3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1333-1340. doi:10.7150/jca.29517 This issue Cite
Research Paper
Knockdown of long non-coding RNA SNHG5 inhibits malignant cellular phenotypes of glioma via Wnt/CTNNB1 signaling pathway
1. Department of Neurobiology, School of Life Science, China Medical University, Shenyang, Liaoning, China
2. Department of Neurosurgery, Shengjing Hospital, China Medical University, Shenyang, Liaoning, China
Received 2018-8-27; Accepted 2019-1-3; Published 2019-1-30
Abstract
Objective: Human brain glioma is the most malignant primary intracranial tumor, which has poor prognosis and high mortality. Long noncoding RNAs are considered to take part in cellular phenotypes and are emerging as diagnostic and prognostic biomarkers of glioma. This study will research the effects of Small Nucleolar RNA Host Gene 5 (SNHG5) gene on malignant cellular phenotypes in glioma and explore the possible mechanisms.
Materials and Methods: The expression level of SNHG5 was examined using quantitative Real-time PCR in glioma tissues and cell lines. Loss-of-function experiments of SNHG5 together with Enhanced Cell Counting Kit-8, flow cytometry and cell invasion assay were used to investigate the effects of SNHG5 on malignant cellular phenotypes of glioma cells. Finally, luciferase assay and western blotting were applied to determine the activity of WNT/CTNNB1 signaling pathway.
Results: SNHG5 gene was high-expressed in glioma tissues and cell lines. Knockdown of SNHG5 gene depressed cell proliferation and invasiveness as well as promoted the apoptosis of U251 and U87 cells. In addition, online database analysis showed SNHG5 was closely related to Wnt/CTNNB1 signaling pathway. Knockdown of SNHG5 inactivated Wnt/CTNNB1 signaling pathway, and the activating of Wnt/CTNNB1 signaling pathway partly restored the influences of SNHG5 knockdown on malignant cellular phenotypes of U251 and U87 cells.
Conclusion: SNHG5 gene was high-expressed in glioma, knockdown of SNHG5 inhibits malignant cellular phenotypes of glioma via Wnt/CTNNB1 signaling pathway.
Keywords: Long non-coding RNA, glioma, Wnt/CTNNB1 signaling pathway, Small Nucleolar RNA Host Gene 5, malignant cellular phenotypes
Introduction
Human brain glioma is the most malignant primary intracranial tumor, which has poor prognosis and high mortality [1,2]. Till now, surgical resection is the preferred therapeutic strategy for glioma. Despite adjuvant chemotherapy and radiation, glioma remains incurable, and the prognosis of patients remains poor due to recurrent gliomas rarely respond to the therapies [3]. Thus, it is important and necessary to elucidate its underlying mechanism of tumorigenesis as well as find novel targeted therapeutic strategy.
Long noncoding RNAs (lncRNAs) implicated in tumorigenesis and progression of numerous tumors through participating in various important cellular biological phenotypes, including proliferation, cell cycle, invasiveness and so on [4-8]. LncRNAs could act as oncogenes or tumor suppressors in glioma, and be diagnostic or prognostic biomarkers for glioma [9-11].
Small Nucleolar RNA Host Gene 5 (SNHG5, Gene ID: 387066) gene belongs to lncRNA gene because it lacks the ability to encode protein, SNHG5 located at chromosome 6q14.3 and consists of 6 exons. The researches about the expression level and functional roles of SNHG5 gene in malignant tumors are less. Of note, SNHG5 was high-expressed in bladder cancer and colorectal cancer, and acted as a potential oncogene [12,13]. However, Zhao L found that SNHG5 was low-expressed in gastric cancer and had close relationship with tumor stage and tumor emboli formation [14,15]. These findings suggested that SNHG5 may have different functions in different types of tumors. Till now, there is no research on the expression level and functional roles of SNHG5 in glioma.
It is well known that Wnt/CTNNB1 signaling pathway participates in the regulation of growth, development and metabolism, and plays important regulatory roles in almost all kinds of malignant tumors [16,17]. CTNNB1 gene (Gene ID: 1499), also named β-catenin, and glycogen synthase kinase 3 beta (GSK3B, Gene ID: 2932) gene are two pivotal effectors in Wnt/CTNNB1 signaling pathway. Accumulating evidences showed Wnt/CTNNB1 signaling pathway was activated frequently during tumorigenesis and progress of glioma [18,19]. Nevertheless, the interaction between Wnt/CTNNB1 signaling pathway and SNHG5 in glioma was poorly demonstrated.
This study will investigate the impacts of SNHG5 gene on malignant cellular phenotypes of glioma cells and explore the possible mechanisms.
Materials and methods
Cell culture
Primary normal human astrocytes (NHA) and astrocyte medium were acquired from Sciencell Research Laboratories (Carlsbad, CA, USA). Human brain glioma cell lines (U251 and U87) were purchased from Shanghai Institutes for Biological Sciences Cell Resource Center and cultured in DMEM medium containing with 10% fetal bovine serum (EXcell, Shanghai, China). All cells were cultured in a 95% air and 5% CO2 incubator at 37°C.
Clinic specimens
47 glioma tissue and corresponding normal brain tissue (NBT) specimens were gathered from Department of Neurosurgery in Shengjing Hospital through tumorectomy in 2013-2014. All patients had no other treatment before operation, they were diagnosed as glioma by two pathologists and their clinical information is clear and complete. The age of the patients ranged from 37 to 73 years. The patients included 22 men and 25 women. All glioma patients included 18 cases of grade Ⅰ-Ⅱ, 29 cases of grade Ⅲ-Ⅳ. This study was approved by Ethics Committee of Shengjing Hospital, all patients provide their written informed consents.
Arraystar LncRNA Array
The TRIzol® Reagent was used to extract total RNA from glioma and NBT specimens according to operating manual. RNasey Mini Kit (Qiagen, Hilden, GER) was used to clean-up the total RNA. The lncRNAs/mRNAs expression profiling was examined with Human 8 x 60K LncRNA expression array (KangchengBio, Shanghai, China). In details, the cleaned RNA was labeled with Quick Amp Labeling Kit One-Color (Agilent, Santa Clara, CA, USA), purified with RNeasy Mini Kit and assessed the labeled cRNA QC by NanoDrop ND-1000, hybridized with Agilent Gene Expression Hybridization Kit (Agilent, Santa Clara, CA, USA). Following exhaustive washing, the arrays were scanned and the data were extracted by using Agilent Feature Extraction Software.
Quantitative real time PCR (qRT-PCR)
Total RNA was extracted with TRNzol Universal reagent (Tiangen, Beijing, China). The A260/A280 ratio of purified RNA was typically between 1.8 and 2.4 and the yield between 80µg and 120µg. RNA samples were stored at -80°C. RNA integrity was assessed by gel electrophoresis.
RNA was reverse transcribed into cDNA with lncRNA cDNA Synthesis Kit (Tiangen, Beijing, China). LncRNA qPCR Detection Kit (Tiangen, Beijing, China) was applied to examine the expression of SNHG5 on a 7500 PCR System (Applied Biosystems, USA) in accordance with manufacturer's instructions. Each reaction contained 2×lnR lncRNA Premix (25 µl), 50×ROX Reference Dye (1 µl), Forward Primer (1.25 µl), Reverse Primer (1.25 µl), RNA template (2 µl) and RNase-Free ddH2O (19.5 µl). The cycling condition is as follows: Stage 1: 42°C for 20 min, 95°C for 3 min,1 Cycle; Stage 2: 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec, 40 Cycles; Stage 3: 72°C for 50 min. SNHG5's forward primer was 5'- CGCTTGGTTAAAACCTGACACT -3' and its reverse primer was 5'- CCAAGACAATCTGGCCTCTATC -3'. Relative expression level of SNHG5 was calculated using 2-ΔΔCT method after normalization with reference gene (GAPDH).
Vectors construction and Transfection
The knockdown plasmid of SNHG5 (pGPU6/GFP/Neo-SNHG5, sh-SNHG5) and the expression plasmid of CTNNB1 (pEGFP-N1-CTNNB1, pE-CTNNB1) were designed and synthesized by GenePharma (Shanghai, China). The empty plasmid pGPU6/GFP/Neo (sh-NC) and pEGFP-N1 (pE-NC) were used as negative controls respectively. The plasmids were transfected respectively or synergistically into U251 and U87 cells via Lipofectamine 3000 (Invitrogen, Foster City, CA, USA) according to protocol. The stable transfected cells were selected using Geneticin (Sigma-Aldrich, St Louis, MO, USA).
Cell proliferation assay
The proliferation ability of U251 and U87 cells was examined by Enhanced Cell Counting Kit-8 (Beyotime, Beijing, China). 2000 cells were suspended in medium and added into one pore of 96-well plate, then 10 μl enhanced CCK-8 solution was added. 96-well plate was incubated for 1 hr at 37°C. The absorbance at 450 nm was recorded using a MK3 microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Flow cytometry assay
The apoptosis of U251 and U87 cells was examined with Annexin V-FITC/PI apoptosis detection kit (BioLegend, San Diego, CA, USA) as previously described [20]. In briefly, cells were harvested and incubated with Annexin V-FITC buffer containing propidium iodide (PI). Cell apoptosis rate was detected by FACScan flow cytometry with Diva 8.0 software (Becton Dickinson, Franklin Lakes, NJ, USA). Cells undergoing apoptosis were FITC Annexin V positive and PI negative in the right lower quadrant.
Cell invasion assay
Cell invasiveness of U251 and U87 cells was examined by Invasion Chamber with Matrigel Matrix (Corning, Corning, NY, USA). 600 μl DMEM medium containing with 10% fetal bovine serum was added into lower chamber, the upper chamber was added to 100 μl serum-free DMEM medium with 5×104 cells. Then the chamber was cultured under 37°C for 12 hr. Cells located at upper chamber were cleaned out. The membrane was fixed with methanol and stained with Giemsa solution. Under the microscope, the number of transmembrane cells was counted in five random regions and averaged.
Luciferase assay
The TOP Flash and FOP Flash luciferase reporter plasmids were purchased from MilliporeSigma company (Burlington, MA, USA). Luciferase plasmids were transiently co-transfected into U251 and U87 cells. 48 hr later, luciferase reporter assays were performed to quantify the activity of Wnt/CTNNB1 signaling pathway with dual-luciferase assay kit (Vigorous Biotech, Beijing, China) in according with protocol.
Western Blotting
Protein Extraction Kit (Beyotime, Beijing, China) was used to extracted proteins of U251 and U87 cells, and Bradford Protein Assay Kit (Beyotime, Beijing, China) was applied to quantify protein concentration. Protein (30 μg) was separated by polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride (PVDF) membrane. Then, membrane was blocked with Tween-Tris-buffered saline (TTBS) containing 5% non-fat milk at 25°C for 2 hr, hybridized at 4°C overnight with GSK3B and CTNNB1 antibody (# 12456S and #8480S, Cell Signaling, Danvers, MA, USA), incubated with second antibody at 25°C for 2 hr, treated with Chemiluminescent reagent (TIANGEN, Beijing, China) to visualize the bands. Finally, the bands were analyzed by ImageJ software (NIH, Bethesda, MD, USA). Protein levels were normalized to GAPDH.
Statistical analysis
All statistical analysis was performed using SPSS version 22.0 (IBM, Armonk, NY, USA) and GraphPad Prism version 5.04 (GraphPad Software, La Jolla, CA, USA). The data are reported as mean ± standard deviation (SD) of five independent experiments. Student's t test and one-way analysis of variance (ANOVA) were used to finish the comparisons. P-value less than 0.05 was considered to have statistically difference (* and #, P<0.05; ** and ##, P<0.01).
Results
SNHG5 was high-expressed in glioma tissues and cell lines
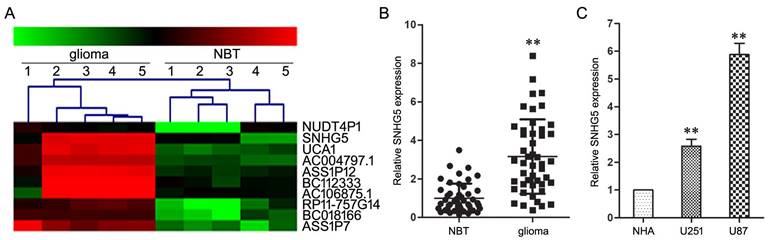
Firstly, lncRNAs microarray assay was used to screen differentially expressed lncRNAs between glioma and NBT specimens, which results displayed SNHG5's expression had an over septuple up-regulation in glioma specimens than that in the NBT specimens (Figure 1A).
Subsequent qRT-PCR assay confirmed the high-expression of SNHG5 in glioma specimens and cell lines (U251 and U87) as compared with NBT specimens and NHA cell line (Figure 1B and 1C, P<0.01). In addition, clinical parameters analysis showed that the increased expression of SNHG5 was correlated with high pathological grade (P<0.01), while was independent of other parameters, including age and gender of glioma patients (P>0.01). These finding suggested SNHG5 was high-expressed in glioma and may be involved in tumorigenesis and progress of glioma.
Knockdown of SNHG5 inhibited malignant cellular phenotypes of glioma cells
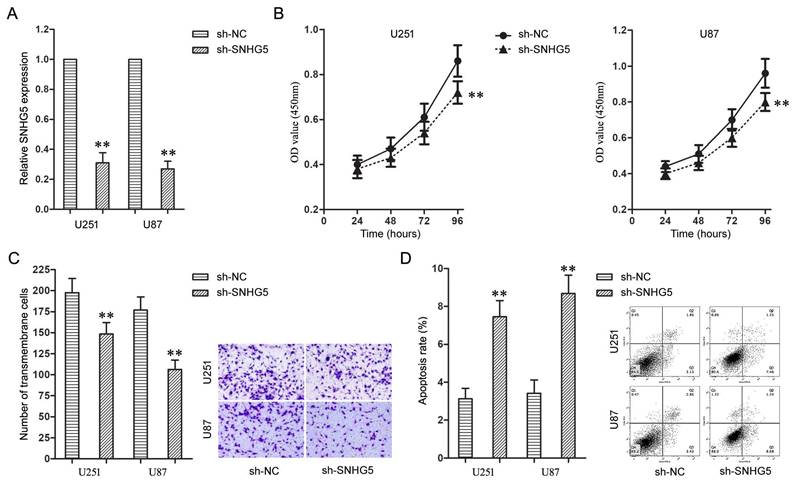
Further, the effects of SNHG5 knockdown on malignant cellular phenotypes of glioma cells were examined with a serious of loss-of-function assays. Firstly, U251 and U87 cells were transfected with sh-SNHG5 to knockdown the expression of SNHG5 (Figure 2A, P<0.01). Enhanced Cell Counting Kit-8 and cell invasion assay found knockdown of SNHG5 significantly depressed cell proliferation and invasiveness in U251 and U87 cells while compared to sh-NC groups (Figure 2B and 2C, P<0.01). In addition, knockdown of SNHG5 advanced apoptosis of U251 and U87 cells (Figure 2D, P<0.01).
SNHG5 was high-expressed in glioma tissues and cell lines. A: Representative microarray analysis of SNHG5 in glioma and NBT specimens. B: The expression levels of SNHG5 in glioma specimens and NBT specimens. ** P<0.01 vs NBT group. C: The expression levels of SNHG5 in NHA, U251 and U87 cell lines. ** P<0.01 vs NHA group.
Knockdown of SNHG5 inhibited malignant cellular phenotypes of glioma cells. A: The expression levels of SNHG5 in U251 and U87 cells. B: The cell proliferation of U251 and U87 cells. C: The cell invasiveness of U251 and U87 cells. D: The cell apoptosis rate of U251 and U87 cells. ** P<0.01 vs sh-NC group.
Knockdown of SNHG5 inactivated Wnt/CTNNB1 signaling pathway
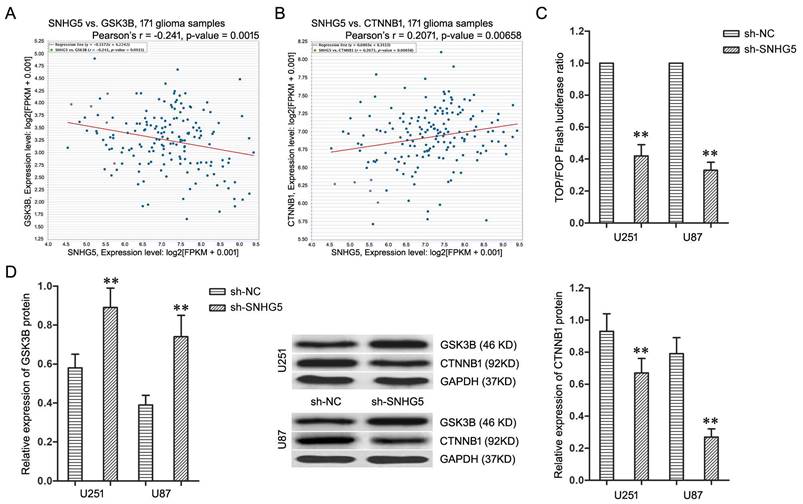
The microarray's data was thoroughly analyzed to explore the underlying regulatory mechanism of SNHG5 in glioma cells, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis found Wnt/CTNNB1 signaling pathway is the most enriched pathway. Furthermore, the co-expression analysis in TCGA Pan-Cancer (PANCAN) database with 171 glioma samples displayed the expression of SNHG5 had significant negative correlation with GSK3B (Figure 3A, r=-0.241, P<0.01) as well as positive correlation with CTNNB1 (Figure 3B, r=0.2071, P<0.01). Therefore, Wnt/CTNNB1 signaling pathway was chosen as a target to highlight SNHG5 associated malignant cellular phenotypes in glioma.
To further confirm, the TOP/FOP ratio in U251 and U87 cells, which represented the transcriptional activity of Wnt/CTNNB1 signaling pathway, was restrained significantly by SNHG5 knockdown (Figure 3C, P<0.01). Moreover, SNHG5 knockdown up-regulated the expression of GSK3B protein and decreased the expression of CTNNB1 protein in U251 and U87 cells (Figure 3D, P<0.01). To sum up, knockdown of SNHG5 inactivated Wnt/CTNNB1 signaling pathway.
Knockdown of SNHG5 inhibited malignant cellular phenotypes of glioma cells via Wnt/CTNNB1 signaling pathway
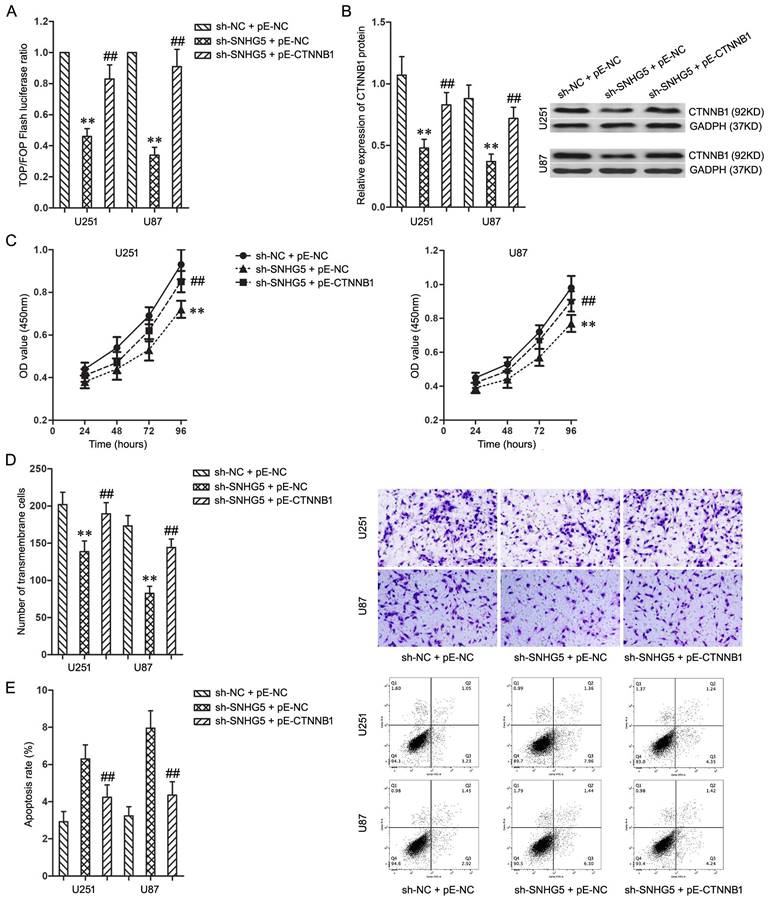
To activate Wnt/CTNNB1 signaling pathway which was inactivated by SNHG5 knockdown, pE-CTNNB1 was co-transfected into U251 and U87 cells with SNHG5 knockdown. The activating of Wnt/CTNNB1 signaling pathway was confirmed using TOP/FOP Flash luciferase reporter assay and western blotting (Figure 4A and 4B, P<0.01).
While combined using sh-SNHG5 and pE-CTNNB1, the cell proliferation and invasiveness of U251 and U87 increased significantly when compared with sh-SNHG5 + pE-NC groups (Figure 4C and 4D, P<0.01). In addition, the cell apoptosis rate in sh-SNHG5 + pE-CTNNB1 groups was much lower than that in sh-SNHG5 + pE-NC groups (Figure 4E, P<0.01). In summary, activating of Wnt/CTNNB1 signaling pathway partly restored the effecting on malignant cellular phenotypes caused by SNHG5 knockdown in U251 and U87 cells, knockdown of SNHG5 depressed malignant cellular phenotypes of glioma cells via Wnt/CTNNB1 signaling pathway.
Discussion
LncRNAs have been well documented to participate in the genesis and progression of various tumors, and are proved to be diagnostic or prognostic biomarkers for almost all kinds of tumors, including glioma. X-inactive specific transcript (XIST) could promote tumorigenesis and angiogenesis of glioma through targeted binding miR-429 as a molecular sponge [21]. Our previous study reported that Tumour suppressor candidate 7 (TUSC7) played the roles of tumor suppressor to restain malignant phenotype of glioma cells, and was a prognostic biomarker of glioma patients [22]; our recent research found the low-expression of TUSC7 in glioma was closely related to chemoresistance with temozolomide (TMZ), TUSC7 inhibited TMZ resistance of glioma through silencing miR-10a [23].
Knockdown of SNHG5 inactivated Wnt/CTNNB1 signaling pathway. A: The co-expression patterns between SNHG5 and GSK3B in TCGA Pan-Cancer (PANCAN) database with 171 glioma samples. B: The co-expression patterns between SNHG5 and CTNNB1 in TCGA Pan-Cancer (PANCAN) database with 171 glioma samples. C: The ratio of TOP/FOP luciferase values in U251 and U87 cells. D: The expression of GSK3B and CTNNB1 protein in U251 and U87 cells. ** P<0.01 vs sh-NC group.
Knockdown of SNHG5 inhibited malignant cellular phenotypes of glioma cells via Wnt/CTNNB1 signaling pathway. A: The ratio of TOP/FOP luciferase values in U251 and U87 cells. B: The expression of CTNNB1 protein in U251 and U87 cells. C: The cell proliferation of U251 and U87 cells. D: The cell invasiveness of U251 and U87 cells. E: The cell apoptosis rate of U251 and U87 cells. ** P<0.01 vs sh-NC + pE-NC group, ## P<0.01 vs sh-SNHG5 + pE-NC group.
In this study, lncRNAs microarray assays firstly found SNHG5 was high-expressed in glioma, and subsequent expression detection in glioma tissues and cell lines confirmed this finding, which suggested SNHG5 might be involved in tumorigenesis of glioma. Ma Z reported that the expression of SNGH5 was up-regulated in bladder cancer and its high-expression level predicted poor prognosis of patients [12]. SNGH5 was high-expressed in colorectal cancer, it was significantly up-regulated both between normal tissues and adenomas as well as from adenomas to carcinoma stage I, suggesting SNHG5 up-regulation as an early event in colorectal cancer development [13].
To verified the roles of SNHG5 in glioma cells, the expression of SNHG5 was knockdown in glioma cells to carry out a series of loss-of-function assays. So far, SNHG5's oncogenic roles were increasingly demonstrated in some kinds of tumors. Recent studies showed that silenced SNHG5 inhibited the proliferation ability of bladder cancer cells and promoted cell apoptosis and arrested cells at G1 phase [12]; knockdown of SNHG5 advanced apoptosis and cell cycle arrest, and limited outgrowth of colorectal cancer in vivo [13]; overexpression of SNHG5 could increase imatinib resistance in chronic myeloid leukemia [24]. In our study, knockdown of SNHG5 inhibited cell proliferation and invasiveness of glioma cells, and advanced cell apoptosis, which showed SNHG5 knockdown restricted malignant cellular phenotypes of glioma cells. Nevertheless, the underlying mechanism is unknown.
The thorough analysis of lncRNA microarray's data and TCGA Pan-Cancer (PANCAN) database predicted a positive regulation model between SNHG5 and Wnt/CTNNB1 signaling pathway, the latter was chosen as a target to highlight SNHG5 associated malignant cellular phenotypes of glioma. As we all known, Wnt/CTNNB1 signaling pathway was activated frequently during tumorigenesis and progress of glioma [25,26]. In our study, the inactivating of Wnt/CTNNB1 signaling pathway caused by SNHG5 knockdown was confirmed by TOP/FOP Flash luciferase reporter assay and western blotting.
Accordingly, we speculated that knockdown of SNHG5 might inhibit malignant cellular phenotypes of glioma through Wnt/CTNNB1 signaling pathway. In subsequent experiments, the activating Wnt/CTNNB1 signaling pathway by up-regulating CTNNB1 reversed the effects of SNHG5 knockdown on malignant cellular phenotypes in glioma cells, Wnt/CTNNB1 signaling pathway mediated effecting induced by SNHG5 knockdown in glioma cells.
However, how SNHG5 activated Wnt/CTNNB1 signaling pathway and the underlying mechanism is still unknown. Recent literatures reported that CTNNB1 was a target gene of miR-5095, SNHG7 could silence miR-5095 and subsequently activate Wnt/CTNNB1 signaling pathway in glioblastoma multiforme [27]; SNHG1 activated Wnt/CTNNB1 signaling pathway via interacting with miR-577, which could targeted silence Wnt Family Member 2B (WNT2B) [28]. These researchers' opinions gave us a nice enlightenment, accordingly, we hypothesized that SNHG5 might activate Wnt/CTNNB1 signaling pathway by targeted regulating some non-coding RNA and this mechanism was involved in modulation of malignant cellular phenotypes in glioma.
In conclusion, our findings clarify SNHG5 is high-expressed in glioma, SNHG5 positively regulates malignant cellular phenotypes of glioma via Wnt/CTNNB1 signaling pathway. Our findings contribute to clarify the molecular mechanism of glioma tumorigenesis, and will provide a promising ncRNA-targeted therapeutic strategy for glioma. However, whether SNHG5 regulates Wnt/CTNNB1 signaling pathway as we speculated, or there are other mechanisms involved in this regulatory process were not investigated in this study, which need further exploring.
Acknowledgements
This study was funded by the National Nature Science Foundation of China (No. 81872067, No. 81301862 and No. 81172408), Natural Science Foundation of Liaoning Province (No. 20180530024) and Basic Scientific Research Projects of Liaoning Higher Education Institutions (LQNK201729).
Authors' contributions
Xuanhao Hu analyzed the data and wrote the manuscript; Chao Shang designed the study and revised the manuscript; Xuanhao Hu and Yang Hong performed all the experiments and analyzed the data.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18(11):1062-1071
2. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432-46
3. Lara-Velazquez M, Al-Kharboosh R, Jeanneret S, Vazquez-Ramos C, Mahato D, Tavanaiepour D, Rahmathulla G, Quinones-Hinojosa A. Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci. 2017:7 (12). pii: E166
4. Zhang H, Guo Y, Song Y, Shang C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol. 2017;79(1):49-55
5. Guo Y, Ma Y, Hu X, Song R, Zhu L, Zhong M. Long non-coding RNA CEBPA-AS1 correlates with poor prognosis and promotes tumorigenesis via CEBPA/Bcl2 in oral squamous cell carcinoma. Cancer Biol Ther. 2018;19(3):205-213
6. Chen J, Hu L, Wang J, Zhang F, Chen J, Xu G, Wang Y, Pan Q. Low Expression LncRNA TUBA4B is a Poor Predictor of Prognosis and Regulates Cell Proliferation in Non-Small Cell Lung Cancer. Pathol Oncol Res. 2017;23(2):265-270
7. Shi B, Wang Y, Yin F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol Ther. 2017;18(10):792-800
8. Xie D, Zhang H, Shang C. Long non-coding RNA CDKN2B antisense RNA 1 gene inhibits Gemcitabine sensitivity in bladder urothelial carcinoma. J Cancer. 2018;9(12):2160-2166
9. Feng W, Li L, Xu X, Jiao Y, Du W. Up-regulation of the long non-coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch Med Sci. 2017;13(6):1315-1321
10. Cai J, Zuo X, Chen Z, Zhao W, Zhu Y, Zhang Z, Ye X. Prognostic value and clinical significance of long noncoding RNA CASC2 in human malignancies: a meta-analysis. Cancer Manag Res. 2018;10:1403-1412
11. Wang R, Li Y, Zhu G, Tian B, Zeng W, Yang Y, Li Z. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/β-catenin signaling pathway. Neuropsychiatr Dis Treat. 2017;13:1805-1813
12. Ma Z, Xue S, Zeng B, Qiu D. lncRNA SNHG5 is associated with poor prognosis of bladder cancer and promotes bladder cancer cell proliferation through targeting p27. Oncol Lett. 2018;15(2):1924-1930
13. Damas ND, Marcatti M, Côme C, Christensen LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF, Seemann SE, Rapin N, Thézenas S, Vang S, Ørntoft T, Andersen CL, Pedersen JS, Lund AH. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875
14. Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, Liu L, Li L, Zhang S, Liu Y, Shi J, Zheng D. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35(44):5770-5780
15. Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31(3):893-903
16. Zuccarini M, Giuliani P, Ziberi S, Carluccio M, Iorio PD, Caciagli F, Ciccarelli R. The Role of Wnt Signal in Glioblastoma Development and Progression: A Possible New Pharmacological Target for the Therapy of This Tumor. Genes (Basel). 2018:9 (2). pii: E105
17. Yang M, Wang M, Li X, Xie Y, Xia X, Tian J, Zhang K, Tang A. Wnt signaling in cervical cancer? J Cancer. 2018;9(7):1277-1286
18. Gao L, Chen B, Li J, Yang F, Cen X, Liao Z, Long X. Wnt/β-catenin signaling pathway inhibits the proliferation and apoptosis of U87 glioma cells via different mechanisms. PLoS One. 2017;12(8):e0181346
19. Wang Y, Jiang M, Yao Y, Cai Z. WWC3 Inhibits Glioma Cell Proliferation Through Suppressing the Wnt/β-Catenin Signaling Pathway. DNA Cell Biol. 2018;37(1):31-37
20. Shang C, Lang B, Meng LR. Blocking NOTCH pathway can enhance the effect of EGFR inhibitor through targeting CD133+ endometrial cancer cells. Cancer Biol Ther. 2018;19(2):113-119
21. Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, Xiao X, Wang Y. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer. 2017;8(19):4106-4116
22. Shang C, Guo Y, Hong Y, Xue YX. Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front Cell Neurosci. 2016;10:235
23. Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother Pharmacol. 2018;81(4):671-678
24. He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. 2017;7(8):1704-1713
25. McCord M, Mukouyama YS, Gilbert MR, Jackson S. Targeting WNT Signaling for Multifaceted Glioblastoma Therapy. Front Cell Neurosci. 2017;11:318
26. Ji ZG, Jiang HT, Zhang PS. FOXK1 promotes cell growth through activating wnt/β-catenin pathway and emerges as a novel target of miR-137 in glioma. Am J Transl Res. 2018;10(6):1784-1792
27. Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, Sun Q. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712-718
28. Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2018;495(1):238-245
Author contact
![]() Corresponding author: Chao Shang, Department of Neurobiology, School of Life Science, China Medical University, No.77 Puhe Road, Shenyang North New Area, Shenyang, 110122, China. Phone/Fax: +86-24-23256666; E-mail address: chaosedu.cn
Corresponding author: Chao Shang, Department of Neurobiology, School of Life Science, China Medical University, No.77 Puhe Road, Shenyang North New Area, Shenyang, 110122, China. Phone/Fax: +86-24-23256666; E-mail address: chaosedu.cn

 Global reach, higher impact
Global reach, higher impact