the first one to identify the association of IL33 with the risk of OS in Han Chinese population. Our study replicates and builds on this previous work by using samples with Chinese Han ancestry. In this study, we have identified SNP rs1048274 to be significantly associated with the disease status of OS. The minor allele of this SNP, the A allele, could significantly decrease the risk of OS by 25%. This SNP, located at the 3' untranslated region of IL-33, was physically closer to IL-33 compared to both rs3765555 and rs55933544, which were identified in the previous GWAS. SNP rs1048274 was missed by this previous GWAS. A potential reason might be its difference in the MAF (0.43 for East Asians and 0.32 for Europeans according to the 1000 Genome Project) and the LD structure around it between Chinese and European populations. In addition to its contribution to OS susceptibility, SNP rs1048274 was also associated with prognosis for OS patients. According to our survival analysis, OS patients with the AA genotype could, in general, exhibit significantly better survival rates compared to those with either the GG or GA genotypes. This pattern was not associated with the Enneking stages of OS or gender. Similar to its effect on OS susceptibility, the A allele of SNP rs1048274 has hazard ratio of 0.35 after adjustment for multiple clinical variables.
In the present study, we have identified that the A allele of SNP rs1048274 significantly increases the survival rate of OS patients compared to the G allele. Our bioinformatics analyses have shown that rs1048274's ancestral G allele is a very conserved miRNA binding site which binds miRNA and inhibits gene expression of IL-33. The A allele is likely to disrupt this binding site and thus increase its expression. Early studies have shown that IL-33 could promote type I antitumor immune responses through CD8+ T cells and NK cells 10,11. In other words, the protein product of the gene IL-33 might be related to the inhibition of tumor cells, and therefore, a higher level of IL-33 expression might be protective against OS. Although a systematic investigation of the expression levels of IL-33 is out of the scope of this study, our findings could still provide some clues for the hypothesis stated above. Nevertheless, since the findings from our bioinformatics analyses were functional predictions based on computer algorithms, and no experimental data has been reported, we need to be careful when interpreting these findings.
With the rapid development of high-throughput sequencing technology, genetic association analysis provides an effective method for studying the molecular genetic mechanism of complex diseases, such as schizophrenia, but it is still unknown for us to understand the genetic risk and molecular mechanism of OS 27-36. Despite our interesting findings on the association between rs1048274 and the susceptibility and prognosis of OS, this study suffered from several limitations. A major issue is that population stratification might be a potential confounder for our association signal. As a candidate gene based study, we did not have enough SNPs to perform some conventional approaches (such as principal component analysis) to address this problem. Nevertheless, we have applied some criteria to restrict the genetic background of our study subjects to minimize their genetic heterogeneity. Thus, due to the subject individuals from the same geographical area, although the potential population stratification cannot be completely excluded, our sampling strategy can effectively avoid significant population stratification to the greatest extent 37-42. Another limitation is that this study is not a randomized clinical trial, thus selection bias might be present. OS patients and controls were not randomly selected individuals and our study subjects may not be a representative sample. To the best of our knowledge, the present study is the first one to identify an association signal for rs1048274 and OS susceptibility and its prognosis. Replication studies are needed in the future, especially those based on different ethnic populations.
In summary, we have showed strong evidence for association between SNP rs1048274 of IL-33 and susceptibility to OS in the Chinese Han population. This SNP was also identified as significantly associated with the prognosis of OS in further analyses of OS patients. This is the first large-scale population based genetic report on polymorphisms of IL-33 using samples with Chinese ancestry. Thus, replication studies based on other populations as well as further investigations of the underlying mechanism responsible for these associations are still needed.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by the Project of Natural Science Fundamental research of Shaanxi province (2018JM7048036). The funding sources had no role in the design of this study, the collection, analysis and interpretation of data, the writing of the report, or the decision to submit the paper for publication.
Authors' Contributions
Authors Kang C and Zhi L conceived and designed the study. Kang C and Zhao J carried out candidate SNPs selection and statistical analyses. Kang C, Zhao J, Wang Y, and Zhi L conducted subject screening. Zhao J, Yang C, and Chen J contributed to the collection and preparation of control DNA samples. Kang C wrote the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Messerschmitt PJ, Garcia RM, Abdul-Karim FW, Greenfield EM, Getty PJ. Osteosarcoma. The Journal of the American Academy of Orthopaedic Surgeons. 2009;17:515-27
2. Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clinical advances in hematology & oncology: H&O. 2010;8:705-18
3. Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert review of anticancer therapy. 2006;6:1075-85
4. Maximov VV, Aqeilan RI. Genetic factors conferring metastasis in osteosarcoma. Future oncology. 2016;12:1623-44
5. Yang M. Prognostic role of pathologic fracture in osteosarcoma: Evidence based on 1,677 subjects. Journal of cancer research and therapeutics. 2015;11:264-7
6. Lu B, Yang M, Wang Q. Interleukin-33 in tumorigenesis, tumor immune evasion, and cancer immunotherapy. Journal of molecular medicine. 2016;94:535-43
7. Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunological reviews. 2008;223:20-38
8. Jia SF, Zwelling LA, Mcwatters A, An T, Kleinerman ES. Interleukin-1 increases the cytotoxic activity of etoposide against human osteosarcoma cells . Journal of Experimental Therapeutics & Oncology. 2010;2:27-36
9. Nakamura Y, Yamada N, Ohyama H, Nakasho K, Nishizawa Y, Okamoto T. et al. Effect of interleukin-18 on metastasis of mouse osteosarcoma cells. Cancer Immunol Immun. 2006;55:1151-8
10. Gao X, Wang XF, Yang QT, Zhao X, Wen W, Li G. et al. Tumoral Expression of IL-33 Inhibits Tumor Growth and Modifies the Tumor Microenvironment through CD8(+) T and NK Cells. Journal of immunology. 2015;194:438-45
11. Gao K, Li XY, Zhang L, Bai L, Dong W, Gao K. et al. Transgenic expression of IL-33 activates CD8(+) T cells and NK cells and inhibits tumor growth and metastasis in mice. Cancer letters. 2013;335:463-71
12. Sun PH, Ben QW, Tu SP, Dong WJ, Qi XG, Wu YL. Serum Interleukin-33 Levels in Patients with Gastric Cancer. Digestive diseases and sciences. 2011;56:3596-601
13. Bergis D, Kassis V, Ranglack A, Koeberle V, Piiper A, Kronenberger B. et al. High Serum Levels of the Interleukin-33 Receptor Soluble ST2 as a Negative Prognostic Factor in Hepatocellular Carcinoma. Transl Oncol. 2013;6:311-8
14. Ishikawa K, Yagi-Nakanishi S, Nakanishi Y, Kondo S, Tsuji A, Endo K. et al. Expression of interleukin-33 is correlated with poor prognosis of patients with squamous cell carcinoma of the tongue. Auris Nasus Larynx. 2014;41:552-7
15. Hu LA, Fu Y, Zhang DN, Zhang J. Serum IL-33 as a diagnostic and prognostic marker in non- small cell lung cancer. Asian Pacific Journal of Cancer Prevention Apjcp. 2013;14:2563
16. Schulze J, Bickert T, Beil FT, Zaiss MM, Albers J, Wintges K. et al. Interleukin-33 Is Expressed in Differentiated Osteoblasts and Blocks Osteoclast Formation From Bone Marrow Precursor Cells. Journal of Bone and Mineral Research. 2011;26:704-17
17. Keller J, Catala-Lehnen P, Wintges K, Schulze J, Bickert T, Ito W. et al. Transgenic over-expression of interleukin-33 in osteoblasts results in decreased osteoclastogenesis. Biochemical and biophysical research communications. 2012;417:217-22
18. Koster R, Panagiotou OA, Wheeler WA, Karlins E, Gastier-Foster JM, Caminada de Toledo SR. et al. Genome-wide association study identifies the GLDC/IL33 locus associated with survival of osteosarcoma patients. Int J Cancer. 2018;142:1594-601
19. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
20. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7
21. R Core Team. R: a language and environment for statistical computing. 2015; 1: 12-21.
22. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S. et al. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
23. Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86-D91
24. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S. et al. A Large-Scale, Consortium-Based Genomewide Association Study of Asthma. New Engl J Med. 2010;363:1211-21
25. Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE. et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nature genetics. 2011;43:887-U103
26. Divaris K, Monda KL, North KE, Olshan AF, Lange EM, Moss K. et al. Genome-wide Association Study of Periodontal Pathogen Colonization. J Dent Res. 2012;91:S21-S8
27. Guan F, Zhang C, Wei S. et al. Association of PDE4B polymorphisms and schizophrenia in Northwestern Han Chinese. Hum Genet. 2012;131(7):1047-1056
28. Guan F, Zhang B, Yan T. et al. Mir137 gene and target gene cacna1c of mir-137 contribute to schizophrenia susceptibility in han chinese. Schizophrenia Research. 2014;152:97-104
29. Chen G, Guan F, Lin H. et al. Genetic analysis of common variants in the HDAC2 gene with schizophrenia susceptibility in Han Chinese. J Hum Genet. 2015;60(9):479-484
30. Guan F, Li L, Qiao C. et al. Evaluation of genetic susceptibility of common variants in CACNA1D with schizophrenia in Han Chinese. Sci Rep. 2015:10 5:12935
31. Zhang B, Guan F, Chen G. et al. Common variants in SLC1A2 and schizophrenia: Association and cognitive function in patients with schizophrenia and healthy individuals. Schizophrenia Research. 2015;169(1-3):128-134
32. Guan F, Zhang T, Liu X. et al. Evaluation of voltage-dependent calcium channel gamma gene families identified several novel potential susceptible genes to schizophrenia. Sci Rep. 2016;6:24914
33. Jia X, Zhang T, Li L. et al. Two-stage additional evidence support association of common variants in the HDAC3 with the increasing risk of schizophrenia susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2016;171:1105-11
34. Guan F, Wei S, Feng J. et al. Association study of a new schizophrenia susceptibility locus of 10q24.32-33 in a Han Chinese population. Schizophrenia Research. 2012;138(1):63-68
35. Guan F, Wei S, Zhang C. et al. A population-based association study of 2q32.3 and 8q21.3 loci with schizophrenia in Han Chinese. J Psychiatr Res. 2013;47(6):712-717
36. Yang H, Zhang B, Zhu J. et al. 4q22.1 Contributes to Bone Mineral Density and Osteoporosis Susceptibility in Postmenopausal Women of Chinese Han Population. PLoS One. 2013;8(11):e80165
37. Guan F, Niu Y, Zhang T. et al. Two-stage association study to identify the genetic susceptibility of a novel common variant of rs2075290 in ZPR1 to type 2 diabetes. Sci Rep. 2016;6:29586
38. Guan F, Zhang T, Li L. et al. Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population. Schizophrenia Research. 2016;176:125-30
39. Guan F, Lin H, Chen G. et al. Evaluation of association of common variants in HTR1A and HTR5A with schizophrenia and executive function. Sci Rep. 2016;6:38048
40. Zhang B, Xu Y, Wei S. et al. Association study identifying a new susceptibility gene (AUTS2) for schizophrenia. International journal of molecular sciences. 2014;15(11):19406-19416
41. Liu X, Hou Y, Yan T. et al. Dopamine D3 Receptor-Regulated NR2B Subunits of N-Methyl-d-Aspartate Receptors in the Nucleus Accumbens Involves in Morphine-Induced Locomotor Activity. CNS neuroscience & therapeutics. 2014;20(9):823-829
42. Zhang T, Zhu L, Ni T. et al. Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population. Journal of psychiatric research. 2018;106:99-105
Author contact
![]() Corresponding author: Liqiang Zhi, M.D. & Ph.D., Department of Joint Surgery, Honghui Hospital, Xi'an Jiaotong University Health Science Center, No.555, Youyi East Road, Xi'an, Shaanxi, China, 710054. Tel: 86-29-88418009; Fax: 86-29-62818386; E-mail: osteolqzhicom
Corresponding author: Liqiang Zhi, M.D. & Ph.D., Department of Joint Surgery, Honghui Hospital, Xi'an Jiaotong University Health Science Center, No.555, Youyi East Road, Xi'an, Shaanxi, China, 710054. Tel: 86-29-88418009; Fax: 86-29-62818386; E-mail: osteolqzhicom

 Global reach, higher impact
Global reach, higher impact