Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(6):1503-1510. doi:10.7150/jca.28541 This issue Cite
Research Paper
Prognostic and Clinicopathological Significance of lncRNA MVIH in Cancer Patients
1. Department of General Surgery, the First People's Hospital of Neijiang, Neijiang 641000, Sichuan Province, P. R. China.
2. Department of Hepatobiliary Surgery, The First Affiliated Hospital of Hainan, Medical College, Haikou 570102, Hainan Province, P. R. China
*These authors contributed equally to this work.
Received 2018-7-15; Accepted 2018-10-24; Published 2019-2-26
Abstract
Objectives: LncRNA associated with microvascular invasion in hepatocellular carcinoma (lncRNA MVIH) is a newly discovered long non-coding RNA that aberrantly up-regulates and holds prognostic value in various tumors. The aim of the review and meta-analysis is to assess its prognostic potential and functions in malignant tumors.
Materials and methods: PubMed, PMC, EMBASE, Web of Science, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang Database were carefully searched for articles published as of Jan. 1, 2018. Hazard ratios (HRs) or odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated to demonstrate prognostic value of lncRNA MVIH using Stata 12.0 software.
Results: Six original studies including 830 cancer patients were ultimately enrolled in this meta-analysis. The pooled results showed that elevated lncRNA MVIH expression was significantly correlated with unfavorable OS (HR=2.17, 95% CI: 1.58-2.76, p < 0.001) and RFS/DFS/PFS (HR=2.21, 95% CI: 1.54-2.87, p < 0.001) in cancer patients. Additionally, high expression of lncRNA MVIH was related to positive lymph node metastasis (OR = 3.04, 95% CI: 1.37-6.75, p = 0.006) and advanced clinical stage (OR = 2.52, 95% CI: 1.68-3.79, p < 0.001).
Conclusions: LncRNA MVIH over-expression was associated with poor clinical outcomes in multiple cancer types. More studies are warranted to verify and strengthen the clinical value of lncRNA MVIH in human cancers.
Keywords: MVIH, lncRNA, cancer, prognosis, meta-analysis
Introduction
In recent years, long non-coding RNAs have drawn increasingly attention and interest [1-3]. By definition, it is one class of non-coding RNA molecules with more than 200 nucleotides in length but no open reading frame [4-5]. In the past several decades, it has been considered as the “junk” of genome. But now, lncRNAs have been rising stars and serving pivotal roles in a variety of biological processes, such as gene regulation, posttranscriptional regulation, and intercellular signaling [6-9]. Importantly, many lncRNAs have been identified as being cancer-specific, which could exert carcinogenic or anti-tumor effects in tumor occurrence and progression [10-13]. Moreover, the abnormal expression of lncRNAs was also reported to be correlated with clinical features and cancer prognosis [14-15]. LncRNAs are currently believed to be utilized as novel therapeutic targets and biomarkers that could aid cancer treatment and prognostication.
Microvascular invasion in hepatocellular carcinoma (MVIH) is a newly found lncRNA. It was first reported to be up-regulated in hepatocellular carcinoma and participated in the process of microvascular invasion [16]. Remarkably, it could serve as a prognostic predictor for patients after hepatectomy [16]. Subsequently, lncRNA MVIH has been reported to be involved in tumor progression, and is suggestive of prognostic potential in multiple human cancers [17-19]. For example, Nie et al [17] reported that lncRNA MVIH was up-regulated in tissues of non-small cell lung cancer, and increased MVIH expression levels could promote cell proliferation and invasion, whereas high lncRNA MVIH levels were associated with malignant biological behavior (such as lymph node metastasis) and poor prognosis. In breast cancer, Lei et al [18] found that elevated MVIH expression levels could promote cell proliferation and the cell cycle, while high lncRNA MVIH was an unfavorable prognostic factor and was associated with Ki67 expression but not with lymph node metastasis. It should be taken into consideration that the individual study consisted of a relatively small sample and some discrepancies existed in these studies for clinical outcomes. In addition, still no study gives a compressive assessment of prognostic values of lncRNA MVIH in human cancers. Therefore, we carried out this systematic review and meta-analysis to clarify the predictive value of lncRNA MVIH in various cancer patients.
Material and Methods
Search Strategy
Two reviewers (Zhang Y and Lin SB) performed the literature retrieval, independently. The following electronic databases were screened: PubMed, PMC, EMBASE, Web of Science, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang Database. The latest search was up to Jan. 1, 2018. The following keywords were used for literature searching: “lncRNA associated with microvascular invasion in hepatocellular carcinoma” OR “lncRNA MVIH” OR “MVIH” OR “Microvascular invasion in hepatocellular carcinoma.” In order to supplement our literature search, the reference lists of relevant articles were observed for additional eligible studies.
Inclusion and Exclusion Criteria
Eligible articles were identified based on the following inclusion criteria: 1) The expression of lncRNA MVIH was detected in any human tumors; 2) Correlation of lncRNA MVIH with prognosis was reported; 3) Sufficient data were available for calculating the hazard ratio (HR) with 95% CI. 4) Patients were categorized as high or low group according to lncRNA MVIH expression level. Articles were excluded when they did not cover the points above.
Data Extraction and Quality Assessment
Two investigators (Zhang Y and Lin SB) independently extracted data from identified studies according to unified form. Extracted data elements included the following records: 1) Name of first author, publication year, country of origin, study design; cancer type, sample size, expression pattern, tumor stage, criterion of high expression, detection method, follow-up time, outcome measures, analysis type; 2) Hazard ratio (HR) with 95% CI for cancer prognosis; 3) Patient number for high expression and low expression, lymph node metastasis (LNM), distant metastasis (DM), microvascular invasion (MVI), and tumor-node-metastasis (TNM) stage.
If a study reported the data in multivariate analysis and univariate analysis, the hazard ratio (HR) with the corresponding 95% CI was directly extracted from multivariate analysis. Those indirectly reported HRs and 95% CIs were extracted from graphical survival plots using Engauge Digitizer version 4.1 (https://sourceforge.net/projects/digitizer/).
The Newcastle-Ottawa quality assessment scale (NOS) with score range from 0-9 was applied to assess the quality of all included studies. A high quality study was identified as having a score of ≥ 6.
Statistical Analysis
In this meta-analysis, Stata version 12.0 (Stata Corporation, College Station, TX, USA) was used to calculate all statistical analyses. Higgins I2 statistics and Cochran's Q-test were applied to assess the heterogeneity among studies. When the percentage of I2 was greater than 50% or Phet less than 0.05, a random-effects model was used. Otherwise, a fixed-effects model was applied to the analyses. Begg's and Egger's test were utilized to detect the publication bias. In order to evaluate the robustness of the results, sensitivity analysis was performed by omitting the study one by one to assess the effects on the pooled results. A p-value < 0.05 was considered to be statistically significant.
Results
Study selection and Characteristics
Initially, there were a total of 220 articles retrieved from a number of online databases after duplications removed. Following, 29 articles were left when 191 articles were excluded after titles and abstracts were screened for the following reasons: reviews, no association between lncRNA MVIH expression and human cancers, no clinical outcomes. After further screening of the remaining 29 studies, 23 were excluded due to lack of survival data, unrelated to specific prognosis. Finally, six available studies were deemed applicable to the meta-analysis [16-21]. The detailed steps for screening eligible articles is presented in Figure 1.
All six studies were published from 2012 to 2017. They were all identified as high-quality. Those studies enrolled a total of 830 cancer patients, with a mean subject size of 138.3, ranging from 42 to 250. All studies measured the expression of lncRNA MVIH in tissue specimens by quantitative real-time polymerase chain reaction (qRT-PCR) and reported the relationship between lncRNA MVIH expression and cancer prognosis.
The steps for screening eligible articles.
Six studies reported patients' OS, and one focused on RFS, DFS, and PFS, respectively. Four studies estimated the relationship between lncRNA MVIH and TNM stage, and four focused on LNM, while one focused on MVI and DM, respectively. The cancers investigated consisted of hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC), breast cancer (BC), gastric cancer (GC), glioma, and colorectal carcinoma (CRC). All included studies were retrospective, conducted in China, and focused on an Asian population. Table 1 and Table 2 summarize the main information and data of all included studies.
Results of the Meta-analysis
MVIH Expression and OS
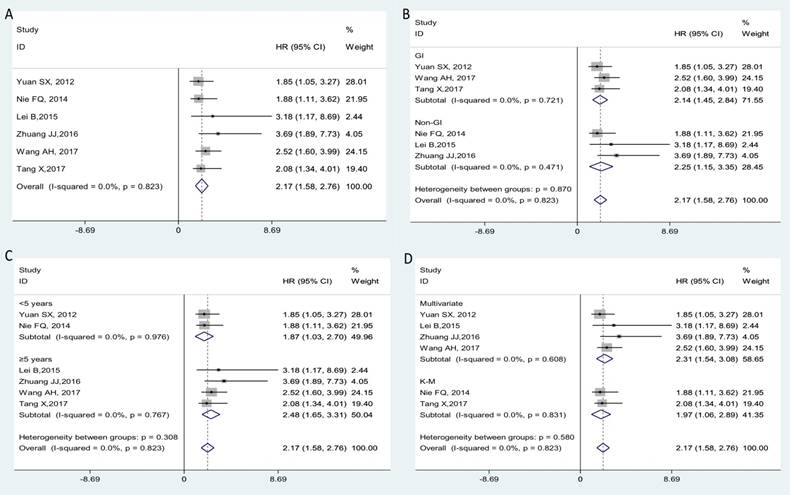
Six clinical studies that comprised a total of 830 cancer patients were conducted to analyze the OS; a fixed-effects model was utilized for no significant heterogeneity (I2=0.0%, Phet=0.823). From the pooled result, we found that increased lncRNA MVIH expression was correlated with poorer OS in cancer patients (HR=2.17, 95% CI: 1.58-2.76; Figure 2A). In the stratified analysis, we found that elevated MVIH expression could be a prognostic biomarker for patients with gastrointestinal (GI) cancers (HR: 2.14, 95% CI: 1.45-2.84, p < 0.001) or patients with non-gastrointestinal cancers (HR: 2.25, 95% CI: 1.15-3.35, p < 0.001). In the subgroup analyses by follow-up time, MVIH could act as a prognostic factor in groups with follow-up time < 5 years (HR: 1.87, 95% CI: 1.03-2.70, p < 0.001) or groups with follow-up time ≥ 5 years (HR: 2.48, 95% CI: 1.65-3.31, p < 0.001). We also found that a high expression of MVIH could act as an unfavorable independent prognostic factor for OS based on multivariate analysis (HR: 2.31, 95% CI: 1.54-3.08, p < 0.001). Stratification analyses for all subgroups are presented in detail in Figure 2B-2D.
MVIH Expression and DFS/RFS/PFS
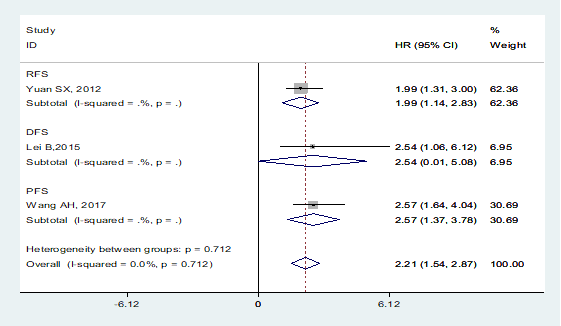
A total of 3 studies including 617 patients investigated the prognostic value of MVIH on cancer progression or recurrence, with a pooled HR of 2.21 (95% CI: 1.54-2.87; p < 0.001, Figure 3). No heterogeneity was observed among the studies, a fixed-effects model was performed to the calculation (I2=0.0%, Phet=0.712). Among them, one focused on the prongostic value of MVIH expression on cancer relapse (HR=1.99, 95% CI: 1.31-3.00), one reported that patients with high MVIH expression levels had poor disease-free survival (HR=2.54, 95% CI: 1.06-6.12). A final one explored the relationship between MVIH and tumor progression (HR=2.57, 95% CI: 1.64-4.04).
MVIH Expression and Clinicopathological Characteristics
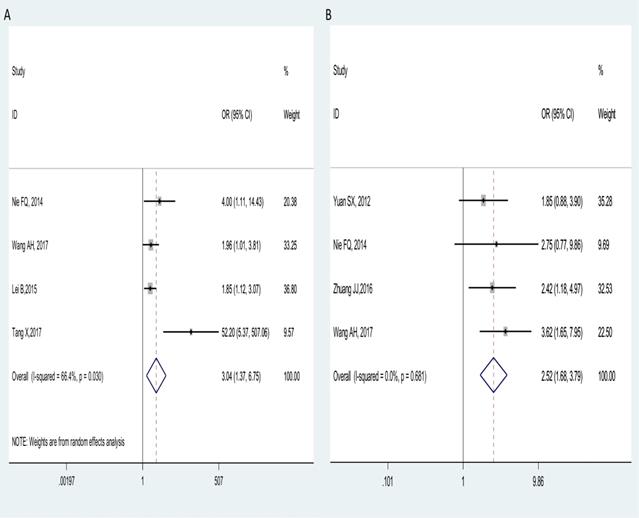
Four studies comprising of 488 cancer patients reported the correlation of MVIH with lymph node metastasis (LNM) in multiple tumors. A random-effect model was used for obvious heterogeneity observed across studies (I2=66.4%, Phet=0.030), the pooled results showed that patients with high MVIH expression in tumor tissues preferentially metastasize to the lymph nodes (OR = 3.04, 95% CI: 1.37-6.75; p = 0.006, Figure 4A).
Furthermore, four studies with 536 patients declared the relationship between MVIH expression levels and clinical stage, with a fixed-effects model on account of no obvious heterogeneity (I2=0.0%, Phet =0.681). The combined results indicated that the patients with high MVIH expression have a more advanced stage (III/IV) (OR = 2.52, 95% CI: 1.68-3.79; p < 0.001, Figure 4B).
Main characteristics of all studies included in the meta-analysis.
| First author, Year | Country | Cancer type | Study design | Sample size | HE (%) | Stage | Criterion of HE | Follow- up | MVIHassay | Detected sample | Outcome measures | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yuan SX, 2012 [16] | China | HCC | R | 215 | 50.20% | I-III | at or above the 50th percentile | < 5 years | qRT-PCR | Tissue | OS, RFS | 8 | |
| Nie FQ, 2014 [17] | China | NSCLC | R | 42 | 50.00% | I -IIIa | MVIH expression ratio ≥ mean ratio((2.12-fold) | < 5 years | qRT-PCR | Tissue | OS | 6 | |
| Lei B, 2015 [18] | China | BC | R | 250 | 50.00% | NR | according to the mean value | ≥ 5 years | qRT-PCR | Tissue | OS, DFS | 9 | |
| Zhuang JJ, 2016 [19] | China | Glioma | R | 127 | 48.80% | I-IV | based on the median expression levels | ≥ 5 years | qRT-PCR | Tissue | OS | 7 | |
| Tang X, 2016 [21] | China | CRC | R | 44 | 77.27% | I-IV | according to the mean value | ≥ 5 years | qRT-PCR | Tissue | OS | 6 | |
| Wang AH, 2017 [20] | China | GC | R | 152 | 44.10% | I-IV | MVIH cut‐off point ( 3.85) | ≥ 5 years | qRT-PCR | Tissue | OS, PFS | 7 |
Abbreviations: R, retrospective; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; BC, breast cancer; GC, gastric cancer; CRC, colorectal carcinoma; NR, not reported; HE, high expression; OS, over survival; DFS, disease-free survival; RFS, recurrence-free survival; PFS, progression‐free survival; qRT-PCR, quantitative real-time polymerase chain reaction.
HRs and 95% CIs of cancer prognosis and progression associated with MVIH expression in all included studies.
| First author, Year | Country | Cancer type | Case number of MVIH expression | HR(95%CI) for OS | HR(95%CI) for RFS/DFS/PFS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | High with LNM | High with DM | High with MVI | High with III/IV | Low | Low with LNM | Low with DM | Low with MVI | Low with III/IV | |||||
| Yuan SX, 2012 [16] | China | HCC | 108 | NR | NR | 40 | 22 | 107 | NR | NR | 23 | 13 | 1.85(1.05-3.27)# | 1.99(1.31-3.00)# |
| Nie FQ, 2014 [17] | China | NSCLC | 21 | 14 | NR | NR | 11 | 21 | 7 | NR | NR | 6 | 1.88(1.11-3.62)* | NR |
| Lei B, 2015 [18] | China | BC | 125 | 77 | NR | NR | NR | 125 | 58 | NR | NR | NR | 3.18(1.17-8.69)# | 2.54(1.06-6.12)# |
| Zhuang JJ, 2016 [19] | China | Glioma | 62 | NR | NR | NR | 41 | 65 | NR | NR | NR | 29 | 3.69(1.89-7.73)# | NR |
| Tang X, 2017 [21] | China | CRC | 34 | 29 | NR | NR | NR | 10 | 1 | NR | NR | NR | 2.08(1.34-4.01)* | NR |
| Wang AH, 2017 [20] | China | GC | 67 | 32 | 4 | NR | 25 | 85 | 27 | 3 | NR | 12 | 2.52(1.60-3.99)# | 2.57(1.64-4.04)# |
Abbreviations: HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; BC, breast cancer; GC, gastric cancer; CRC, colorectal carcinoma; OS, over survival; DFS, disease-free survival; RFS, recurrence-free survival; PFS, progression‐free survival; NR, not reported; LNM, lymph nodes metastasis; DM, distant metastasis; MVI, microvascular invasion; *HR calculated from survival curves; #HR directly from multivariate analysis.
Subgroup analysis for OS. Over-all (A); stratified by the cancer type (B), by follow-up time (C) and by analysis type (D).
Meta-analysis of the pooled HRs of DFS/RFS/PFS of patients with high MVIH expression.
Forest plots of combined analyses associated with MVIH expression. (A): LNM; (B): clinical stage.
The studies by Yuan et al [16] and Wang et al [20] conducted the microvascular invasion (MVI) and distant metastasis (DM) analysis of cancer patients, respectively. Those data suggested that the overexpression of MVIH was associated with frequent MVI (p=0.016), but was unable to accurately predict the occurrence of DM (p=0.476).
Publication Bias
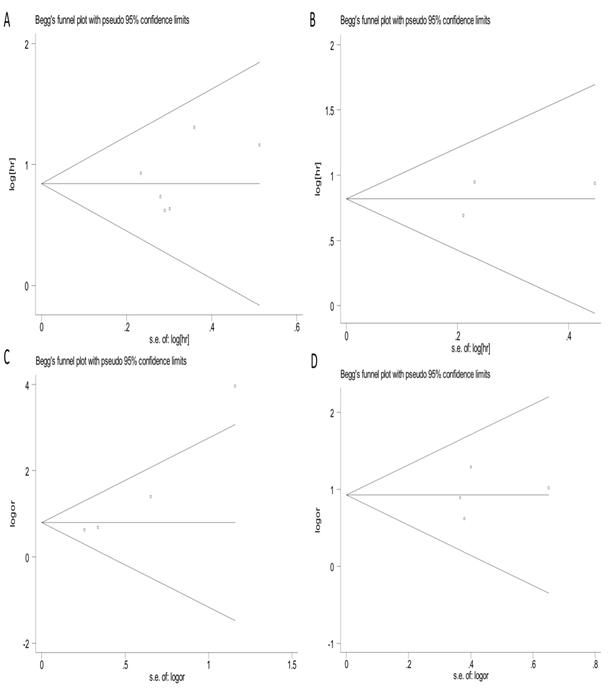
Funnel plots and Begg's and Egger's test were used to evaluate the publication bias in this meta-analysis. As shown in Figures 5A-5D, the Begg's funnel plots with pseudo 95% CIs were symmetric. The test results also indicated no significant publication bias in the meta-analysis.
Sensitivity Analysis
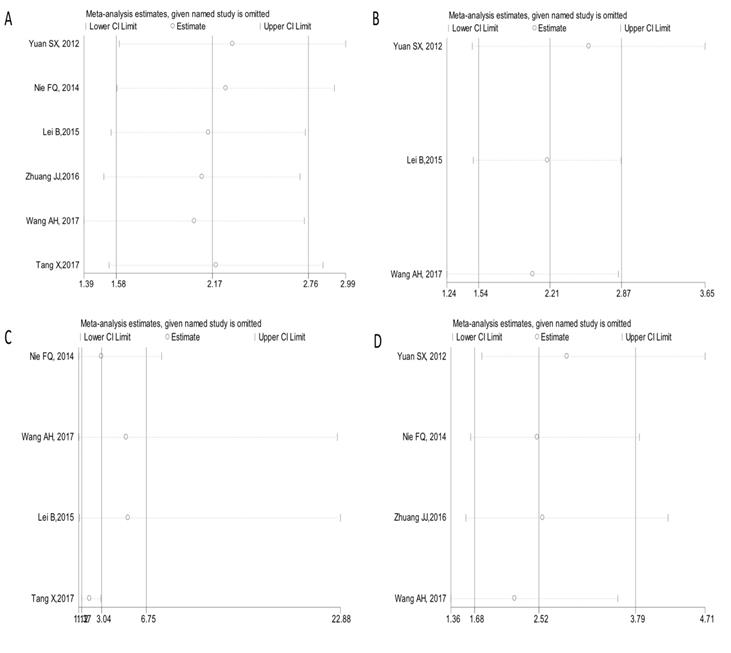
As illustrated in Figures 6A-6D, the analyzed results suggested that our results were comparatively credible and stable.
Begg's funnel plots of publication bias test. (A) OS; (B) RFS/DFS/PFS; (C) LNM; (D) clinical stage.
Discussion
MVIH is a novel identified lncRNA transcript, the majority of the MVIH sequence was located within the intron of the RPS24 gene, but both the MVIH and RPS24 genes were independently transcribed [16, 22]. As a cancer-associated lncRNA, MVIH was discovered in several cancers and showed carcinogenic roles in tumor progression. In hepatocellular cancer, MVIH could activate tumor-inducing angiogenesis by inhibiting the secretion of phosphor-glycerate kinase 1, while MVIH expression was inversely correlated with cancer prognosis. Therefore, deregulation of lncRNA MVIH could be utilized as a potential target for new adjuvant therapies and serves as a predictor for poor recurrence-free survival after hepatectomy [16]. Furthermore, Shi et al [23] found mechanistically MVIH could promote cell growth and inhibit cell apoptosis of HCC via inhibiting miR-199a expression. Finally, Cheng et al [24] reported that ARID1A could interact with lncRNA MVIH and ARID1A suppressed HCC cells proliferation and migration by inhibiting MVIH. In breast cancer, the knockdown of MVIH expression by siRNA could suppress cell proliferation and cell cycle, while increasing cell apoptosis, whereas up-regulated MVIH expression showed the converse [18]. MVIH also displayed oncogenic functions in NSCLC, high levels of MVIH could partly promote cell proliferation and invasion via regulating MMP2 and MMP9 protein expression in vitro [17]. MVIH also showed the same oncogenic effect in glioma cell migration and invasion [19]. Although MVIH has been discovered in several different kinds of cancers, the exact functional roles of it in carcinogenesis remain unclear and need to be further elucidated.
Sensitivity analysis under specific model. (A): for OS; (B): for RFS/DFS/PFS; (C): for LNM; (D): for clinical stage.
As we all know, this is the first meta-analysis that elaborated the clinical value of lncRNA MVIH in human cancers. We demonstrated that the high expression of lncRNA MVIH was significantly related with unsatisfactory clinical outcomes; the patients with high MVIH level in tissues had a worse OS when compared with those with low MVIH expression (HR=2.17, 95% CI: 1.58-2.76), and the prognostic value of MVIH on cancer progression or recurrence was also revealed (HR=2.21, 95% CI: 1.54-2.87). Furthermore, the clinical role of MVIH expression was also exhibitted in regard to LNM (OR = 3.04, 95% CI: 1.37-6.75) and tumor stages (OR = 2.52, 95% CI: 1.68-3.79). In this meta-analysis, no significant heterogeneity was observed when we carried out OS and DFS/RFS/PFS analysis of MVIH, as well as comparison for TNM stage. The results from the sensitivity analysis and publication bias evaluation also further indicated our analyses were credible and stable.
Although this meta-analysis was conducted with standard procedure and rigorous statistics, our conclusion still has several limitations that should be clearly interpreted. First of all, the total sample size was relatively small and the number of enrolled cancer types still needs to be enlarged. Secondly, the quantity of included studies was not sufficient for a more comprehensive analysis for other prognostic indicators and clinic-pathological features, such as DFS, DM or MVI. Thirdly, all included were retrospective researches, and only a Chinese population was applicable in this meta-analysis, potentially minimizing the analyzing value to some extent and affecting the broader applicability of our conclusions. Fourthly, studies with positive results are more easily published than negative results, which might cause potential publication bias and some missing data. At last, the cut-off value of high lncRNA MVIH expression is needed to be unified before its clinical application. Regarding these deficiencies, the role of lncRNA MVIH in multiple human carcinomas might be exceedingly evaluated. Therefore, further multi-center studies that are well-designed and at a large-scale are needed to verify the function of lncRNA MVIH in various cancers.
To summarize, the present meta-analysis highlighted the clinical effect of lncRNA MVIH in predicting prognosis, metastasis, and clinical stage in multiple cancers. To better evaluate the prognostic role of lncRNA MVIH expression in human cancers, higher quality studies with larger sample sizes are required for a more complete confirmation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yamashita A, Shichino Y, Yamamoto M. the long non-coding RNA world in yeasts. Biochim Biophys Acta. 2016;1859:147-154
2. Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58:116-141
3. Kazemzadeh M, Safaralizadeh R, Feizi MA, Ravanbakhsh R, Somi MH, Hashemzadeh S. LOC100287225, novel long intergenic non-coding RNA, misregulates in colorectal cancer. Cancer Biomark. 2016;16:499-505
4. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17-29
5. Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930-939
6. He A, Chen Z, Mei H, Liu Y. Decreased expression of LncRNA MIR31HG in human bladder cancer. Cancer Biomark. 2016;17:231-236
7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159
8. He A, Chen Z, Mei H, Liu Y. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18:95-104
9. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756-770
10. Luo P, Jing W, Zhu M, Li ND, Zhou H, Yu MX. et al. Decreased expression of LncRNA SRA1 in hepatocellular carcinoma and its clinical significance. Cancer Biomark. 2017;18:285-290
11. Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187-22202
12. Sun W, Mo X, Li T, Xie Y, Guo J. Clinical significance of the long noncoding RNA RP11-19P22.6-001 in gastric cancer. Cancer Biomark. 2017;18:397-403
13. Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2017;48:8-15
14. Liu FT, Dong Q, Gao H, Zhu ZM. The prognostic significance of UCA1 for predicting clinical outcome in patients with digestive system malignancies. Oncotarget. 2017;8:40620-40632
15. Liu F, Dong Q, Huang J. Overexpression of lncRNA PVT1 predicts advanced clinic-pathological features and serves as an unfavorable Risk Factor for Survival of Patients with Gastrointestinal Cancers. Cell Physiol Biochem. 2017;43:1077-1089
16. Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G. et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231-2241
17. Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M. et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35:7587-7594
18. Lei B, Xu SP, Liang XS, Li YW, Zhang JF, Zhang GQ. et al. Long non-coding RNA MVIH is associated with poor prognosis and malignant biological behavior in breast cancer. Tumour Biol. 2016;37:5257-5264
19. Zhuang JJ, Yue M, Zheng YH, Li JP, Dong XY. Long non-coding RNA MVIH acts as a prognostic marker in glioma and its role in cell migration and invasion. Eur Rev Med Pharmacol Sci. 2016;20:4898-4904
20. Wang A, Du L, Jiang K, Kong Q, Zhang X, Li L. Long noncoding RNA microvascular invasion in hepatocellular carcinoma is an indicator of poor prognosis and a potential therapeutic target in gastric cancer. J Can Res Ther. 2017 doi: 10.4103/0973-1482.204882
21. Tang X, Xu YH, Wang CL, Qiu P, Hu Y. The expression of MVIH-lncRNA in colorectal cancer. Genomics and Applied Biology. 2017;36:4414-4418
22. He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW. et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20-27
23. Shi Y, Song Q, Yu S, Hu D, Zhuang X. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. Onco Targets Ther. 2015;8:2303-2310
24. Cheng S, Wang L, Deng CH, Du SC, Han ZG. ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem Biophys Res Commun. 2017;491:178-182
Author contact
![]() Corresponding authors: Xianjin Yang, E-mail: 2439015338com, Tel +86 138 9059 3536; Xu Zhang, E-mail: 1990486976com, Tel +86 139 8143 2513
Corresponding authors: Xianjin Yang, E-mail: 2439015338com, Tel +86 138 9059 3536; Xu Zhang, E-mail: 1990486976com, Tel +86 139 8143 2513

 Global reach, higher impact
Global reach, higher impact