Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(7):1772-1780. doi:10.7150/jca.26722 This issue Cite
Research Paper
Five P53 SNPs Involved in Low Rectal Cancer Risk and Prognosis in a Chinese Population
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang 110001, China
Received 2018-4-17; Accepted 2018-10-5; Published 2019-4-20
Abstract
Although the impact and potential mechanisms of p53 polymorphisms on human malignancies have been intensively studied, analyses for association between p53 polymorphisms and colorectal cancer (CRC) risk were still limited to some common variants. Moreover, the majority of previous studies did not classify the specimens of CRC based on tumor location. This case-control study aimed to evaluate the association of five p53 polymorphisms (rs1042522, rs12947788, rs1625895, rs2909430 and rs12951053) with the risk of low rectal cancer (LRC) and investigate the prognostic significance. A total of 347 cases and 353 controls from a Chinese population were recruited and genotyped using KASP assay. Individuals carrying the variant rs12947788 A allele were observed to associate with an increased risk of LRC. After stratification for clinicopathological parameters, rs12947788 was significantly co-related with the histological type of LRC under a dominant model. Although none of the selected p53 polymorphisms was significantly associated with patient prognosis in total population, significant associations with the overall survival were revealed in the heterozygosis carriers vs. wild type carriers model through subgroup analyses based on clinical characteristics. Moreover, haplotype analyses showed that C-A-G-A-A haplotype was associated with a significantly higher LRC risk as compared to the other haplotypes. In low rectal cancer, P53 protein expression was obviously higher in p53 rs1042522 mutant carriers than in other genotypes. Our study further proves the involvement of p53 polymorphisms in pathogenesis of LRC and may provide potential therapeutic implications.
Keywords: p53, polymorphisms, low rectal cancer, risk, prognosis
Introduction
Colorectal cancer (CRC) ranks as the third most common type of tumor around the world and accounts for more than 7% of overall cancer-related death in China1, 2. According to their anatomical locations, CRCs are generally separated into upper-, mid- and low- rectal cancers. Low rectal cancer (LRC) refers to CRC that is situated within 8 cm of the anal verge. As compared with mid- and upper- rectal cancers, LRC shows wide differences in surgical difficulties, pathological features and outcomes3, 4. Despite the LRC treatment has been improved greatly the cancer still remains a serious challenge to health care due to the high risk of local recurrence5. In view of the facts, an effective risk prediction and prognostic assessment are needed, which may help for us to take intervention measures to reduce the incidence and mortality of LRC as well as improve the life quality of patients.
Genetic variants in cancer have provided a greater understanding of differences in individual disease prediction and prognosis. It has been found that the abnormal changes of many genes are related to CRC, such as K-ras, BRAF, PIK3CA, APC, PTEN and p536, 7. The p53 tumor suppressor gene, which is located on chromosome 17p13, plays a crucial role in various cellular processes including cell cycle, apoptosis, senescence, genomic stability, and inhibition of angiogenesis8. As a key element of anti-cancer defense mechanism, p53 is commonly mutated in human cancers to facilitate the progression of tumorigenesis. Mutations of p53 occur at a frequency of nearly 40%-50% in CRC9. In addition to the mutation, several groups have reported that p53 polymorphisms in germline also contribute to the aggressiveness, invasiveness and transformation of CRC and might therefore serve as a clinically useful marker of prognosis10-13.
Till now, the roles of p53 polymorphisms in modulation of CRC risk have been investigated in different ethnic groups14-20. However, despite the large number of polymorphisms identified in p53 gene, analyses for association with CRC risk were still limited to some common variants such as rs1042522 and rs1787836213. Moreover, the majority of previous studies did not classify the specimens of CRC based on tumor location or just simply divided into colon cancer and rectal cancer, and the results are often inconsistent and conflicting. For instance, rs1042522, which results in an Arg to Pro change in codon 72, is one of the most frequently studied functional p53 single nucleotide polymorphisms (SNPs) in CRC. Gemignani et al. showed no statistically significant association between rs1042522 and CRC21. Conversely, two Asian groups both confirmed that the Pro/Pro genotype of rs1042522 was related to increased CRC risk. Results from these two groups were also not completely consistent, with Song et al. revealed no significant interactions between rs1042522 and smoking or drinking22, whereas Zhu et al. proved that 72P allele conferred a more pronounced increase risk of CRC among alcohol consumers23. Through updated meta-analysis based on 32 published data, a very recent study demonstrated that rs1042522 was not significantly associated with CRC risk in the overall population24. It is noteworthy that a significant association of CC genotype of rs1042522 with CRC was observed when the study population was limited to patients of rectal cancer, suggesting that the site of the origin of tumor may have a huge impact on the association between p53 SNPs and CRC risk.
Thus, the current study is specifically interested in LRC, one of the most aggressive types of CRC, and studies on its relationship with functional p53 SNPs rs1042522, rs12947788, rs1625895, rs2909430 and rs12951053 in a Chinese population. Among the selected SNPs, rs1042522 and rs12947788 have been widely studied in CRC with inconsistent results, whereas the contributions of rs1625895, rs2909430 and rs12951053 to CRC risk have never been reported before. To our best knowledge, it is the first time to evaluate relationship between p53 polymorphisms and LRC cancer risk in a Chinese population. Furthermore, the prognostic significance of these SNPs and the effect of different combination of p53 SNPs on LRC risk were also explored in the present study.
Materials and Methods
Ethics statement
The research protocol was approved by the medical ethics committee of the first affiliated hospital of China Medical University. All methods were performed in accordance with the relevant guidelines and regulations. Additionally, written informed consent was obtained from each study participant.
Study population
This study initially consisted of 347 LRC patients and 353 cancer-free controls. In brief, rectal cancer patients with the location of tumor within 8 cm of the anal verge were consecutively recruited when presented for treatment at The First Affiliated Hospital of China Medical University, Shenyang, China, between December 2011 and June 2016. Hospital-based individuals admitted to gastroenterological departments for treatment of anal benign diseases, who were found to be free from CRC by digital rectal examination and other related examinations, were consecutively recruited as controls during the same period. Subsequently, a peripheral blood sample from each recruited individual was obtained for genomic DNA extraction.
For all cases and controls, the basic information regarding gender, age and smoking and alcohol use was collected using self-reported questionnaires. Detailed clinical information about the LRC patients including tumor-node-metastasis (TNM) system classification, depth of invasion, growth mode, lymphatic metastasis status, histological type, peritumor lymphocyte infiltration status, perineural invasion status, vascular cancer embolus status and extra nodal tumor deposits (ENTD) status was collected from their medical records. In this study, the outcome variable measured was overall survival (OS) and information about the date of death was collected with a follow-up period until 21st, August 2016. Due to the loss of follow-up visit or unwelcomed by relatives, death information was only obtained from 304 case individuals.
SNP Genotyping
Identification and measurement of the selected p53 SNPs in this study were achieved through the KASP genotyping assay. In brief, total genomic DNA was extracted from cryopreserved clotted blood samples of cases and controls obtained from the participants through the standard processes of phenol and chloroform extraction method. DNA samples were then randomly placed on batches of 1,536-well plate where an equal number of cases and controls were run simultaneously. Genotyping of these elected SNPs was performed according to the manufacturer's instructions (http://www.lgcgenomics.com/genotyping/kasp-genotyping-reagents/kasp-technical-resources/). For the purpose of quality control, duplicate samples (5% of the total numbers of samples) were repeated for each p53 SNP and non-template controls were set in each plate.
Immunohistochemical assay
Each paraffin specimen of the included 347 patients diagnosed with LRC and 353 controls was sectioned into five continuous slices (4-μm thick). One slice was used for hematoxylin and eosin (H&E) staining, while the remaining slices were used for immunohistochemical staining, according to manufacturer's protocol.
P53 protein expression was independently read and scored by two pathologists, in accordance with the double-blind principle. Inconsistent scores were consulted with a senior pathologist to arrive at a consensus. The positive P53 protein expression was located in the nuclei of cancer cells, and appeared as stronger brown granules under a microscope with high magnification. Then, the positive P53 protein expression area was detected under a microscope with low magnification. Ten fields of each slide were randomly selected under a microscope with high magnification, and 100 cancer cells were counted in each field. Next, the percentage of cancer cells with a positive P53 protein expression was calculated. The scores for the positive expression of p53 were determined through the percentage of P53 positive cells in each sample, as follows: <10% negative (-); 10% to 30% (+); 30% to 50% (++); 50% -100 % (+++).
Statistical analyses
For each polymorphism, Hardy-Weinberg Principle (HWP) was calculated in the control groups to check for the expected frequency using a Chi-square test. The association between the selected p53 polymorphisms and LRC risk was assessed using the χ2 test, as well as variables of clinicopathological parameters. Considering the major allele as the reference, we performed logistic regression model to calculate the odds ratios (ORs) and their 95% confidence intervals (CI) of each genotype and genetic model. Interactions of SNPs with classic risk factors including gender, age, smoking, and alcohol use were estimated using a logistic regression model. The subjects for who there were missing data for these variables were excluded in the subgroup analysis. A Cox proportional hazards model was used for univariate and multivariate analysis of the relative risk of death. Haplotype analysis was performed by SHEsis online software (http://analysis.bio-x.cn/myAnalysis.php). Kaplan-Meier analysis was adopted to draw survival curves concerning each p53 polymorphism. The log-rank test was used to evaluate the significance of differences in OS among various patient subgroups. Statistical analyses were performed using SPSS version 20.0(SPSS Inc. Chicago, IL, USA) software. The significance levels of all tests were set at p<0.05 and were two-sided.
Results
Study population
This study included 347 cases and 353 controls (detailed information in Supplementary Table S1). The median age of all patients at diagnosis was 61 years, with a range from 24 to 91 years, while the median age of all cancer-free controls at recruitment was 61 years, with a range from 28 to 90 years. No significant differences in gender, age and smoking and alcohol use were detected between LRC patients and controls (all p>0.05, Supplementary Table S1). The clinical characteristics and outcomes of patients with LRC were listed in Table 1. As shown in Table 1, the TNM stage Ⅲ-Ⅳ, positive status of lymphatic metastasis and histological type of low differentiation are among the most significant clinical characteristics associated with poor prognosis in LRC.
Relationship between the selected p53 polymorphisms and LRC risk
Genotypic frequencies of the selected p53 SNPs among patients and controls were shown in Table 2. In the control group, the distributions of the genotypes of all five p53 polymorphisms did not deviate from HWP (all p>0.05, Table 2). In brief, the distributions of the analyzed genotypes demonstrated no significant relation with the risk of LRC, except for individuals with variant A allele of p53 rs12947788 which were associated with an increased LRC risk (AA+GA vs. GG: OR = 1.393, 95% CI 1.030-1.884, p=0.032) (Table 2). We next investigated the relationship between some classic risk factors (gender, age and smoking and alcohol use) of patients with LRC and the selected p53 SNPs. As presented in Supplementary Table S2, associations were observed in three of five selected SNPs (rs1042522, rs12947788, rs12951053), when the study population was stratified based on these risk factors. To be specific, rs1042522 was associated with the increased risk of LRC in cigarette smokers under the recessive model (CC vs. CG+GG: OR = 2.561, 95% CI 1.146-5.721, p=0.022). Under dominant model, significant association of rs12947788 with a higher LRC susceptibility was observed in people with drinking history (AA+GA vs. GG: OR = 3.235, 95% CI 1.158-9.040, p=0.025) or less than 60 years old (AA+GA vs. GG: OR = 1.749, 95% CI 1.113-2.748, p=0.015), respectively. Relative to the heterozygosis and wild type carriers in a recessive model, carriers of the homozygous variant CC genotype in rs12951053 was co-related with an increased risk of LRC in males (CC vs. CA+AA: OR = 2.473, 95% CI 1.061-5.765, p=0.036).
Clinical characteristics and outcomes of patients with LRC.
| Variables | Casesa n (%) (N=304) | Deatha n (%) (N=44) | Median Survival Time | P-value | |
|---|---|---|---|---|---|
| Age | 0.827 | ||||
| ≤60 | 138 (45.4) | 20 (45) | 39.656 | ||
| >60 | 166 (54.6) | 24 (55) | 38.362 | ||
| Gender | 0.193 | ||||
| Male | 178 (58.6) | 29 (65.9) | 38.504 | ||
| Female | 126 (41.4) | 15 (34.1) | 38.908 | ||
| TNM stage | 3.246E-10 | ||||
| Ⅰ-Ⅱ | 181 (59.7) | 8 (18.2) | 43.197 | ||
| Ⅲ-Ⅳ | 122 (40.3) | 36 (81.8) | 33.081 | ||
| Depth of invasion | 0.003 | ||||
| T1+T2 | 87 (28.6) | 4 (9.1) | 42.988 | ||
| T3+T4 | 217 (71.4) | 40 (90.9) | 37.261 | ||
| Lymphatic metastasis | 2.406E-11 | ||||
| Negative | 193 (63.7) | 8 (18.2) | 43.288 | ||
| Positive | 110 (36.3) | 36 (81.8) | 32.490 | ||
| Histological type | 4.1911E-8 | ||||
| High-Middle differentiation | 195 (64.1) | 13 (29.5) | 42.412 | ||
| Low differentiation | 109 (35.9) | 31 (70.5) | 32.060 | ||
| Peritumor lymphocyte infiltration | 0.619 | ||||
| Negative | 24 (8.5) | 4 (9.8) | 40.125 | ||
| Positive | 259 (91.5) | 37 (90.2) | 35.165 | ||
| Perineural invasion | 0.002 | ||||
| Negative | 72 (27.1) | 4 (10.3) | 38.586 | ||
| Positive | 194 (72.9) | 35 (89.7) | 32.952 | ||
| Vascular cancer embolus | 0.001 | ||||
| Negative | 222 (75.8) | 25 (56.9) | 40.574 | ||
| Positive | 71 (24.2) | 19 (43.1) | 33.467 | ||
| ENTD | 0.000005 | ||||
| Negative | 246 (93.2) | 30 (76.9) | 35.886 | ||
| Positive | 18 (6.8) | 9 (23.1) | 23.202 | ||
a In some subgroups, the total number of cases or death may not up to 100% of subjects due to the lack of corresponding information for some patients. ENTD is the abbreviation for extranodal tumor deposits.
Relationship between the selected p53 polymorphisms and clinicopathological parameters
The association of the selected p53 SNPs with clinicopathological parameters of LRC including tumor-node-metastasis (TNM) system classification, depth of invasion, growth mode, lymphatic metastasis status, histological type, peritumor lymphocyte infiltration status, perineural invasion status, vascular cancer embolus status and extra nodal tumor deposits (ENTD) status was next assessed. The only significant association was observed for the rs12947788, which was significantly co-related with the histological type of LRC in a dominant model (p=0.044, Table 3). To confirm this result, we further evaluated the association of the rs12947788 with the histological type of LRC based on epidemiological risk factors. As shown in Table 3, rs12947788 was significantly associated with the histological type of LRC in patients older than 60 years (p=0.043), male (p=0.022), with no drinking history (p=0.048) and having smoking habit (p=0.039), respectively.
Genotype distribution of the investigated p53 SNPs in cases and controls and the associations of these SNPs with the risk of LRC.
| SNPs | Cases n (%) | Controls n (%) | Cases vs Controls | HWP | |
|---|---|---|---|---|---|
| (N=347) | (N=353) | pa | OR(95%CI) | Pa,b | |
| P53 rs1042522 | 0.9294 | ||||
| GG | 101(30) | 116(33) | 1(Ref) | ||
| CG | 178(51) | 176(49) | 0.399 | 1.158(0.824-1.628) | |
| CC | 68(19) | 61(18) | 0.307 | 1.258(0.810-1.953) | |
| CC+CG vs GG | 0.3 | 1.186(0.859-1.638) | |||
| CC vs CG+GG | 0.507 | 1.139(0.775-1.674) | |||
| C VS.G | |||||
| P53 rs12947788 | 0.9528 | ||||
| GG | 142(41) | 172(49) | 1(Ref) | ||
| GA | 167(48) | 149(42) | 0.054 | 1.365(0.994-1.873) | |
| AA | 38(11) | 32(9) | 0.141 | 1.484(0.877-2.510) | |
| AA+GA VS.GG | 0.032 | 1.393(1.030-1.884) | |||
| AA VS.GA+GG | 0.368 | 1.257(0.764-2.070) | |||
| A VS. G | |||||
| P53 rs1625895 | 0.2988 | ||||
| GG | 313(90.2) | 316(90) | 1(Ref) | ||
| GA | 34(9.8) | 37(10) | 0.612 | 0.880(0.536-1.443) | |
| G VS. A | |||||
| P53 rs2909430 | 0.3127 | ||||
| AA | 307(88) | 317(90) | 1(Ref) | ||
| GA | 40(12) | 36(10) | 0.71 | 1.095(0.678-1.770) | |
| G VS. A | |||||
| P53 rs12951053 | 0.3476 | ||||
| AA | 170(49) | 174(49) | 1(Ref) | ||
| CA | 143(41) | 155(44) | 0.668 | 0.934(0.781-2.384) | |
| CC | 34(10) | 27(7) | 0.275 | 1.364(0.781-2.384) | |
| CA+CC VS.AA | 1 | 1.0(0.742-1.348) | |||
| CC VS.CA+AA | 0.218 | 1.402(0.819-2.399) | |||
| C VS. A | |||||
Abbreviations: SNPs, single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; HWP, Hardy-Weinberg principle. aP-value obtained from the Chi-squared test. bP-value obtained from Hardy-Weinberg principle of the control group.
Relationship between the selected p53 polymorphisms and LRC prognosis
Prognostic significance of the selected p53 polymorphisms was analyzed through univariate and multivariate analysis using the Cox proportional hazards model and further evaluated by Kaplan-Meier survival analysis. As depicted in Supplementary Table S3, none of the genotypes of the selected p53 polymorphisms showed a significant association with LRC prognosis. However, for the heterozygosis carriers relative to the wild type carriers, many associations between the selected p53 polymorphisms and prognosis were observed according to the subgroup analyses based on some clinical characteristics (Supplementary Table S4). Interestingly, rs12947788 was co-related with good prognosis in LRC patients with nest growth mode (HT vs. WT: HR = 0.204, 95% CI 0.042-0.980, p=0.047), whereas rs1625895 and rs2909430 have significant poor prognosis (HT vs. WT: HR = 26.189, 95% CI 2.399-285.91, p=0.007; HT vs. WT: HR = 7.284, 95% CI 1.361-38.996, p=0.020) (Supplementary Table S4 and Figure 1). In addition, rs12951053 was associated with poor prognosis in LRC patients with female gender (HT vs. WT: HR = 4.170, 95% CI 1.017-17.106, p=0.047), younger age (less than 60 years old) (HT vs. WT: HR = 3.561, 95% CI 1.192-10.638, p=0.023), history of smoking (HT vs. WT: HR = 4.940, 95% CI 1.178-20.718, p=0.029), positive status of lymphatic metastasis (HT vs. WT: HR = 2.367, 95% CI 1.028-5.453, p=0.043) or invasion state of growth mode (HT vs. WT: HR = 3.007, 95% CI 1.271-7.115, p=0.012) (Supplementary Table S4 and Figure 2).
Relationship between the selected p53 SNP-SNP interaction and LRC risk
Haplotype analyses were further conducted to assess the effect of different combination of these selected SNPs on the LRC risk. As shown in Table 4, the most frequent haplotypes in the current study were C-A-G-A-C and G-G-G-A-A, which account for 81.5% of cases and 82.3% of controls. However, the only significant association was observed for C-A-G-A-A haplotype, which was associated with a significantly increased risk of LRC as compared to the other haplotypes (OR=4.15, 95%CI=1.63-10.58, p=0.001). The relationship between P53 haplotypes and clinicopathological parameters or prognosis of LRC was also studied but no significant interaction was revealed (data not shown).
The association between p53 gene polymorphism and P53 protein expression
In order to study the influence of p53 gene polymorphism on the protein expression of P53, five polymorphic locus of the p53 gene (rs1042522, rs12947788, rs1625895, rs2909430 and rs12951053) and the protein expression of P53 in the 347 patients diagnosed with LRC were synchronously detected. Results revealed that the p53 rs1042522 polymorphism could affect P53 protein expression (CG vs. GG, P=0.027, CC + CG vs. GG, P=0.032), indicating that positive P53 protein expression was significantly higher than other genotypes in the heterozygous and dominant models (Table 5). The other four polymorphic loci were not found to be relevant.
Discussion
Although the impact and potential mechanisms of p53 polymorphisms on human malignancies have been intensively studied, analyses for association between p53 polymorphisms and CRC risk were still limited to some common variants. Moreover, the majority of previous studies did not classify the specimens of CRC based on tumor location. It has been well accepted that CRC from different sites have different biological behaviors. Distinguishing the special biological characteristics of CRC at different sites has important value and guiding significance for clinical diagnosis and treatment. In the present study, we classified the specimens of CRC based on tumor location, strictly grouped the patients based on the site of CRC tumors, and assessed five p53 polymorphisms. This new approaches uncovered some new significant findings which otherwise would not have been discovered in the previous studies.
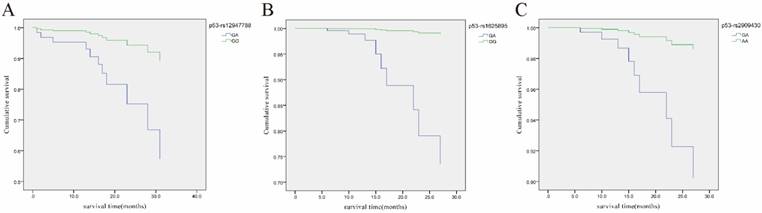
Cox regression analysis of the association of three p53 SNPs with overall survival (OS) of LRC patients with growth mode of nest. A, Cumulative OS curve of the LRC patients with growth mode of nest for the two genotypes of p53 rs12947788 (p=0.049 for GA vs. GG); B, Cumulative OS curve of the LRC patients with growth mode of nest for the two genotypes of p53 rs1625895 (p=0.007 for GA vs. GG); C, Cumulative OS curve of the LRC patients with growth mode of nest for the two genotypes of p53 rs2909430 (p=0.049 for GA vs. AA).
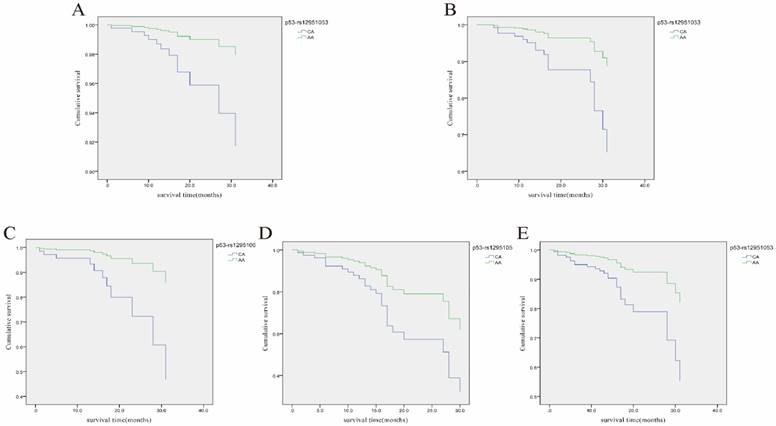
Cox regression analysis of the association of rs12951053 with overall survival (OS) of LRC patients with different clinical characteristics. A, Cumulative OS curve of the female LRC patients for the two genotypes of p53 rs12951053 (p=0.047 for CA vs. AA); B, Cumulative OS curve of the LRC patients younger than 60 years old for the two genotypes of p53 rs12951053 (p=0.023 for CA vs. AA); C, Cumulative OS curve of the LRC patients with smoking habit for the two genotypes of p53 rs12951053 (p=0.029 for CA vs. AA); D, Cumulative OS curve of the LRC patients with a positive status of lymphatic metastasis for the two genotypes of p53 rs12951053 (p=0.043 for CA vs. AA); E, Cumulative OS curve of the LRC patients with invasion growth mode for the two genotypes of p53 rs12951053 (p=0.012 for CA vs. AA).
Associations of rs12947788 polymorphism with the histological type of LRC.
| Histological type | High-Middle Differentiation | Low Differentiation | Dominant Model | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Wild type | Heterozygous type | Mutant type | Wild type | Heterozygous type | Mutant type | p-value | ||
| Total | 99 | 101 | 21 | 43 | 66 | 17 | 0.044 | ||
| Age | ≥60 | 59 | 55 | 10 | 24 | 39 | 11 | 0.043 | |
| <60 | 40 | 46 | 11 | 19 | 27 | 6 | 0.481 | ||
| Gender | Male | 65 | 50 | 14 | 25 | 40 | 9 | 0.022 | |
| Female | 34 | 51 | 7 | 18 | 26 | 8 | 0.844 | ||
| Drinking | Yes | 13 | 18 | 1 | 5 | 7 | 2 | 0.517 | |
| No | 86 | 83 | 20 | 38 | 59 | 15 | 0.048 | ||
| Smoking | Yes | 43 | 27 | 5 | 13 | 18 | 4 | 0.039 | |
| No | 56 | 74 | 16 | 30 | 48 | 13 | 0.423 | ||
P53 haplotype distribution between LRC patients and controls.
| Haplotypea | Case(%)b | Control(%)b | Cases vs Controls | |
|---|---|---|---|---|
| (N=694) | (N=706) | P | OR(95%CI) | |
| C-A-G-A-A | 22.02(3.2) | 5.57(0.8) | 0.004 | 4.26(1.57-11.51) |
| C-A-G-A-C | 201.32(29.0) | 190.13(26.9) | 0.391 | 1.11(0.87-1.42) |
| C-G-A-G-A | 32.48(4.7) | 31.98(4.5) | 0.725 | 0.90(0.49-1.65) |
| C-G-G-A-A | 49.62(7.1) | 66.09(9.4) | 0.375 | 0.85(0.59-1.22) |
| G-G-G-A-A | 364.23(52.5) | 391.30(55.4) | 0.348 | 0.90(0.72-1.12) |
aLoci rs1042552-rs12947788-rs1625895-rs2909430-rs12951053. b Numbers of alleles are listed. The total number of alleles is twice the total number of individuals because of the existence of two alleles in each individual. c P was adjusted by gender, age, drinking and smoking status.
A novel finding based on our data is the co-relation of p53 rs12947788 with a higher risk of LRC. Furthermore, a significant association of p53 rs12947788 with the histological type of LRC was observed under a dominant model. SNP of rs12947788, also known as IVS7+72C>T polymorphism, located in intron 7 of chromosome 17 and has been reported to be associated with some human diseases such as gastric cancer25, oral neoplasm26 and breast cancer27. Relatively fewer studies have investigated the role of rs12947788 in CRC as compared with the most popular studied rs1042522. One Czech group demonstrated a lower frequency of rs12947788 in CRC cases than in controls, but this association was not significant28. Another observation showed that the rs12947788 polymorphism dominant model showed significant relationship with a decreased CRC risk in Lynch syndrome patients in Taiwan20, which is opposite to our current finding that individuals carrying variant A allele of p53 rs12947788 were related with increased LRC risk. The discrepancy can be mainly attributed to the differences between study populations. Although both studies used the Chinese population as the subjects of study, Kamiza et al. collected the clinical data of Lynch syndrome patients as cases, with only approximately half of them (46.2%) developed histologically confirmed CRC in follow-up period. In sharp contrast, the study population in our work was strictly the patients with LRC.
Many efforts have been made to reveal the link between p53 polymorphisms and the clinical outcome of CRC. A large population-based study showed that colon tumors with p53 mutations were associated with a significantly worse 5-year survival than those with wild type p53. However, G245 hot spot mutations and p53 mutations in proximal tumors were the only significant predictors of poor prognosis found in this study29. Another study suggested that Pro72 allele of rs1042522 demonstrated significant association with a lower survival rate in patients with Duke's B stage of CRC30. In a study on sporadic CRC patients, mutation in codon 175 of exon 5 conferred a better prognosis and alterations of exon 8 were related to a worse outcome in different population subgroups including male, patients younger than 71 years old, tumors located in proximal colon, moderately differentiated and mucinous31. Compared with the former studies, some novel and intriguing findings were observed from our work. Firstly, four of the five selected p53 SNPs were found to be associated with prognosis of LRC in subgroups classified by tumor growth mode. To be specific, p53 rs12947788 was related with low survival in LRC patients with nest growth mode, whereas rs2909430 and rs1625895 had poor prognosis. On the contrary, p53 rs12951053 was associated with poor clinical outcome in LRC patients with invasion growth mode. Secondly, rs12951053 was significantly associated with a short overall survival in LRC patients after stratification for various risk factors such as female, younger age than 60 years old, history of smoking, positive status of lymphatic metastasis and invasion state of growth mode. These results suggest that p55 SNPs such as rs12951053 could be served as a clinically useful prognostic factor of LRC. The relationship between p53 polymorphisms and the growth mode of LRC deserve to be further investigated.
In comparison with analysis of single SNP, haplotype analysis comes up with more convincing and powerful results due to its involvement of combined effect of multiple SNPs in the pathogenesis of disease. Few studies have reported the relationship between p53 SNP-SNP interaction and CRC risk. Polakova et al. performed haplotype analysis on p53 SNPs of rs17878362: A1>A2, rs1042522: G>C, rs12947788:C>T, and rs17884306:G>A in a Czech population. It was revealed that haplotype A2CCG was associated with a higher CRC risk as compared with the most common haplotype A1GCG, while the four other haplotypes A1CCG, A2GCG, A1GTG and A1GCA were associated with a lower risk28. Another study showed an increased CRC risk for the Pro72/Ser47 haplotype of p53 in south Indian population32. Recently, Kamiza et al. found that the CT haplotype (rs1042522/rs12947788) of p53 was related with a reduced CRC risk as compared to the common GC haplotype20. In the current study, we found a relatively rare haplotype C-A-G-A-A was associated with a significantly increased risk of LRC. This previously unknown connection may facilitate understanding the pathogenesis of LRC after further verification.
The association between the five polymorphism of p53 gene and P53 protein expression
| Protein expression | Genotype | Heterozygous vs. Wild | Mutant vs. Wild | Dominant model | Recessive model | ||
|---|---|---|---|---|---|---|---|
| Wild type | Heterozygous type | Mutant type | P-value | P-value | P-value | P-value | |
| rs1042522 | 0.027 | 0.239 | 0.032 | 0.905 | |||
| Positive | 64 | 135 | 49 | ||||
| Negative | 37 | 43 | 19 | ||||
| rs12947788 | 0.203 | 0.990 | 0.278 | 0.659 | |||
| Positive | 97 | 125 | 26 | ||||
| Negative | 45 | 42 | 12 | ||||
| rs1625895 | 0.280 | ||||||
| Positive | 221 | 27 | |||||
| Negative | 92 | 7 | |||||
| rs2909430 | 0.204 | ||||||
| Positive | 216 | 32 | |||||
| Negative | 91 | 8 | |||||
| rs12951053 | 0.154 | 0.787 | 0.191 | 0.905 | |||
| Positive | 116 | 108 | 24 | ||||
| Negative | 54 | 35 | 10 | ||||
The association between these SNPs and P53 protein expression was evaluated, and results revealed that p53 rs1042522 polymorphism was associated with P53 protein expression, which was evidenced by the significantly higher P53 protein expression in p53 rs1042522 mutant carriers compared with the other genotypes, while the remainders were not found to be relevant. As it is known, SNPs exist in any region of the gene, including both coding and non-coding region. In general, SNPs from coding sequences of the gene may affect the expression of its encoded protein, whereas SNPs from the non-coding sequences of these genes do not necessarily affect the expression of its encoded protein. Among the five polymorphic loci selected in the present study, only p53_rs1042522 was located in the exon region, while other four polymorphic loci were located in the intron region. This indicates that the p53 rs1042522 mutant may fall into the coding sequence of the p53 gene, thereby affecting the protein expression of P53. However, this does not mean that other polymorphisms are not functionally important, since SNPs that do not fall into these protein coding regions could possibly affect other processes, such as gene splicing, which needs to be further investigated.
A major strength of our study is that all cases collected in the current study were restricted to tumors growing within 8 cm of the anal verge which was classified as low rectal cancer, while most previous studies addressed total colorectal cancer29-32. Limited but significant evidences have revealed the influence of tumor sites on the frequency, type and prognostic and predictive role of p53 mutation in CRC10, 33, 34. It was reported that p53 mutation was observed in 34% of the proximal colon tumors, while the proportion was up to 45% in distal CRC10. Mutations in p53 were co-related with lymphatic invasion in proximal CRC, while significant association of p53 mutation with both lymphatic and vascular invasion was shown in distal CRC10. Clinical outcome analysis also indicated a poorer prognosis for proximal colon patients with mutant p53 in exon 510. Moreover, a significantly higher frequency of p53 protein accumulation in the nucleus was found in distal CRC as compared with proximal CRC34. All these data suggests that precise definition of different subtypes of CRC based on tumor location is needed to acquire more accurate and reliable information in case-control studies.
There were some limitations in our study. Foremost, the clinical information was not obtained from patients with mid- or upper- rectal cancers and thus comparisons between different types of rectal cancer could not be performed, which should be further studied. Secondly, a number of significant associations were found out of p53 polymorphisms with risk and prognosis of LRC, but few results remained significant by Bonferroni correction, which required stricter significance. Therefore, the main findings in this study need to be further confirmed by studies based on various ethnicities and large populations. Thirdly, the molecular mechanism involved has not been thoroughly investigated in the present study. The “validation experiments” by knock-in and knock-out strategies and both cultured cell and transgenic mice model will be carried out in the further.
In summary, this case-control study evaluated the association of five p53 polymorphisms (rs1042522, rs12947788, rs1625895, rs2909430 and rs12951053) with the risk of LRC and investigate their prognostic significance. The exploratory and preliminary findings have not been reported in the previous studies. First, individuals carrying the variant rs12947788 A allele were observed to associate with an increased risk of LRC; second, rs12947788 was significantly co-related with the histological type of LRC under a dominant model; third, significant associations with the overall survival were revealed in the heterozygosis carriers vs. wild type carriers model; fourth, different combinations of SNPs may have different function; fifth, different SNPs differently affect the expression of p53 protein. All these new findings provide enlightening clues for future study of LRC target prevention and treatment.
Supplementary Material
Supplementary tables.
Abbreviations
LRC: low rectal cancer; CRC: colorectal cancer; SNPs: single nucleotide polymorphisms; TNM: tumor-node metastasis; ENTD: extranodal implantation; OS: overall survival; ORs: odds ratio; CI: confidence interval; MAF: minimum allele frequency; KASP: Kompetitive Allele Specific PCR.
Acknowledgements
This work is supported by grants from the National Science and technology support project (2015BAI13B07).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:104-17
2. Liu S, Zheng R, Zhang M, Zhang S, Sun X, Chen W. Incidence and mortality of colorectal cancer in China, 2011. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2015;27:22-8
3. Nagtegaal ID, van de Velde CJH, Marijnen CAM, van Krieken JHJM, Quirke P. Low rectal cancer: A call for a change of approach in abdominoperineal resection. Journal of Clinical Oncology. 2005;23:9257-64
4. West NP, Finan PJ, Anderin C, Lindholm J, Holm T, Quirke P. Evidence of the oncologic superiority of cylindrical abdominoperineal excision for low rectal cancer. Journal of Clinical Oncology. 2008;26:3517-22
5. Rullier E, Denost Q, Vendrely V, Rullier A, Laurent C. Low rectal cancer: classification and standardization of surgery. Diseases of the colon and rectum. 2013;56:560-7
6. Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. International journal of molecular sciences. 2017:18
7. Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745-55
8. May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621-36
9. Iacopetta B. TP53 mutation in colorectal cancer. Human mutation. 2003;21:271-6
10. Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:7518-28
11. Iacopetta B, Russo A, Bazan V, Dardanoni G, Gebbia N, Soussi T. et al. Functional categories of TP53 mutation in colorectal cancer: results of an International Collaborative Study. Annals of oncology: official journal of the European Society for Medical Oncology. 2006;17:842-7
12. Lopez I, L PO, Tucci P, Alvarez-Valin F, R AC, Marin M. Different mutation profiles associated to P53 accumulation in colorectal cancer. Gene. 2012;499:81-7
13. Naccarati A, Polakova V, Pardini B, Vodickova L, Hemminki K, Kumar R. et al. Mutations and polymorphisms in TP53 gene-an overview on the role in colorectal cancer. Mutagenesis. 2012;27:211-8
14. Sjalander A, Birgander R, Athlin L, Stenling R, Rutegard J, Beckman L. et al. P53 germ line haplotypes associated with increased risk for colorectal cancer. Carcinogenesis. 1995;16:1461-4
15. Perez LO, Abba MC, Dulout FN, Golijow CD. Evaluation of p53 codon 72 polymorphism in adenocarcinomas of the colon and rectum in La Plata, Argentina. World J Gastroenterol. 2006;12:1426-9
16. Koushik A, Tranah GJ, Ma J, Stampfer MJ, Sesso HD, Fuchs CS. et al. p53 Arg72Pro polymorphism and risk of colorectal adenoma and cancer. Int J Cancer. 2006;119:1863-8
17. Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S, Harris CC. Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. Int J Cancer. 2006;118:1790-7
18. Tan XL, Nieters A, Hoffmeister M, Beckmann L, Brenner H, Chang-Claude J. Genetic polymorphisms in TP53, nonsteroidal anti-inflammatory drugs and the risk of colorectal cancer: evidence for gene-environment interaction? Pharmacogenet Genom. 2007;17:639-45
19. Li XL, Zhou JB, Chen ZR, Chng WJ. p53 mutations in colorectal cancer- molecular pathogenesis and pharmacological reactivation. World J Gastroentero. 2015;21:84-93
20. Kamiza AB, Hsieh LL, Tang R, Chien HT, Lai CH, Chiu LL. et al. TP53 Polymorphisms and Colorectal Cancer Risk in Patients with Lynch Syndrome in Taiwan: A Retrospective Cohort Study. Plos One. 2016;11:e0167354
21. Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutierrez-Enriquez S. et al. A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene. 2004;23:1954-6
22. Song HR, Kweon SS, Kim HN, Piao JM, Yun WJ, Choi JS. et al. p53 codon 72 polymorphism in patients with gastric and colorectal cancer in a Korean population. Gastric Cancer. 2011;14:242-7
23. Zhu ZZ, Wang AZ, Jia HR, Jin XX, He XL, Hou LF. et al. Association of the TP53 codon 72 polymorphism with colorectal cancer in a Chinese population. Jpn J Clin Oncol. 2007;37:385-90
24. Tian X, Dai S, Sun J, Jiang S, Jiang Y. The association between the TP53 Arg72Pro polymorphism and colorectal cancer: An updated meta-analysis based on 32 studies. Oncotarget. 2016
25. Shen H, Solari A, Wang X, Zhang Z, Xu Y, Wang L. et al. P53 codon 72 polymorphism and risk of gastric cancer in a Chinese population. Oncology reports. 2004;11:1115-20
26. Li YQ, Li YL, Gu QH, Ye AH, Wu TS. [p53 gene intron 7 polymorphism and its association with oral neoplasms]. Zhonghua kou qiang yi xue za zhi = Zhonghua kouqiang yixue zazhi = Chinese journal of stomatology. 2005;40:386-9
27. Vymetalkova V, Soucek P, Kunicka T, Jiraskova K, Brynychova V, Pardini B. et al. Genotype and Haplotype Analyses of TP53 Gene in Breast Cancer Patients: Association with Risk and Clinical Outcomes. Plos One. 2015;10:e0134463
28. Polakova V, Pardini B, Naccarati A, Landi S, Slyskova J, Novotny J. et al. Genotype and haplotype analysis of cell cycle genes in sporadic colorectal cancer in the Czech Republic. Human mutation. 2009;30:661-8
29. Samowitz WS, Curtin K, Ma KN, Edwards S, Schaffer D, Leppert MF. et al. Prognostic significance of P53 mutations in colon cancer at the population level. Int J Cancer. 2002;99:597-602
30. Csejtei A, Tibold A, Varga Z, Koltai K, Ember A, Orsos Z. et al. GSTM, GSTT and p53 polymorphisms as modifiers of clinical outcome in colorectal cancer. Anticancer Res. 2008;28:1917-22
31. Vidaurreta M, Maestro ML, Sanz-Casla MT, Rafael S, Veganzones S, de la Orden V. et al. Colorectal carcinoma prognosis can be predicted by alterations in gene p53 exons 5 and 8. Int J Colorectal Dis. 2008;23:581-6
32. Singamsetty GK, Malempati S, Bhogadhi S, Kondreddy R, Govatati S, Tangudu NK. et al. TP53 alterations and colorectal cancer predisposition in south Indian population: a case-control study. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:2303-11
33. Catalano T, Curia MC, Aceto G, Verginelli F, Cascinu S, Cama A. et al. Mutations in the p53 and Ki-ras genes, microsatellite instability and site of tumor origin in colorectal cancer. Oncology reports. 2005;14:625-31
34. Mahdavinia M, Bishehsari F, Verginelli F, Cumashi A, Lattanzio R, Sotoudeh M. et al. P53 mutations in colorectal cancer from northern Iran: Relationships with site of tumor origin, microsatellite instability and K-ras mutations. Journal of cellular physiology. 2008;216:543-50
Author contact
![]() Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; Fax: +86-024-83282383; Email: yuanyuanedu.cn
Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, North Nanjing Street 155#, Heping District, Shenyang110001, China. Telephone: +86-024-83282153; Fax: +86-024-83282383; Email: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact