3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(8):1781-1793. doi:10.7150/jca.31699 This issue Cite
Review
Bacteria in Cancer Therapeutics: A Framework for Effective Therapeutic Bacterial Screening and Identification
1. Department of Microbiology & Immunology, Schulich School of Medicine & Dentistry, University of Western Ontario, London, Ontario, Canada.
2. London Research and Development Centre, Agriculture and Agri-Food Canada, London, Ontario, Canada.
3. Department of Biology, McMaster University, Hamilton, Ontario, Canada.
Received 2018-11-21; Accepted 2019-2-21; Published 2019-4-21
Abstract
By 2030, the global incidence of cancer is expected to increase by approximately 50%. However, most conventional therapies still lack cancer selectivity, which can have severe unintended side effects on healthy body tissue. Despite being an unconventional and contentious therapy, the last two decades have seen a significant renaissance of bacterium-mediated cancer therapy (BMCT). Although promising, most present-day therapeutic bacterial candidates have not shown satisfactory efficacy, effectiveness, or safety. Furthermore, therapeutic bacterial candidates are available to only a few of the approximately 200 existing cancer types. Excitingly, the recent surge in BMCT has piqued the interest of non-BMCT microbiologists. To help advance these interests, in this paper we reviewed important aspects of cancer, present-day cancer treatments, and historical aspects of BMCT. Here, we provided a four-step framework that can be used in screening and identifying bacteria with cancer therapeutic potential, including those that are uncultivable. Systematic methodologies such as the ones suggested here could prove valuable to new BMCT researchers, including experienced non-BMCT researchers in possession of extensive knowledge and resources of bacterial genomics. Lastly, our analyses highlight the need to establish and standardize quantitative methods that can be used to identify and compare bacteria with important cancer therapeutic traits.
Keywords: cancer, bacterium, therapeutics, bacterium-mediated cancer therapy, screening, microbiology
Introduction
Cancer is a group of diseases caused by disproportionately dividing cells that grow into invasive lumpy masses, commonly referred to as tumors. Approximately 200 human cancers are currently recognised by the National Cancer Institute (http://www.cancer.gov/types/). Some cancers are capable of spreading from their tissue of origin to distant body parts in a process called metastasis. Despite their metastatic ability, cancers are mainly categorized based on their tissue and/or organ of origin. For example, cancers that begin in tissues that line or cover body organs are known as carcinomas. Sarcomas, melanomas, lymphomas, and leukemias are other well-known examples of cancers. It is worth noting that not all cancers are tumorous, for example, blood cancers such as leukemia, lymphoma, and multiple myeloma are known to be non-solid cancers. In addition to pathologically categorizing cancers based on their tissues or organs of origin, molecular taxonomies based on recurrent genetic and epigenetic alterations in human tissue have been suggested.(1) Regardless of the cancer type in question, cancers can indeed have devastating effects on affected and surrounding body organs and are thus deemed a global public health issue.
It is estimated that approximately 18.1 million new cancer cases occurred globally in 2018, with 92.8% of these cases occurring in Asia, Europe and the Americas. Of these 18.1 million cases, an estimated 6.56 million individuals were newly diagnosed with lung, breast, colorectal, or prostate cancer.(2) An estimated 9.6 million individuals died from cancer in 2018, with approximately 5.5 million of these deaths solely occurring in Asia. It is estimated that lung, liver and stomach cancer were the most fatal cancers, jointly leading to approximately 3.33 million deaths.(2) Other cancers of note estimated to have caused significant global mortality in 2018 are breast and colorectal cancers. They jointly caused a total of about 1.18 million deaths. When compared to their counterparts in developing regions, the cumulative risk of dying from cancer was 7.55% higher in men in developed regions. However, there was no notable difference in the estimated cumulative risk of dying from cancer between women in developing and developed regions.(2) Overall, cancer mortality and incidence trends in 2018 were shown to substantially vary at country and regional levels.(2)
Similar to global cancer mortality and incidence trends, cancer risk factors can often vary by region and country. A wide range of factors including genetic background, age, smoking, alcohol consumption, body weight, diet, exposure to pollutants, and microbial infection have been linked to the risk of developing certain cancer types. Of these risk factors, carcinogenic microbial infection stands out. Together, infectious microbial agents were responsible for causing 2.2 million new cancer cases in 2012.(3) Interestingly, the link between microbial infection and cancer was hypothesized and investigated as early as the nineteenth century.(4) Today, approximately 16% of the global cancer burden is attributed to microbial infections. (3, 5) Curiously, not only can bacteria and other microbes enhance the risk of getting cancer but they can also enhance its treatment.(6) In fact, infection mediated cancer therapy is an age-old therapy that pre-dates the seventeenth century discovery of microorganisms and can be traced to as early as 2600 BC. (7, 8)
Aside from infection mediated cancer therapy, several therapeutic options are currently available to cancer patients. These therapeutic options can vary greatly based on a host of factors, including the locations of the cancer, its size, and the patient's health status. Together, cancer therapies can be broadly classified into two main types: systemic and localised therapies. In cases of metastasized cancers, systemic therapies are often the treatment of choice. Systemic therapies refer to treatments that target the entire body through the bloodstream. Three well-known examples of systemic therapies include chemo-, hormone-, and immuno- therapy. Like everything else, each of these therapies has its advantages and limitations. Generally speaking, one of the main advantages in using systemic therapies is that resection can at times be avoided. On the other hand, systemic therapies can often have unintended consequences on healthy tissues and organs. Furthermore, these treatments rely mostly on blood circulation and could have limited effectiveness in quiescent tumor regions with limited vascularizetion.(9) Unlike systemic therapies, localised therapies have limited effectiveness against metastasized cancers. However, they can instantly limit cancer or its symptoms by reducing the mass effect of tumors, and in some cases, they can cure cancer. Furthermore, localised therapies, such as surgeries, are able to remove cancers from the body which are inaccessible to certain chemotherapies, the brain for example. Despite recent and impressive advances, there are still many unmet needs in cancer therapeutics.
Coupled to revolutionary cancer therapies such as robot assisted surgery, checkpoint inhibitors and monoclonal antibodies,(10, 11) the scientific community has witnessed the renaissance of bacterium-mediated cancer therapy (BMCT) over the last two decades.(9, 12-26) However, despite notable advances made in improving BMCT in recent years, most bacterial candidates have yet to attain satisfactory efficacy, effectiveness, or safety. The current but rather few therapeutic bacterial candidates are relatively limited in tumor cytotoxicity, immunogenicity, chemotacticcity, or safety. Furthermore, the possibility of an occurrence of septicemia caused by antibiotic resistance and/or the reversion of attenuated pathogenic phenotypes in these candidate bacteria are still causes for concern. Similarly, the impairment of therapeutic effectiveness due to previous bacterial immunization is also possible. As a result, the search for optimal bacterial candidates is still ongoing.
Recent advances in environmental microbiology, in particular, our understanding of microbiomes from diverse ecological niches are contributing to the interest in and the renaissance of BMCT. It is estimated that there may be 1 trillion microbial species on earth, approximately 99% of which cannot be cultured.(27) The candidate pool is therefore huge. Could it be possible that some of these microbes, including the uncultivable ones, are potential therapeutic bacterial candidates? With so many prospective candidates to consider but limited resources to screen every single bacterium, this paper aims to provide a necessary practical guide to screening and identifying bacteria with significant cancer therapeutic potential.
A brief history of bacteria-mediated cancer therapy
Although it was implicitly used prior to the nineteenth century, BMCT was only explicitly used and brought to the forefront of cancer therapeutics in 1891 by Dr. William Coley.(8, 28) Dr. Coley, a bone surgeon, used both attenuated and unattenuated mixtures of Streptococcus pyogenes and Serratia marcescens to treat sarcoma patients. In spite of his relative success in treating inoperable sarcomas, his treatments, known as Coley's toxins, were met with much skepticism due to their inconsistencies and the extent of their side effects. Also, the emergence of radiotherapy at the time provided a less controversial therapeutic option for cancer treatment.(28, 29) Despite the initial drawbacks and skepticism, research on BMCT persisted. Clostridium filtrates and spores were used in cancer treatment for the first time approximately half a century later in 1935 and 1947. (30, 31) In 1988, the very first recombinant Clostridium was developed for BCMT,(32) that was followed by the development of an auxotrophic Salmonella about a decade later in 1997.(33) The year 2002 marked a monumental milestone in the field of BMCT as the very first clinical trial in recent times was carried out.(34) Despite the limited success of the aforementioned clinical trial, the field of BMCT has generated an unprecedented amount of interest, mainly due to the abundance of potential microbial candidates and the diversity of recombinant DNA techniques being used to further explore relevant bacterial traits. (9-24)
To facilitate BMCT research, we sought to answer the following questions in this paper: (1) what makes a bacterium a good BMCT candidate? (2) How would a good BMCT prospect be identified? (3) How can different BMCT candidates be compared? Building on previously published literature, we discuss key bacterial traits useful in screening for new or better prospects.
Tips necessary for screening promising bacteria prospects
Traditionally, cancer therapeutic bacterial screening was solely a wet lab experimental process. However, recent advances to our understanding of bacterial genomes have added a secondary dimension to the process of cancer therapeutic bacterial screening. Presently, there are over 22,000 complete bacterial genome sequences deposited in the National Center for Biotechnology Information's genome database. In silico analyses of these and other data can significantly speed up the screening for cancer therapeutic bacteria. Ideally, a good BMCT prospect should show: (1) cancer selectivity, (2) cancer cytotoxicity and/or immunogenicity, (3) limited toxicity to normal cells, and (4) stability within the human body. Although many bacteria are known to secrete a wide array of cancer cytotoxic substances,(6) little is known about the genes responsible for synthesizing and secreting these substances. Similarly, some bacteria are known to hinder tumor growth through inflammasome and effector T-cell pathways,(6) but little attention has been given to genes necessary for their cancer immunogenicity. Aside from their inherent tumor cytotoxicity and immunogenicity, bacteria can also be used as vectors for the delivery of other anti-cancer drugs as they are able to localise in hypoxic tumor regions. (6, 35) However, to make BMCT more successful, many questions need to be further addressed, including (1) what makes bacteria localise within tumors? (2) Are there niche-specific genes necessary for preferential tumor growth? (3) How can a bacterium prospect with tumor specificity, cytotoxicity, immunogenicity and stability be mined? (4) Does such a bacterium even exist naturally? Or, can it be engineered? And (5) are there better cancer therapeutic bacteria combinations that could improve patient clinical outcomes?
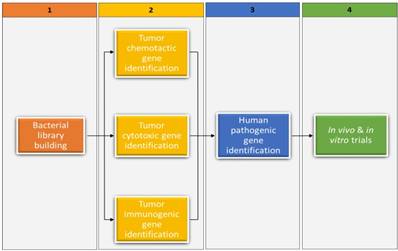
The rationale for further bacterial screening is embedded in the above questions and the fact that therapeutic bacterial candidates have been identified for only a few of the approximately 200 existing cancer types. In Figure 1, we suggest a screening framework that entails mining genes important to bacterial cytotoxicity, chemotacticity, immunogenicity, and pathogenicity in currently available therapeutic bacterial candidates, followed by finding analogs or orthologs to those genes in prospect candidates. Fortunately, most present-day cancer therapeutic bacterial candidates have been sequenced and some genes pertaining to cancer therapeutics have been reported. For example, the niche-specific genes necessary for preferential growth of Salmonella typhimurium in solid tumors have been identified.(36) In light of the suggested framework, we have provided below a nonexclusive list of cancer cytotoxic bacteria reported in literature, highlighting the cancer's cytotoxic substance they produce and their synthesizing gene(s). We have also provided a list of cancer immunogenic and chemotactic bacteria, highlighting relevant genes whenever possible. It is worth noting that in currently available therapeutic bacterial candidates whose relevant gene clusters are unknown, bioinformatical methods such as gene network, probabilistic algorithms, and metagenomic islands can be used to predict gene function. (37-40) Once predicted, algorithms such as BLAST can be used to identify analogs or orthologs in bacterial libraries. Our suggested framework could be valuable in advancing BMCT, especially in the current genomic era. More importantly, it could prove to be valuable to new BMCT researchers, including experienced non- BMCT researchers in possession of extensive bacterial genomic resources.
Cancer cytotoxic traits
Cancer cytotoxic traits are attributes that enable bacteria to secrete substances that are toxic to cancerous cells. Given the number of currently identified cancer cytotoxic bacteria, advancing BMCT requires identifying therapeutic bacterial candidates with better selective cytotoxicity and/or other useful cancer therapeutic traits. In view of characterizing cytotoxic gene analogs and orthologs in prospective candidates, we provided a non-exhaustive list of previously identified cancer cytotoxic bacteria, their secreted substances, the chemical nature of these substances, and their synthesizing genes (Table 1). The contents of this table could be useful to researchers seeking to identify the genes responsible for the secretion of certain cancer cytotoxic substances and to those screening necessary genes for bacterial vectors. As shown in Table 1, the main focus of this review was to demonstrate the diversity of bacteria capable of secreting anti-cancer substances. Consequently many compounds produced by Actinomycetes and their corresponding biosynthetic genes were omitted. Furthermore, although toxicities to normal cells are not shown in Table 1, they can be referenced accordingly. We note that many bacteria with anti-cancer abilities also produce antimicrobial substances. We hereby encourage researchers working in the field of antimicrobial development to screen their current bacterial libraries for potential anti-cancer activities.
Within the context of identifying bacteria with better or new cancer cytotoxic traits, uncultivable bacteria have been under-explored and represent a huge potential source of anti-cancer substances. This is mostly attributed to our failure to reproduce important aspects of their natural environments under laboratory conditions. It has been previously shown that uncultivable microbes constitute the majority of bacterial genetic diversity in nature and could represent an important BMCT source. (41) The framework suggested in Figure 1 takes into account uncultivable bacteria; however, step 4 for uncultivable bacteria involves processes such as partial genomic digestion and cloning of functional gene fragments into appropriate vectors. Amongst other considerations, isolation and purification of metagenomic DNA from soil, fragment size selection, and choice of vector and host have been previously discussed by Dr. Robin Pettit.(41) Similar and more recent techniques such as primer restriction are also applicable to metagenomic DNA from aquatic environments. More importantly, aside from cloning functional genes belonging to uncultivable bacteria, growing uncultivable bacteria can be achieved through recent advances in bacterial culture, including co-culture with other bacteria, recreating bacterial environment in laboratories, and combining these approaches with micro-cultivation technology.(42)
Cancer immunogenic traits
Cancer immunogenic traits are attributes that enable certain bacteria to induce human immune responses against cancerous cells. Bacteria's inherent ability to elicit the immune system makes them crucial to BMCT. Immune system stimulation within the context of cancer treatment can be achieved in several ways including: (1) inflammasome activation, (2) tumor associated macrophage repolarization, (3) tumor-associated myeloid derived suppressor cell alteration and, (4) effector T-cell responses. (6, 170-172) Of major interest to the process of cancer therapeutic bacterial screening is identifying previously studied immunogenic bacteria and genes necessary for their immunogenicity. In Table 2, we provided a non- exclusive list of cancer immunogenic bacteria, their respective immunogen(s) and synthesizing gene(s). Furthermore, within the context of exploring uncultivable bacteria, analogous or orthologous immunogenic genes from uncultivable bacteria can be cloned into suitable vectors and tested in vivo, or grown by means of recent advances in bacterial culture.(42)
A four-step framework for screening and identifying bacteria with cancer therapeutic potential. Bacterial library building within the context of this review entails all procedures involved in obtaining various bacterial DNAs (prospective candidates) from diverse sources (step 1). The procedure depicted in step 2 entails identifying genes important to bacterial cytotoxicity, chemotacticity, and immunogenicity in currently available therapeutic bacterial candidates, and then identifying the analogs or orthologs of those genes in prospective candidates. Step 3 identifies whether the prospects from step 2 are pathogenic to humans that need attenuation. Lastly, step 4 tests the prospects from step 3 in vivo and in vitro trials, including in cell lines that have not been previously investigated.
Representative list of bacteria with cancer cytotoxic traits
| Bacteria | Cytotoxic substance | Chemical nature | Active against | Synthesizing gene(s) | Growth Inhibition | Reference(s) |
|---|---|---|---|---|---|---|
| Actinoalloteichus cyanogriseus | Caerulomycin F-K | Bipyridines | K562, HL-60 (leukemia), KB(epidermoid carcinoma), and A549 (alveolar adenocarcinoma) | The caerulomycin A gene cluster is known | IC50 = 0.37 and 25.7 μM | (43, 44) |
| Actinomadura sp. | Chandrananimycin A-C | Phenoxazines | CCL HT29 (colon carcinoma), MEXF 514L (melanoma), LXFA 526L (lung carcinoma), LXFL 529L (lung carcinoma), CNCL SF268, LCL H460, MACL MCF-7 (breast carcinoma), PRCL PC3M, and RXF 631L (kidney tumor) | The chandrananimycin gene cluster | IC70 values down to 1.4μg/mL | (45, 46) |
| Actinomadura verrucosospora | Esperamicin A | Enediyne | B16-F1O (melanoma), HCT116, MOSER (colorectal carcinoma), H298, and SW900 (lung adenocarcinoma) | Although unknown in A. verrucosospora, esperamicin A gene analogs have been localised in Actinomadura madurae and Streptomyces carzinostaticus | IC50 =0.3-8.3ng/mL | (47, 48) |
| Actinomyces sp. | Chlorinated dihydroquinones | Terpene | Human colon adenocarcinona (HCT-116) | Unknown | IC50 = 0.97-2.40μg/mL | (49) |
| Bacillus amyloliquefaciens | Exopolysaccharide | Polysaccharide | MC-4 and SGC-7901(gastric carcinoma) | Unknown | IC50 = 19.7 and 26.8µg/µL | (50) |
| Bacillus licheniformis, Nocardiopsis alba, Enterobacter cloacae and several other bacteria including Escherichia coli, Pseudomonas otitidis, Streptomyces sp., Erwinia carotovora etc. | L-asparaginases | Peptide | Jurkat clone E6-1, K-562, HL-60, MOLT-4 (leukemia), MCF-7, MDA-MB-231 (breast carcinoma), and Caco2(colorectal adenocarcinoma) | The ansA1 and ansA3 genes in Bacillus licheniformis have been identified. The ansA in Nocardiopsis alba NIOT-VKMA08 is partially analogous to that of several unreported actinobacteria | IC50 = 0.15-11.2 IU/mL | (51-55) |
| Bacillus mojavensis | Iso-C16 fengycin B, and anteiso-C17 fengycin B | Lipopeptides | HL-60 (leukaemia) | Although unknown in B.mojavensis , the fengycin gene cluster in sister species Bacillus subtilis have been localised | IC50 = 1.6 and100µM | (56, 57) |
| Bacillus silvestris | Bacillistatin 1 and 2 | Peptide | P388 (leukemia), BXPC-3 (pancreatic carcinoma), MCF-7 (breast carcinoma), SF-268 (glioblastoma), NCI-H460(lung cancer), KM20L2 (colon cancer), and DU-145 (prostate cancer) | Unknown | IC50 = 0.26-13 ng/mL | (58) |
| Bacillus sp. | Mixirin A-C | Acylpeptide | HCT116 (colorectal carcinoma) | Unknown | IC50 = 0.65-1.6 µg/ml | (59) |
| Bacillus sp. | Halobacillin | Peptide | HCT116 (colorectal carcinoma) | Unknown | IC50 = 0.98 µg/mL | (60) |
| Bacillus subtilis and Streptomyces albulus | Epsilon-poly-L- lysin (ε-PL) | L-lysine homopolymer | HeLa (cervical adenocarcinoma), and HepG2 (hepatocellular carcinoma) | The Pls gene | The culture supernatant of Bacillus subtilis SDNS inhibited 56.2 and 77.2 % of HepG2 and HeLaS3 after 72 hrs. The IC50 for S. albulus derived ε-PL against HepG2 is 13.49 and 8.664 µg/mL at 24 and 48h, respectively | (61-63) |
| Brevibacillus sp. | Laterosporulin 10 | Peptide | MCF-7(breast cancer), HEK293T (embryonic kidney cancer), HT1080 (fibrosarcoma), HeLa (cervical carcinoma), and H1299(lung carcinoma) | The laterosporulin10 gene cluster | A minimum of 70% cytotoxicity was observed at 10 μM in all tested cells | (64, 65) |
| Chromobacterium violaceum | Romidepsin (FK228) | Peptide | In over 20 cell lines including nueroblastoma, T-cell lymphomas, lung, mammary, stomach, and colon adenocarcinoma. | The romidepsin gene cluster | IC50 = 0.3-6.3 ng/mL | (66-69) |
| Clostridium botulinum | Botulinum neurotoxin type A | Peptide | T47D (breast cancer), PC-3, LNCaP, (prostate cancer), and SH-SY5Y(neuroblastoma) | The boNT/A gene | IC50 = 0.54-300 nM | (70, 71) |
| Corynebacterium diphtheriae | Diphtheria toxin | Peptide | MCF 7(breast carcinoma), H295R (adrenocortical carcinoma), HeLa (cervical adenocarcinoma), CTCL (cutaneous T cell lymphomas), U118MG, U373MG, and U87MG(glioblastomas) | The tox gene | IC50 =0.55-2.08 µg/mL | (70, 72) |
| Dermacoccus abyssi | Dermacozine F and G | Phenazine | K562 (leukemia) | Unknown | IC50 = 7 and 9 μM | (73) |
| Enterococcus sp. | Enterococcal anti-proliferative peptide | Peptide | MDA-MB-231(breast adenocarcinoma), HeLa(cervical adenocarcinoma), and AGS (gastric adenocarcinoma) | Unknown | 29.1-38.4% reduction in proliferative activity | (74, 75) |
| Escherichia coli | Colicin A and E1 | Peptide | MCF7, MDA-MB-231, ZR75, BT474BT549, SKBR3, T47D (breast carcinoma), SKUT-1 (leiomyosarcoma), HOS (osteosarcoma), and HS913T (fibrosarcoma) | The caa and cea genes | Inhibition of cellular growth was 17-40% for Colicin E1 and 16-56% for Colicin A | (76, 77) |
| Escherichia coli | Cytosine deaminase (often used with 5-fluorocytosine) | Peptide | A549 (alveolar adenocarcinoma), C6, U251 (glioma), HCT116 (colorectal carcinoma), and DU145 (prostrate carcinoma) | The codA gene | IC50 =0.3mM - 30 mM | (78-81) |
| Geitlerinema sp. | Ankaraholide A | Macrolide | NCI-H460 (lung cancer), Neuro-2a (neuroblastoma), and MDA-MB-435(breast cancer) | Unknown | IC50 = 8.9-262 nM | (82) |
| Klebsiella pneumoniae | Microcin E492 | Peptide | RJ2.25 (B-lymphoblastoid cells), HeLa (cervical adenocarcinoma), and Jurkat (acute T cell Leukaemia) | The microcin E492 gene cluster | 4 ± 3 to 57 ± 11% cell survival | (83, 84) |
| Lactococcus lactis | Nisin A | Peptide | UM-SCC-17B, UM-SCC-14A, HSC-3 (head and neck carcinoma), MCF 7(breast carcinoma), HepG2 (hepatocellular carcinoma), and Jurkat (T cell leukaemia) | The nisin A gene cluster | IC50 =105.5-225 µM | (70, 85) |
| Listeria monocytogenes | Listeriolysin O | Peptide | SKBR-3, MCF 7(breast carcinoma), and Jurkat (T cell leukaemia) | The hly gene | IC50 = 50 pM to 0.1 nM | (70, 86) |
| Lyngbya majuscula | Aurilides B & C | Depsipeptide | NCI-H460 (lung cancer) and neuro-2a (neuroblastoma) | Unknown | IC50 = 0.01-0.13 μM | (87) |
| Lyngbya majuscula | Hermitamide A and B | Alkaloid | CCL-131(neuroblastoma) | Unknown | 2.2-5.5 μM | (88) |
| Lyngbya majuscula | Lyngbyabellin E-I | Peptide | NCI-H460 (lung tumor) and neuro-2a (neuroblastoma) | Unknown | IC50 = 0-2-4.8 μM | (89) |
| Lyngbya semiplena | Wewakpeptin A and B | Peptide | H460 (lung tumor) and neuro-2a (neuroblastoma) | Unknown | IC50 =0.2-0.65 μM | (90) |
| Marinispora sp. | Marinomycin A-C | Polyketide | NCI's panel of 60 cancer cell lines | Unknown | Mean IC50 = 0.2-2.7μM | (91) |
| Mechercharimyces asporophorigenens | Urukthapelstatin A | Peptide | A549, DMS114, NCIH460(lung cancer), MCF-7 (breast cancer), and HCT-116 (colorectal carcinoma) | Unknown | log GI50 = 3.5-5.2 nM | (92) |
| Microbispora rosea | Hibarimicin A-D | polyketide | 16-F10(skin melanoma) and HCT-116(colorectal carcinoma) | Unknown | IC50 = 0.7-3.6 µg/mL | (93) |
| Microcystis aeruginosa | Microcyclamide | Peptide | P-388 (leukemia) | The microcyclamide gene cluster | IC50 = 1.2 μg/mL | (94, 95) |
| Micromonospora sp. | IB-96212 | Polyketide | P-388 (leukemia), A-549(alveolar adenocarcinoma), HT-29 (colorectal adenocarcinoma), and MEL-28(melanoma) | Unknown | IC50 = 0.0001-1 µg/mL | (96) |
| Micromonospora sp. | Arisostatin A and B | Tetrocarcin group spirotetronate | Myeloid leukemia U937 | The tetrocarcin A gene cluster in M. chalcea NRRL 11289 has been localised | IC50 = 0.4 and 4 μg/mL | (97, 98) |
| Nocardia dassonvillei | N-(2-hydroxyphenyl)-2-phenazinamine | Phenazine | HepG2(hepatocellular carcinoma), A549(alveolar adenocarcinoma), HCT-116 (colorectal carcinoma), and COC1(ovarian cancer) | Unknown | IC50 = 28.11 and 40.33 μg/mL | (99) |
| Nocardiopsis lucentensis | Lucentamycin A and B | Peptide | HCT116 (colorectal carcinoma) | Unknown | IC50 = 0.2 and 11µM | (100) |
| Nostoc sp. | Cryptophycins-1 | Depsipeptide | KB(cervical carcinoma), B16V(murine melanoma), and LoVo (colon carcinoma) | The cryptophycin gene cluster | IC50 =3 pM -I.3nM | (101-103) |
| Paenibacillus profundus | Heptapeptide | Peptide | SK-MEL-28 (human melanoma) | Unknown | IC50 = 3.07µM | (104) |
| Pediococcus acidilactici | Rec-pediocin | Peptide | HepG2 ( hepatocarcinoma), HeLa (cervical adenocarcinoma), MCF7 (mammary gland adenocarcinoma), and Sp2/0-Ag14 (spleen lymphoblast) | The CP2 gene | 25µg/ml of rec-pediocin reduced cell viability by at least 89% in all cell line tested | (105, 106) |
| Pelagiobacter variabilis | Pelagiomicin A | Phenazine | HeLa (cervical carcinoma) | The pelagiomicin gene cluster | IC50 = 0.04-0.2 μg/mL | (107, 108) |
| Pseudomonas aeruginosa | Exotoxin A | Peptide | PaCa-2 (pancreatic cancer, FEMX, Melmet-1, Melmet-5, Melmet-44, MelRM, MM200 (melanomas), Daudi, CA46 (Burkitt's lymphoma), EHEB, MEC1(leukemias), head and neck squamous carcinomas | The PE gene | 0.3-8.6 ng/mL | (70, 109) |
| Pseudomonas aeruginosa | Azurin | Peptide | MCF7, MDA-MB-157, T- 47D and ZR-75-1 (breast carcinoma) HCT-116 (colon adenocarcinona) and UISO-Mel-2 (melanoma) | The azurin gene | IC50 = 32 and 53 μM for MCF-7 and MDA-MB-157, respectively. In T- 47D and ZR-75-1, IC50 = 72± 3µg/mL. Approximately 40% of HCT-116 were inhibited by 10ug/uL | (110-113) |
| Pseudomonas aeruginosa | Pyocin S2 | Peptide | HepG2 (hepatocellular carcinoma) and Im9 (lymphoblast myeloma), | The pys2 gene | IC50 = 3.5- 50 U/mL for both crude and purified pyocin S2 in HepG2 and Im9 | (114, 115) |
| Pseudomonas aeruginosa, Sphingobacterium sp., Streptococcus pyogenes | Arginine deiminase | Peptide | HCT-116(colon adenocarcinona), K-562(leukemic cell lines),PC-3(prostrate carcinoma), T47D(breast carcinoma) HROG02, HROG05, HROG10, and HROG17 (Glioblastoma) | The arcA gene in Pseudomonas aeruginosa and Streptococcus pyogenes have been identified. | IC50 in Streptococcus pyogenes is ~ 35mU/mL in HROG05 cell lines; in Pseudomonas aeruginosa PS2 it ranges from 0.8-1.4 IU/mL in various cell lines, while in Sphingobacterium sp. the same ranges from 0.8-1.8 IU/mL | (116-119) |
| Pseudomonas chlororaphis subsp. aurantiaca | Phenazine-1-carboxylic acid | Phenazine | HCT-116 (colon adenocarcinona) | The phenazine gene cluster (phzXYFABCD genes) | IC50 =15.6 μM | (120,121) |
| Pseudomonas sp. | DOB-41 | Phenazine | Leukemia P388 in mice. | Unknown | T/C: 113-153% | (120) |
| Saccharothrix aerocolonigenes | Bromo analog of rebeccamycin | Indolocarbazole | P388(murine lymphocytic leukemia) | The rebeccamyacin gene cluster | T/C: 115-165% | (121, 122) |
| Salinispora arenicola | Arenamides A-C | Peptide | HCT-116 (colorectal carcinoma) | Unknown | IC50 = 13.2 and 19.2 μg/mL | (123) |
| Salinispora tropica | Salinosporamide A | Polyketide | NCI's 60-cell-line panel and HCT-116 (colorectal carcinoma) | The sal gene cluster | Mean GI50 (NCI's cell-line) = < 10 nM and IC50 = 11 ng/mL in HCT-116 | (124, 125) |
| Salmonella enterica and Escherichia coli | Cytolysin A | Peptide | CT26 (colon cancer) | ClyA gene | 4.5 × 107 colony-forming units of Salmonella typhimurium vector suspended in 100 μL of PBS mediated complete tumor eradication. | (126, 127) |
| Serratia marcescens | Prodigiosin | Pyrrolylpyrromethane | A2780, A2780RCIS (ovarian cancer), EPG85-257, EPG85-257RDB, EPG85-257RNOV(gastric carcinoma),SW-620 (colorectal cancer), HeLa (cervical adenocarcinoma), Hep2 (laryngeal carcinoma), B-CLL (leukemia), LNCaP, DU145(prostate cancer) and MCF-7 (breast carcinoma) | The pig cluster | IC50 = ~116 nM -127 μM | (128-133) |
| Serratia surfactantfaciens | Serrawettin W2 | Cyclic lipopeptide | Hela (cervical carcinoma) and Caco2 (colorectal adenocarcinoma) | A hybrid polyketide synthases-non-ribosomal peptide synthetases gene cluster | IC50= 20.9 and 54.1 μM in the Hela and Caco2 cell line, respectively | (134) |
| Sorangium cellulosum | Epothilone A &B | Lactones | CCRF-CEM/VBL100 (multidrug-resistant lymphoblastic leukemia) | The epothilone gene cluster | IC50 = 0.02 and 0.002 μM for epothilon A and B in CCRF-CEM/VBL100 | (135-137) |
| Staphylococcus aureus | α-hemolysin | Protein | MCF-7 (mammary carcinoma) | The α-hemolysin gene | A reduction in cell viability of 7.1% per min and a saturation constant of 0.14 | (138, 139) |
| Streptococcus pneumoniae | Pep27anal2 (pep27 analog) | Peptide | AML-2, HL-60, Jurkat (leukemia), SNU-601 (gastric cancer), and (MCF-7) breast cancer | The Pep27 gene | IC50 = <10-29 μM | (140, 141) |
| Streptococcus bovis | Bovicin | Peptide | HepG2 (hepatocellular carcinoma) and MCF 7(breast carcinoma) | The bovicin gene cluster | IC50 = 279.39 and 289.3 µM in MCF-7 and HepG2 | (142, 143) |
| Streptomyces verticillus | Bleomycin | Polyketide | Has been tested against over 30 different cancer type including leukemias, lymphomas, myelomas, and carcinomas | The bleomycin gene cluster | IC50 =25.2 nM - 2.93 mM | (70, 144) |
| Streptomyces peucetius var. caesius | Doxorubicin (Adriamycin) | Anthracycline | Tested in over 900 cell types including various carcinomas, sarcomas, melanomas, lymphomas, and leukemias | The doxorubicin gene cluster | IC50 = 0.0044 - 44.7µM | (70, 145, 146) |
| Streptomyces caespitosus or Streptomyces lavendulae | Mitomycin C | Aziridine | Tested in over 900 different cell lines | The mitomycin C gene cluster | IC50 = 0.00948-249 μM | (147, 148) |
| Streptomyces griseofuscus | Azinomycin A & B | Aziridine | L5178Y (leukemia) | The azinomycin gene cluster | IC50 = 0.07 and 0.11μg/mL for azinomycins A and B, respectively | (149) |
| Streptomyces iakyrus | Actinomycin G2 | Chromopeptide | HM02 (gastric adenocarcinoma), HepG2 (hepatocellular carcinoma), and MCF7 (breast adenocarcinoma) | The actinomycin G gene cluster | IC50= 0.0013 - 0.0039 μM | (150, 151) |
| Streptomyces macromomyceticus | Auromomycin | Polypeptide | Ehrlich ascites (carcinoma), ascites sarcoma 180, L1210 (leukemia), and LEWIS lung carcinoma | The macromomycin gene (auromomycin apoprotein) has been sequenced | IC50 =3mg/kg | (152, 153) |
| Streptomyces peucetius | Daunorubicin | Anthracycline | L3.6 (pancreatic carcinoma) and HeLa (cervical adenocarcinoma) | The daunorubicin gene cluster | IC50 =0.02 - 0.4µM | (146, 154, 155) |
| Streptomyces sp. | Chromomycin | Polyketide | Wide range of cancer cells including ovary, breast, prostrate, pancreas, skin, lung, | The chromomycin gene cluster | IC50 =0.26 nM - >50µM | (156-159) |
| Streptomyces sp. | IT-62-B | Baumycin group anthracycline | LI210 (murine leukemia cells), P388(murine lymphocytic leukemia), P388/ ADR (doxorubicin resistant), and KB(human nasopharyngeal carcinoma) | The baumycin gene cluster (dox) within the Streptomyces genus is known | IC50 = 0.006-0.04 μg/mL | (160, 161) |
| Streptomyces sp. | Diketopiperazine derivative | Piperazine | HCT-116 (colon adenocarcinona) and HepG-2(hepatocellular carcinoma) | Type II polyketide gene cluster | IC50=3.3 and 1.1 µg/mL against HCT-116 and HepG-2 cell lines | (162) |
| Streptomyces sp. | SF2575 | Tetracycline | P388(murine lymphocytic leukemia) | Unknown | IC50 = 7.7 ng/mL | (163) |
| Streptomyces sp. | 1, 2- benzene dicarboxylic acid, mono 2- ethylhexyl ester | Dicarboxylic acid | HepG 2 (hepatocellular carcinoma) and MCF-7 (breast adenocarcinoma) | Unknown | IC50 = 42 and 100 μg/mL for HepG 2 and MCF-7, respectively | (164) |
| Streptomyces sp. | 2-bromo-1-hydroxyphenazine | Phenazine | Human colon adenocarcinona (HCT-116) | Unknown | IC50 = 0.1 μM | (165) |
| Streptomyces violaceochromogenes | Arugomycin | Anthracycline | Sarcoma S-180 | Unknown | I.7mg/kg of interperi toneal injection | (166) |
| Symploca sp. | Belamide A | Tetrapeptide | HCT-116 colon cancer | Unknown | IC50=0.74-1.6 μM | (167) |
| Thermoactinomyces sp. | Mechercharmycin A (Mechercharstatin) | Peptide | A549 (lung cancer), and Jurkat (T cell leukaemia) | Unknown | IC50 = 0.04-0.046µM | (168) |
| Verrucosispora spp. | Proximicin A-C | Peptide | MCF-7(breast carcinoma) AGS (gastric adenocarcinoma), and HepG2 (hepatocellular carcinoma) | Unknown | GI50 = 0.25-9.5 μg/mL | (169) |
IU: the amount of enzyme that liberates 1 µM; T/C: the ratio of mean survival days of the treated group divided by that of the control group; IC50: the drug concentration that inhibits biological activity by 50%; GI50: the drug concentration that inhibits the growth of cancer cells by 50%.
Traits necessary for preferential tumor growth
The most important and well explored bacterial traits for efficient BMCT are the abilities to differentiate cancerous cells from healthy cells or recognize the peculiar bacterial growth environment provided by the cancer cells. This is typically achieved by bacteria recognizing specific chemical signals emitted by cancer cells. Due to recent advances in recombinant DNA techniques, these traits have been leveraged by engineering bacterial vectors for the precise delivery of diverse anti-cancerous proteins to tumors. This has been mostly achieved by cloning genes coding for diverse anti-cancerous proteins into bacterial vectors, including those coding for immunogenic antigens, cytokines, cell cycle check-point inhibitors, antibodies, and cytotoxic agents. (186) Although bacterial vector systems are typically designed to enable the direct expression of these anti-cancerous proteins, they have also been used for bactofection or gene transfer to mammalian cancer cells. (186) Within the context of cancer therapeutic bacterial screening, we provided examples of bacteria that are currently used as vectors for the delivery of diverse anti-cancerous molecules (Table 3). We however note that, for most of these bacterial vectors, genes responsible for preferential growth around tumor environment are either unreviewed or unknown. Advancing BMCT within the context of bacterial screening would not only entail identifying these genes in currently used vectors for the delivery of diverse anti-cancerous molecules, but also randomly screening both facultative and obligate anaerobes in order to identify new prospects with better tumor discriminatory abilities or other useful cancer therapeutic traits in addition to their tumor discriminatory abilities. This is particularly important as the future of BMCT hinges on bacteria with improved tumor discriminatory abilities.
Representative list of bacteria with cancer immunogenic traits
| Bacteria | Immunogen(s)/ Immunogenic properties | Active against | Synthesizing gene(s) | Reference(s) |
|---|---|---|---|---|
| Clostridium novyi | Phospholipases | Sarcomas, gliomas, squamous and colon carcinomas | The phospholipase C gene (NT01CX0979) | (170, 173, 174) |
| Escherichia coli | Lipopolysaccharide | Colon and breast carcinomas | Lipid A, core polysaccharide, and O-antigen genes | (175, 176) |
| Mycobacterium bovis | α antigen | Cervical adenocarcinoma and bladder cancer | The α antigen gene | (177-179) |
| Streptococcus pyogenes | Emm55, speA, speB and speC | Pancreatic carcinoma and lymphosarcoma | speA, speB, speC, and Emm55 genes | (180-182) |
| Listeria monocytogenes | Listeriolysin O | Ovarian and breast carcinomas. | The hly gene | (171, 172, 183, 184) |
| Salmonella typhimurium | Lipopolysaccharide/ survival within macrophages | Colon carcinoma | Lipid A, core polysaccharide, and O-antigen genes; slyA, STM3120 and htrA genes | (16, 185) |
Representative list of bacteria that are known to preferential accumulate in tumors
| Bacteria | Comments | Effective against | Important gene(s) | Reference(s) |
|---|---|---|---|---|
| Bifidobacterium longum | At 168 hours, tumors had 60,000 bacilli per gram of tumor tissue in contrast to no germination in livers, spleens, kidneys, or lungs. | Diverse solid tumors including B16-F10 melanoma and Lewis lung carcinoma | Unknown | (187) |
| Clostridium novyi | Within 16 hours the bacterium had floridly germinated within the tumors in contrast to no germination in livers, spleens, kidneys, lungs, or brains | Diverse solid tumors including HCT116 colon and B16 skin carcinomas | Unknown | (188) |
| Escherichia coli | Preferential accumulated in tumor at a ratio of >108:1 | Diverse solid tumors including gliomas, breast, skin and colon and carcinoma | Unknown | (189, 190) |
| Listeria monocytogenes | Selectively infected, survived and multiplied in tumors; 27 hours after injection, the bacterium was only detected in tumors | Diverse solid tumors including 4T1 mammary and PC-3 prostate tumors | Unknown | (189, 191) |
| Magnetococcus marinus | Aerotactic bacteria with the ability to swim along magnetic field lines. Approximately 55% of the bacterium penetrated into hypoxic regions | HCT116 colorectal carcinoma | Unknown | (192, 193) |
| Salmonella typhimurium | Preferential accumulated in tumor at a ratio of >1,000:1 | Multiple solid tumors | cheY, motAB,and eutC genes | (36, 189) |
| Vibrio cholerae | Selectively infected and multiplied in tumors and metastases, with titres reaching approximately 5.97 × 108 after 32 hours | Bladder carcinomas, gliomas and fibrosarcoma | Unknown | (189) |
Pathogenic traits
The rationale for identifying bacterial pathogenic traits or genes as proposed in our four-step framework (Figure 1), lies in the fact that pathogenicity of the candidate bacteria has been one of the major drawbacks to BMCT. Nonetheless, a significant number of bacteria strains used in BMCT are pathogenic to human. A critical consideration after the in silico screening of bacterial prospects with aforementioned traits should be that of pathogenicity. For instance, Clostridium histolyticum spores preferentially germinate in hypoxic tumor regions, making it an excellent BMCT candidate. However, Clostridium histolyticum can also produce significant amount of exotoxins consequently causing pathophysiological changes to healthy tissue and organs. (31) In an attempt to circumvent some of the issues pertaining to toxicity while maintaining their efficacy, attenuated bacterial strains with less or no toxicity are being used in BMCT. L. monocytogenes, S. typhimurium, and C. novyi are some of the well-known examples of bacterial candidates that have been attenuated to improve safety. (173, 194, 195) For example, the L. monocytogenes cancer vaccine was rendered safer by the deletion of virulence factors such as ActA and Internalin B (ΔactA/ΔinlB), leading to >1,000-fold reduction in toxicity. (173) Similarly, S. typhimurium defective in the synthesis of guanosine tetraphosphate virtually resulted in an avirulent strain.(31) Thus, within the context of advancing BMCT, identifying non- pathogenic analogs to pathogenic strains of key importance is vital to BMCT. Nonetheless, pathogenic bacterial prospects with good BMCT potential cannot be disregarded since there is always the potential for attenuation. However, if issues pertaining to attenuation reversion and septicemia are to be permanently addressed, then identifying non-pathogenic bacteria with aforementioned anti-cancerous traits must be further explored.
Conclusion and perspective
In this review we (1) highlighted bacterial traits that make them good therapeutic candidates for the treatment of cancer, (2) suggested a four-step framework that can be used to identify bacteria with good cancer therapeutic potential, including uncultivable strains, and (3) touched on quantifiable attributes such as growth inhibition, cytotoxicity to normal cells, and preferential accumulation ratio that can be used to compare and contrast important cancer therapeutic traits for BMCT.
The singular most important bacterial trait to cancer therapeutics is their ability to specifically target tumors or cancerous cells. The future of BMCT lies in being able to find bacteria that can target cancerous cells, secrete cytotoxic and/or immunogenic substances, and be tolerated and are stable in the targeted tissue and cancer environment. Recent advances in recombinant DNA techniques significantly advanced BMCT. Engineering a bacterium that targets tumors or cancerous cells, produces cytotoxic or immunogenic proteins, self-propels and responds to triggering signals, senses the local environment and produces externally detectable signals is not so far-fetched anymore.(196) However, despite the overall enthusiasm about the future of BMCT, using multi-layered genetically modified bacteria could result in stability issues. Identifying bacterial prospects with better cancer therapeutic potential, which require minimum genetic modifications, would go a long way to improve BMCT. Lastly, our analyses highlight the need to establish and standardize quantitative methods to identify and characterize bacteria with important cancer therapeutic traits.
Acknowledgements
This work was supported by Mitacs-Accelerate [Fund IT07941-Yuan_OGVG], Ontario Greenhouse Vegetable Growers (OGVG), Agriculture and Agri- Food Canada Growing Forward-II [project J-001589] and Natural Sciences and Engineering Research Council of Canada (NSERC) [Discovery grant RGPIN-2015-06052]. We also thank Michelle Bargel for help with literature search, Nanyikun (Nancy) Zhao and Michelle S.M. Li for critically reading and editing this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929-44
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136(5):E359-E86
3. Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. The Lancet Global Health. 2016;4(9):e609-e16
4. Blackadar CB. Historical review of the causes of cancer. World J Clin Oncol. 2016;7(1):54
5. De Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D. et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The lancet oncology. 2012;13(6):607-15
6. Song S, Vuai MS, Zhong M. The role of bacteria in cancer therapy-enemies in the past, but allies at present. Infect Agents Cancer. 2018;13(1):9
7. Gest H. The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes and Records of the Royal Society. 2004;58(2):187-201
8. Kucerova P, Cervinkova M. Spontaneous regression of tumour and the role of microbial infection - possibilities for cancer treatment. Anti-Cancer Drugs. 2016;27(4):269-77
9. Łukasiewicz K, Fol M. Microorganisms in the Treatment of Cancer: Advantages and Limitations. Journal of immunology research. 2018:2018
10. Voena C, Chiarle R. Advances in cancer immunology and cancer immunotherapy. Discovery medicine. 2016;21(114):125-33
11. Zanagnolo V, Achilarre MT, Maruccio M, Garbi A. Might robotic-assisted surgery become commonplace in endometrial cancer treatment?: Taylor & Francis; 2018.
12. Hoffman RM. Back to the future: are tumor-targeting bacteria the next-generation cancer therapy?. Gene Therapy of Solid Cancers: Springer. 2015 p. 239-60
13. Zhang Y, Zhang N, Zhao M, Hoffman RM. Comparison of the selective targeting efficacy of Salmonella typhimurium A1-R and VNP20009 on the Lewis lung carcinoma in nude mice. Oncotarget. 2015;6(16):14625
14. Hoffman R. Preface: a brief history of bacterial therapy of cancer. Methods in Molecular Biology. 2016:1409
15. Kim JE, Phan TX, Nguyen VH, Dinh-Vu HV, Zheng JH, Yun M. et al. Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1β. Theranostics. 2015;5(12):1328-42
16. Phan TX, Nguyen VH, Duong MTQ, Hong Y, Choy HE, Min JJ. Activation of inflammasome by attenuated Salmonella typhimurium in bacteria-mediated cancer therapy. Microbiol Immunol. 2015;59(11):664-75
17. Murakami T, Igarashi K, Kawaguchi K, Kiyuna T, Zhang Y, Zhao M. et al. Tumor-targeting Salmonella typhimurium A1-R regresses an osteosarcoma in a patient-derived xenograft model resistant to a molecular-targeting drug. Oncotarget. 2017;8(5):8035
18. Murakami T, DeLong J, Eilber FC, Zhao M, Zhang Y, Zhang N. et al. Tumor-targeting Salmonella typhimurium A1-R in combination with doxorubicin eradicate soft tissue sarcoma in a patient-derived orthotopic xenograft (PDOX) model. Oncotarget. 2016;7(11):12783
19. Zheng JH, Nguyen VH, Jiang S-N, Park S-H, Tan W, Hong SH. et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med. 2017;9(376):eaak9537
20. Yamamoto M, Zhao M, Hiroshima Y, Zhang Y, Shurell E, Eilber FC. et al. Efficacy of tumor-targeting Salmonella A1-R on a melanoma patient-derived orthotopic xenograft (PDOX) nude-mouse model. PLoS ONE. 2016;11(8):e0160882
21. Gontero P, Bohle A, Malmstrom P-U, O'Donnell MA, Oderda M, Sylvester R. et al. The role of bacillus Calmette-Guérin in the treatment of non-muscle-invasive bladder cancer. European urology. 2010;57(3):410-29
22. Murphy C, Rettedal E, Lehouritis P, Devoy C, Tangney M. Intratumoural production of TNFα by bacteria mediates cancer therapy. PLoS ONE. 2017;12(6):e0180034
23. Kocijancic D, Felgner S, Schauer T, Frahm M, Heise U, Zimmermann K. et al. Local application of bacteria improves safety of Salmonella-mediated tumor therapy and retains advantages of systemic infection. Oncotarget. 2017;8(30):49988
24. Mehta N, Lyon JG, Patil K, Mokarram N, Kim C, Bellamkonda RV. Bacterial carriers for glioblastoma therapy. Molecular Therapy-Oncolytics. 2017;4:1-17
25. Luo C-H, Huang C-T, Su C-H, Yeh C-S. Bacteria-mediated hypoxia-specific delivery of nanoparticles for tumors imaging and therapy. Nano lett. 2016;16(6):3493-9
26. Fan J-X, Li Z-H, Liu X-H, Zheng D-W, Chen Y, Zhang X-Z. Bacteria-Mediated Tumor Therapy Utilizing Photothermally-Controlled TNF-α Expression via Oral Administration. Nano lett. 2018;18(4):2373-80
27. Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 2016;113(21):5970-5
28. Wei MQ, Mengesha A, Good D, Anné J. Bacterial targeted tumour therapy-dawn of a new era. Cancer Lett. 2008;259(1):16-27
29. McCarthy EF. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. The Iowa orthopaedic journal. 2006;26:154
30. Connell HC. The study and treatment of cancer by proteolytic enzymes: preliminary report. Canadian Medical Association Journal. 1935;33(4):364
31. Parker RC, Plummer HC, Siebenmann CO, Chapman MG. Effect of histolyticus infection and toxin on transplantable mouse tumors. Proceedings of the Society for Experimental Biology and Medicine. 1947;66(2):461-7
32. Schlechte H, Elbe B. Recombinant plasmid DNA variation of Clostridium oncolyticum—model experiments of cancerostatic gene transfer. Zentralblatt für Bakteriologie, Mikrobiologie und Hygiene Series A: Medical Microbiology, Infectious Diseases, Virology, Parasitology. 1988;268(3):347-56
33. Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57(20):4537-44
34. Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ. et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20(1):142
35. Patyar S, Prakash A, Medhi B. Bacteria as a therapeutic approach in cancer therapy. Bacteria and Cancer. 9789400725850: Springer Netherlands. 2012 p. 185-208
36. Silva-Valenzuela CA, Desai PT, Molina-Quiroz RC, Pezoa D, Zhang Y, Porwollik S. et al. Solid tumors provide niche-specific conditions that lead to preferential growth of Salmonella. Oncotarget. 2016;7(23):35169
37. Lehtinen S, Lees J, Bähler J, Shawe-Taylor J, Orengo C. Gene function prediction from functional association networks using kernel partial least squares regression. PLoS ONE. 2015;10(8):e0134668
38. Belda-Ferre P, Cabrera-Rubio R, Moya A, Mira A. Mining virulence genes using metagenomics. PLoS ONE. 2011;6(10):e24975
39. Massjouni N, Rivera CG, Murali T. VIRGO: computational prediction of gene functions. Nucleic Acids Res. 2006;34(suppl_2):W340-W4
40. Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R. et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237-W43
41. Pettit RK. Soil DNA libraries for anticancer drug discovery. Cancer Chemotherapy and Pharmacology. 2004;54(1):1-6
42. Stewart EJ. Growing unculturable bacteria. Journal of bacteriology. 2012 JB. 00345-12
43. Fu P, Wang S, Hong K, Li X, Liu P, Wang Y. et al. Cytotoxic bipyridines from the marine-derived actinomycete Actinoalloteichus cyanogriseus WH1-2216-6. J Nat Prod. 2011;74(8):1751-6
44. Zhu Y, Fu P, Lin Q, Zhang G, Zhang H, Li S. et al. Identification of caerulomycin A gene cluster implicates a tailoring amidohydrolase. Organic letters. 2012;14(11):2666-9
45. Maskey RP, Li FC, Qin S, Fiebig HH, Laatsch H. Chandrananimycins AC: production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. The Journal of antibiotics. 2003;56(7):622-9
46. Hou YH, Wang QF, Ding L, Li FC, Qin S. attB site disruption in marine Actinomyces sp. M048 via DNA transformation of a site-specific integration vector. Biotechnology and applied biochemistry. 2008;50(1):11-6
47. Long BH, Golik J, Forenza S, Ward B, Rehfuss R, Dabrowiak JC. et al. Esperamicins, a class of potent antitumor antibiotics: mechanism of action. Proceedings of the National Academy of Sciences. 1989;86(1):2-6
48. Van Lanen SG, Shen B. Biosynthesis of enediyne antitumor antibiotics. Current topics in medicinal chemistry. 2008;8(6):448-59
49. Soria-Mercado IE, Prieto-Davo A, Jensen PR, Fenical W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J Nat Prod. 2005;68(6):904-10
50. Chen YT, Yuan Q, Shan LT, Lin MA, Cheng DQ, Li CY. Antitumor activity of bacterial exopolysaccharides from the endophyte bacillus amyloliquefaciens sp.isolated from ophiopogon japonicus. Oncol Lett. 2013;5(6):1787-92
51. El-Naggar NE-A, Deraz SF, Soliman HM, El-Deeb NM, El-Ewasy SM. Purification, characterization, cytotoxicity and anticancer activities of L-asparaginase, anti-colon cancer protein, from the newly isolated alkaliphilic Streptomyces fradiae NEAE-82. Sci Rep. 2016;6:32926
52. Mahajan RV, Kumar V, Rajendran V, Saran S, Ghosh PC, Saxena RK. Purification and characterization of a novel and robust L-asparaginase having low-glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS ONE. 2014;9(6):e99037
53. Meena B, Anburajan L, Vinithkumar NV, Shridhar D, Raghavan RV, Dharani G. et al. Molecular expression of l-asparaginase gene from Nocardiopsis alba NIOT-VKMA08 in Escherichia coli: A prospective recombinant enzyme for leukaemia chemotherapy. Gene. 2016;590(2):220-6
54. Husain I, Sharma A, Kumar S, Malik F. Purification and characterization of glutaminase free asparaginase from Enterobacter cloacae: in-vitro evaluation of cytotoxic potential against human myeloid leukemia HL-60 cells. PLoS ONE. 2016;11(2):e0148877
55. Husain I, Sharma A, Kumar S, Malik F. Purification and characterization of glutaminase free asparaginase from Pseudomonas otitidis: Induce apoptosis in human leukemia MOLT-4 cells. Biochimie. 2016;121:38-51
56. Ma Z, Wang N, Hu J, Wang S. Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. The Journal of antibiotics. 2012;65(6):317
57. Steller S, Vollenbroich D, Leenders F, Stein T, Conrad B, Hofemeister J. et al. Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus subtilis b213 and A1/3. Chemistry & biology. 1999;6(1):31-41
58. Pettit GR, Knight JC, Herald DL, Pettit RK, Hogan F, Mukku VJ. et al. Antineoplastic agents. 570. Isolation and structure elucidation of bacillistatins 1 and 2 from a marine Bacillus silvestris. J Nat Prod. 2009;72(3):366-71
59. Zhang HL, Hua HM, Pei YH, Yao XS. Three new cytotoxic cyclic acylpeptides from marine Bacillus sp. Chemical and pharmaceutical bulletin. 2004;52(8):1029-30
60. Trischman JA, Jensen PR, Fenical W. Halobacillin: a cytotoxic cyclic acylpeptide of the iturin class produced by a marine Bacillus. Tetrahedron Lett. 1994;35(31):5571-4
61. El-Sersy NA, Abdelwahab AE, Abouelkhiir SS, Abou-Zeid DM, Sabry SA. Antibacterial and Anticancer activity of ε-poly-L-lysine (ε-PL) produced by a marine Bacillus subtilis sp. Journal of basic microbiology. 2012;52(5):513-22
62. Shi C, Zhao X, Liu Z, Meng R, Chen X, Guo N. Antimicrobial, antioxidant, and antitumor activity of epsilon-poly-L-lysine and citral, alone or in combination. Food & nutrition research. 2016;60(1):31891
63. Geng W, Yang C, Gu Y, Liu R, Guo W, Wang X. et al. Cloning of ε-poly-L-lysine (ε-PL) synthetase gene from a newly isolated ε-PL-producing S treptomyces albulus NK 660 and its heterologous expression in S treptomyces lividans. Microbial biotechnology. 2014;7(2):155-64
64. Baindara P, Gautam A, Raghava G, Korpole S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci Rep. 2017;7:46541
65. Baindara P, Singh N, Ranjan M, Nallabelli N, Chaudhry V, Pathania GL. et al. Laterosporulin10: A novel defensin like class iid bacteriocin from brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology. 2016;162(8):1286-99
66. Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T. et al. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968. The Journal of antibiotics. 1994;47(3):301-10
67. Panicker J, Li Z, McMahon C, Sizer C, Steadman K, Piekarz R. et al. Romidepsin (FK228/depsipeptide) controls growth and induces apoptosis in neuroblastoma tumor cells. Cell cycle. 2010;9(9):1830-8
68. Piekarz RL, Robey RW, Zhan Z, Kayastha G, Sayah A, Abdeldaim AH. et al. T-cell lymphoma as a model for the use of histone deacetylase inhibitors in cancer therapy: impact of depsipeptide on molecular markers, therapeutic targets, and mechanisms of resistance. Blood. 2004;103(12):4636-43
69. Cheng Y-Q, Yang M, Matter AM. Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968. Applied and environmental microbiology. 2007;73(11):3460-9
70. Karpiński TM, Adamczak A. Anticancer Activity of Bacterial Proteins and Peptides. Pharmaceutics. 2018;10(2):54
71. Franciosa G, Floridi F, Maugliani A, Aureli P. Differentiation of the gene clusters encoding botulinum neurotoxin type A complexes in Clostridium botulinum type A, Ab, and A (B) strains. Applied and environmental microbiology. 2004;70(12):7192-9
72. Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. Journal of Infectious Diseases. 2000;181(Supplement_1):S156-S67
73. Abdel-Mageed WM, Milne BF, Wagner M, Schumacher M, Sandor P, Pathom-aree W. et al. Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Organic & biomolecular chemistry. 2010;8(10):2352-62
74. Szkaradkiewicz A, Karpinski T, Gamian A, Brzozowska E, Zeidler A. Identification of carcinoma suppressive factor produced by bacteria of Enterococcus genus. Clinical Microbiology & Infection. 2012;18:355
75. Karpiński TM, Szkaradkiewicz AK. Anticancer peptides from bacteria. Banladesh J Pharm. 2013;8(3):343-8
76. Chumchalová J, Šmarda J. Human tumor cells are selectively inhibited by colicins. Folia microbiologica. 2003;48(1):111-5
77. Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubes R, Postle K. et al. Colicin biology. Microbiology and molecular biology reviews. 2007;71(1):158-229
78. Raza A, Kohila V, Ghosh SS. Redesigned Escherichia coli cytosine deaminase: a new facet of suicide gene therapy. The journal of gene medicine. 2015;17(6-7):132-9
79. Austin EA, Huber BE. A first step in the development of gene therapy for colorectal carcinoma: cloning, sequencing, and expression of Escherichia coli cytosine deaminase. Molecular pharmacology. 1993;43(3):380-7
80. Fuchita M, Ardiani A, Zhao L, Serve K, Stoddard BL, Black ME. Bacterial Cytosine Deaminase Mutants Created by Molecular Engineering Show Improved 5-Fluorocytosine-Mediated Cell Killing In vitro and In vivo. Cancer Res. 2009;69(11):4791-9
81. Lv S-Q, Yang H, He J-Q, Wang B, Yoshimura I, Liu Y-S. Effects of CD/5-FC suicide gene therapy system on human malignant glioma cells in vitro. Acta Biochimica Et Biophysica Sinica-Chinese Edition-. 2003;35(5):430-4
82. Andrianasolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail K, Leal RM. et al. Isolation of swinholide A and related glycosylated derivatives from two field collections of marine cyanobacteria. Organic letters. 2005;7(7):1375-8
83. Hetz C, Bono MR, Barros LF, Lagos R. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proceedings of the National Academy of Sciences. 2002;99(5):2696-701
84. Lagos R, Baeza M, Corsini G, Hetz C, Strahsburger E, Castillo JA. et al. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Molecular microbiology. 2001;42(1):229-43
85. Van der Meer J, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, De Vos W. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. Journal of bacteriology. 1993;175(9):2578-88
86. Kayal S, Charbit A. Listeriolysin O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol Rev. 2006;30(4):514-29
87. Han B, Gross H, Goeger DE, Mooberry SL, Gerwick WH. Aurilides B and C, Cancer Cell Toxins from a Papua New Guinea Collection of the Marine Cyanobacterium Lyngbya m ajuscula. J Nat Prod. 2006;69(4):572-5
88. Tan LT, Okino T, Gerwick WH. Hermitamides A and B, toxic malyngamide-type natural products from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63(7):952-5
89. Han B, McPhail KL, Gross H, Goeger DE, Mooberry SL, Gerwick WH. Isolation and structure of five lyngbyabellin derivatives from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. Tetrahedron. 2005;61(49):11723-9
90. Mayer AMS, Gustafson KR. Marine pharmacology in 2005-2006: Antitumour and cytotoxic compounds. European Journal of Cancer. 2008;44(16):2357-87
91. Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A- D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J Am Chem Soc. 2006;128(5):1622-32
92. Matsuo Y, Kanoh K, Yamori T, Kasai H, Katsuta A, Adachi K. et al. Urukthapelstatin A, a novel cytotoxic substance from marine-derived Mechercharimyces asporophorigenens YM11-542. The Journal of antibiotics. 2007;60(4):251
93. Kajiura T, Furumai T, Igarashi Y, Hori H, Higashi K, Ishiyama T. et al. Signal Transduction Inhibitors, Hibarimicins A, B, C, D and G Produced by Microbispora. The Journal of antibiotics. 1998;51(4):394-401
94. Ishida K, Nakagawa H, Murakami M. Microcyclamide, a Cytotoxic Cyclic Hexapeptide from the Cyanobacterium Microcystis a eruginosa. J Nat Prod. 2000;63(9):1315-7
95. Ziemert N, Ishida K, Quillardet P, Bouchier C, Hertweck C, de Marsac NT. et al. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Applied and environmental microbiology. 2008;74(6):1791-7
96. Fernandez-Chimeno RI, Canedo L, Espliego F, Gravalos D, De La Calle F, FernAndez-Puentes JL. et al. IB-96212, a Novel Gytotoxic Macrolide Produced by a Marine Micromonospora. The Journal of antibiotics. 2000;53(5):474-8
97. Furumai T, Takagi K, Igarashi Y, Saito N, Oki T. Arisostatins A and B, new members of tetrocarcin class of antibiotics from Micromonospora sp. TP-A0316: I. Taxonomy, fermentation, isolation and biological properties. Journal of Antibiotics (Tokyo). 2000;53(3):227-32
98. Fang J, Zhang Y, Huang L, Jia X, Zhang Q, Zhang X. et al. Cloning and characterization of the tetrocarcin A gene cluster from Micromonospora chalcea NRRL 11289 reveals a highly conserved strategy for tetronate biosynthesis in spirotetronate antibiotics. Journal of bacteriology. 2008;190(17):6014-25
99. Gao X, Lu Y, Xing Y, Ma Y, Lu J, Bao W. et al. A novel anticancer and antifungus phenazine derivative from a marine actinomycete BM-17. Microbiological research. 2012;167(10):616-22
100. Cho JY, Williams PG, Kwon HC, Jensen PR, Fenical W. Lucentamycins A- D, cytotoxic peptides from the marine-derived actinomycete Nocardiopsis lucentensis. J Nat Prod. 2007;70(8):1321-8
101. Trimurtulu G, Ohtani I, Patterson GM, Moore RE, Corbett TH, Valeriote FA. et al. Total structures of cryptophycins, potent antitumor depsipeptides from the blue-green alga Nostoc sp. strain GSV 224. J Am Chem Soc. 1994;116(11):4729-37
102. Weiss C, Figueras E, Borbely AN, Sewald N. Cryptophycins: cytotoxic cyclodepsipeptides with potential for tumor targeting. Journal of Peptide Science. 2017;23(7-8):514-31
103. Magarvey NA, Beck ZQ, Golakoti T, Ding Y, Huber U, Hemscheidt TK. et al. Biosynthetic characterization and chemoenzymatic assembly of the cryptophycins. Potent anticancer agents from Nostoc cyanobionts. ACS chemical biology. 2006;1(12):766-79
104. Kalinovskaya NI, Romanenko LA, Kalinovsky AI, Dmitrenok PS, Dyshlovoy SA. A new antimicrobial and anticancer peptide producing by the marine deep sediment strain" Paenibacillus profundus" sp. nov. Sl 79. Natural product communications. 2013;8(3):381-4
105. Kumar B, Balgir P, Kaur B, Mittu B, Chauhan A. In vitro cytotoxicity of native and rec-pediocin CP2 against cancer cell lines: a comparative study. Pharmaceutical Analytical Acta. 2012:1 (6)
106. Kaur B, Balgir P. Pediocin CP2 gene localization to plasmid pCP289 of Pediococcus acidilactici MTCC 5101. Internet Journal of Microbiology. 2007:3 (2)
107. Imamura N, Nishijima M, Takadera T, Adachi K, Sakai M, Sano H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. Journal of Antibiotics (Tokyo). 1997;50(1):8-12
108. Mukherjee S, Seshadri R, Varghese NJ, Eloe-Fadrosh EA, Meier-Kolthoff JP, Göker M. et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nature biotechnology. 2017;35(7):676
109. Gray GL, Smith DH, Baldridge JS, Harkins RN, Vasil ML, Chen EY. et al. Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 1984;81(9):2645-9
110. Mohamed MS, Fattah SA, Mostafa HM. Azurin as antitumor protein and its effect on the cancer cell lines. Current Research Journal of Biological Sciences. 2010;2(6):396-401
111. Punj V, Bhattacharyya S, Saint-Dic D, Vasu C, Cunningham EA, Graves J. et al. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene. 2004;23(13):2367
112. Osman YA, El-Deep DR, Younis SA. Azurin: A powerful anticancer from" A" local Pseudomonas aeruginosa isolate. J Am Sci. 2013;9(12):755-64
113. Yamada T, Goto M, Punj V, Zaborina O, Chen ML, Kimbara K. et al. Bacterial redox protein azurin, tumor suppressor protein p53, and regression of cancer. Proceedings of the National Academy of Sciences. 2002;99(22):14098-103
114. Abdi-Ali A, Worobec E, Deezagi A, Malekzadeh F. Cytotoxic effects of pyocin S2 produced by Pseudomonas aeruginosa on the growth of three human cell lines. Canadian journal of microbiology. 2004;50(5):375-81
115. Denayer S, Matthijs S, Cornelis P. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. Journal of bacteriology. 2007;189(21):7663-8
116. Sharma A, Bala K, Husain I. Preliminary evaluation of arginine deiminase activity of indigenous bacterial strains for suitable chemotherapeutic applications. Biocatal Agric Biotechnol. 2017;12:66-77
117. Fiedler T, Strauss M, Hering S, Redanz U, William D, Rosche Y. et al. Arginine deprivation by arginine deiminase of Streptococcus pyogenes controls primary glioblastoma growth in vitro and in vivo. Cancer Biol Ther. 2015;16(7):1047-55
118. Mercenier A, Stalon V, Simon J, Haas D. Mapping of the arginine deiminase gene in Pseudomonas aeruginosa. Journal of bacteriology. 1982;149(2):787-8
119. Kloosterman TG, Kuipers OP. Regulation of Arginine Acquisition-and Virulence Gene Expression in the Human Pathogen Streptococcus pneumoniae by Transcription Regulators ArgR1 and AhrC. J Biol Chem. 2011 jbc. M111. 295832
120. Shoji J, Sakazaki R, Nakai H, Terui Y, Hattori T, Shiratori O. et al. Isolation of a New Phenazine Antibiotic Dob-41 from Pseudomonas Species. Journal of Antibiotics (Tokyo). 1988;41(5):589-94
121. Lam KS, Schroeder DR, Veitch JM, Matson JA, Forenza S. Isolation of a Bromo Analog of Rebeccamycin from Saccharothrix-Aerocolonigenes. Journal of Antibiotics (Tokyo). 1991;44(9):934-9
122. Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chemistry & biology. 2002;9(4):519-31
123. Asolkar RN, Freel KC, Jensen PR, Fenical W, Kondratyuk TP, Park E-J. et al. Arenamides A- C, cytotoxic NFκB inhibitors from the marine actinomycete Salinispora arenicola. J Nat Prod. 2008;72(3):396-402
124. Olano C, Méndez C, Salas JA. Antitumor compounds from marine actinomycetes. Mar Drugs. 2009;7(2):210-48
125. Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G. et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proceedings of the National Academy of Sciences. 2009;106(30):12295-300
126. Nguyen VH, Kim H-S, Ha J-M, Hong Y, Choy HE, Min J-J. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2009:0008-5472 CAN-09-3453
127. Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J. et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115(1):25-35
128. Elahian F, Moghimi B, Dinmohammadi F, Ghamghami M, Hamidi M, Mirzaei SA. The anticancer agent prodigiosin is not a multidrug resistance protein substrate. DNA and cell biology. 2013;32(3):90-7
129. Montaner B, Pérez-Tomás R. Prodigiosin-induced apoptosis in human colon cancer cells. Life sciences. 2001;68(17):2025-36
130. Maheswarappa G, Kavitha D, Vijayarani K, Kumanan K. Prodigiosin as anticancer drug Produced from bacteria of termite gut. Indian Journal of Basic and Applied Medical Research. 2013;1:257-66
131. Athavale MA, Somaiya SS, More GA, Satish S, Srivastava SA. Comparitive In-Vitro Cytotoxicity of Red Pigment Extract of Serratia. Marcescens on Breast and Prostrate Cancer Cell Lines. International Journal of Current Pharmaceutical Research. 2013;5:140-3
132. Campas C, Dalmau M, Montaner B, Barragan M, Bellosillo B, Colomer D. et al. Prodigiosin induces apoptosis of B and T cells from B-cell chronic lymphocytic leukemia. Leukemia. 2003;17(4):746
133. Harris AK, Williamson NR, Slater H, Cox A, Abbasi S, Foulds I. et al. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species-and strain-dependent genome context variation. Microbiology. 2004;150(11):3547-60
134. Su C, Xiang Z, Liu Y, Zhao X, Sun Y, Li Z. et al. Analysis of the genomic sequences and metabolites of Serratia surfactantfaciens sp. nov. YD25T that simultaneously produces prodigiosin and serrawettin W2. BMC Genomics. 2016:17 (1)
135. Gerth K, Bedorf N, HÖFLE G, Irschik H, Reichenbach H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). The Journal of antibiotics. 1996;49(6):560-3
136. Molnar I, Schupp T, Ono M, Zirkle R, Milnamow M, Nowak-Thompson B. et al. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chemistry & biology. 2000;7(2):97-109
137. Chou T-C, Zhang X-G, Balog A, Su D-S, Meng D, Savin K. et al. Desoxyepothilone B: an efficacious microtubule-targeted antitumor agent with a promising in vivo profile relative to epothilone B. Proceedings of the National Academy of Sciences. 1998;95(16):9642-7
138. Swofford CA, St. Jean AT, Panteli JT, Brentzel ZJ, Forbes NS. Identification of Staphylococcus Aureus α-hemolysin as a protein drug that is secreted by anticancer bacteria and rapidly kills cancer cells. Biotechnol Bioeng. 2014;111(6):1233-45
139. Walker B, Krishnasastry M, Zorn L, Kasianowicz J, Bayley H. Functional expression of the alpha-hemolysin of Staphylococcus aureus in intact Escherichia coli and in cell lysates. Deletion of five C-terminal amino acids selectively impairs hemolytic activity. J Biol Chem. 1992;267(15):10902-9
140. Lee DG, Hahm K-S, Park Y, Kim H-Y, Lee W, Lim S-C. et al. Functional and structural characteristics of anticancer peptide Pep27 analogues. Cancer cell international. 2005;5(1):21
141. Kim E-H, Choi S-Y, Kwon M-K, Tran TD-H, Park S-S, Lee K-J. et al. Streptococcus pneumoniae pep27 mutant as a live vaccine for serotype- independent protection in mice. Vaccine. 2012;30(11):2008-19
142. Paiva AD, de Oliveira MD, de Paula SO, Baracat-Pereira MC, Breukink E, Mantovani HC. Toxicity of bovicin HC5 against mammalian cell lines and the role of cholesterol in bacteriocin activity. Microbiology. 2012;158(11):2851-8
143. Liu G, Zhong J, Ni J, Chen M, Xiao H, Huan L. Characteristics of the bovicin HJ50 gene cluster in Streptococcus bovis HJ50. Microbiology. 2009;155(2):584-93
144. Du L, Sánchez C, Chen M, Edwards DJ, Shen B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chemistry & biology. 2000;7(8):623-42
145. Institute WS. Genomics of Drug Sensitivity in Cancer (Doxorubicin) [cited 2018 10 September 2018]. Available from: https://www.cancerrxgene.org/ translation/Drug/133.
146. Lomovskaya N, Otten SL, Doi-Katayama Y, Fonstein L, Liu X-C, Takatsu T. et al. Doxorubicin Overproduction in Streptomyces peucetius: Cloning and Characterization of the dnrU Ketoreductase anddnrV Genes and the doxA Cytochrome P-450 Hydroxylase Gene. Journal of bacteriology. 1999;181(1):305-18
147. Institude WS. Genomics of Drug Sensitivity in Cancer (Mitomycin-C ) 2018 [cited 2018 27/07/2018]. Available from: https://www.cancerrxgene.org/ translation/Drug/136.
148. Mao Y, Varoglu M, Sherman DH. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chemistry & biology. 1999;6(4):251-63
149. Nepal KK, Lee RP, Rezenom YH, Watanabe CMH. Probing the Role of N -Acetyl-glutamyl 5-Phosphate, an Acyl Phosphate, in the Construction of the Azabicycle Moiety of the Azinomycins. Biochemistry. 2015;54(29):4415-8
150. Bitzer J, Gesheva V, Zeeck A. Actinomycins with altered threonine units in the β-peptidolactone. J Nat Prod. 2006;69(8):1153-7
151. Wang X, Tabudravu J, Rateb ME, Annand KJ, Qin Z, Jaspars M. et al. Identification and characterization of the actinomycin G gene cluster in Streptomyces iakyrus. Molecular BioSystems. 2013;9(6):1286-9
152. Yamashita T, Naoi N, Hidaka T, Watanabe K, Kumada Y, Takeuchi T. et al. Studies on Auromomycin. Journal of Antibiotics (Tokyo). 1979;32(4):330-9
153. Sakata N, Kanbe T, Tanabe M, Hayashi H, Hori M, Hotta K. et al. Nucleotide sequence of the macromomycin apoprotein gene and its expression in Streptomyces macromomyceticus. The Journal of antibiotics. 1989;42(11):1704-13
154. Gervasoni JE, Hindenburg AA, Vezeridis MP, Schulze S, Wanebo HJ, Mehta S. An effective in vitro antitumor response against human pancreatic carcinoma with paclitaxel and daunorubicin by induction of both necrosis and apoptosis. Anticancer Res. 2004;24(5A):2617-26
155. Arican GÖ, Soy NN. Effects of epirubicin and daunorubicin on cell proliferation and cell death in HeLa cells. Journal of Cell & Molecular Biology. 2005:4 (1)
156. Guimarães LA, Jimenez PC, Sousa TdS, Freitas HPS, Rocha DD, Wilke DV. et al. Chromomycin A2 induces autophagy in melanoma cells. Mar Drugs. 2014;12(12):5839-55
157. Aftab U, Zechel DL, Sajid I. Antitumor compounds from Streptomyces sp. KML-2, isolated from Khewra salt mines, Pakistan. Biological research. 2015;48(1):58
158. Hu Y, Espindola APD, Stewart NA, Wei S, Posner BA, MacMillan JB. Chromomycin SA analogs from a marine-derived Streptomyces sp. Bioorganic & medicinal chemistry. 2011;19(17):5183-9
159. Menéndez N, Nur-e-Alam M, Braña AF, Rohr J, Salas JA, Méndez C. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogs. Chemistry & biology. 2004;11(1):21-32
160. Kawauchi T, Sasaki T, Yoshida K-I, Matsumoto H, Chen R-X, Huang M-Y. et al. A new anthracycline antibiotic, IT-62-B, converts the morphology of ras-transformed cells back to normal: Taxonomy, fermentation, isolation, structure elucidation and biological characterization. Journal of Antibiotics (Tokyo). 1997;50(4):297-303
161. Al-Mestarihi A, Romo A, Liu H-w, Bachmann BO. Nitrososynthase-Triggered Oxidative Carbon-Carbon Bond Cleavage in Baumycin Biosynthesis. J Am Chem Soc. 2013;135(31):11457-60
162. Ahmad MS, El-Gendy AO, Ahmed RR, Hassan HM, El-Kabbany HM, Merdash AG. Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 isolated from Egyptian soil. Front Microbiol. 2017:8 (MAR)
163. Hatsu M, Sasaki T, Watabe H-O, Miyadoh S, Nagasawa M, Shomura T. et al. A New Tetracycline Antibiotic with Antitumor Activity I. Taxonomy and Fermentation of the Producing Strain Isolation and Characterization of Sf2575. Journal of Antibiotics (Tokyo). 1992;45(3):320-4
164. Krishnan K, Mani A, Jasmine S. Cytotoxic activity of bioactive compound 1, 2-benzene dicarboxylic acid, mono 2-ethylhexyl ester extracted from a marine derived Streptomyces sp. VITSJK8. International journal of molecular and cellular medicine. 2014;3(4):246
165. Conda-Sheridan M, Marler L, Park E-J, Kondratyuk TP, Jermihov K, Mesecar AD. et al. Potential chemopreventive agents based on the structure of the lead compound 2-bromo-1-hydroxyphenazine, isolated from Streptomyces species, strain CNS284. Journal of medicinal chemistry. 2010;53(24):8688-99
166. Kawai H, Hayakawa Y, Nakagawa M, Furihata K, Furihata K, Shimazu A. et al. Arugomycin a New Anthracycline Antibiotic I. Taxonomy Fermentation Isolation and Physico-Chemical Properties. Journal of Antibiotics (Tokyo). 1987;40(9):1266-72
167. Simmons TL, McPhail KL, Ortega-Barría E, Mooberry SL, Gerwick WH. Belamide A, a new antimitotic tetrapeptide from a Panamanian marine cyanobacterium. Tetrahedron Lett. 2006;47(20):3387-90
168. Kanoh K, Matsuo Y, Adachi K, Imagawa H, Nishizawa M, Shizuri Y. Mechercharmycins A and B, cytotoxic substances from marine-derived Thermoactinomyces sp. YM3-251. The Journal of antibiotics. 2005;58(4):289
169. Fiedler H-P, Bruntner C, Riedlinger J, Bull AT, Knutsen G, Goodfellow M. et al. Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. The Journal of antibiotics. 2008;61(3):158
170. Agrawal N, Bettegowda C, Cheong I, Geschwind J-F, Drake CG, Hipkiss EL. et al. Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proceedings of the National Academy of Sciences. 2004;101(42):15172-7
171. Lizotte PH, Baird JR, Stevens CA, Lauer P, Green WR, Brockstedt DG. et al. Attenuated Listeria monocytogenes reprograms M2-polarized tumor-associated macrophages in ovarian cancer leading to iNOS-mediated tumor cell lysis. OncoImmunology. 2014;3(5):e28926
172. Chandra D, Jahangir A, Quispe-Tintaya W, Einstein M, Gravekamp C. Myeloid-derived suppressor cells have a central role in attenuated Listeria monocytogenes-based immunotherapy against metastatic breast cancer in young and old mice. Br J Cancer. 2013;108(11):2281
173. Roberts NJ, Zhang L, Janku F, Collins A, Bai R-Y, Staedtke V. et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci Transl Med. 2014;6(249):249ra111-249ra111
174. Bettegowda C, Huang X, Lin J, Cheong I, Kohli M, Szabo SA. et al. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nature biotechnology. 2006;24(12):1573
175. Stern C, Kasnitz N, Kocijancic D, Trittel S, Riese P, Guzman CA. et al. Induction of CD 4+ and CD 8+ anti-tumor effector T cell responses by bacteria mediated tumor therapy. International journal of cancer. 2015;137(8):2019-28
176. Weibel S, Stritzker J, Eck M, Goebel W, Szalay AA. Colonization of experimental murine breast tumours by Escherichia coli K-12 significantly alters the tumour microenvironment. Cellular microbiology. 2008;10(6):1235-48
177. Kitamura A, Mastumoto S, Asahina I. Growth inhibition of HeLa cell by internalization of Mycobacterium bovis Bacillus Calmette-Guérin (BCG) Tokyo. Cancer cell international. 2009;9(1):30
178. Kuromatsu I, Matsuo K, Takamura S, Kim G, Takebe Y, Kawamura J. et al. Induction of effective antitumor immune responses in a mouse bladder tumor model by using DNA of an α antigen from mycobacteria. Cancer Gene Ther. 2001;8(7):483
179. Matsuo K, Yamaguchi R, Yamazaki A, Tasaka H, Yamada T. Cloning and expression of the Mycobacterium bovis BCG gene for extracellular alpha antigen. Journal of bacteriology. 1988;170(9):3847-54
180. Lawman M, Kane C, Eidizadeh S, Shaw N, Lawman P. A novel cancer vaccine meets canine trial endpoints. American Association for Cancer Research. 2007
181. Maletzki C, Linnebacher M, Kreikemeyer B, Emmrich J. Pancreatic cancer regression by intratumoural injection of live Streptococcus pyogenes in a syngeneic mouse model. Gut. 2008;57(4):483-91
182. Babbar A. Streptococcal superantigens. Streptococcal Superantigens: Springer. 2015 p. 1-41
183. Kim SH, Castro F, Paterson Y, Gravekamp C. High efficacy of a Listeria-based vaccine against metastatic breast cancer reveals a dual mode of action. Cancer Res. 2009;69(14):5860-6
184. Bahjat KS, Prell RA, Allen HE, Liu W, Lemmens EE, Leong ML. et al. Activation of immature hepatic NK cells as immunotherapy for liver metastatic disease. The Journal of Immunology. 2007;179(11):7376-84
185. Zhang X, Xu Q, Yang L, Lai Y, Zhang Z, Han C. et al. The genes slyA, STM3120 and htrA are required for the anticancer ability of VNP20009. Oncotarget. 2016;7(49):81187
186. Liu S, Xu X, Zeng X, Li L, Chen Q, Li J. Tumor-targeting bacterial therapy: A potential treatment for oral cancer. Oncol Lett. 2014;8(6):2359-66
187. Yazawa K, Fujimori M, Amano J, Kano Y, Taniguchi Si. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 2000;7(2):269
188. Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B. Combination bacteriolytic therapy for the treatment of experimental tumors. Proceedings of the National Academy of Sciences. 2001;98(26):15155-60
189. Yong AY, Shabahang S, Timiryasova TM, Zhang Q, Beltz R, Gentschev I. et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nature biotechnology. 2004;22(3):313
190. Min J-J, Kim H-J, Park JH, Moon S, Jeong JH, Hong Y-J. et al. Noninvasive real-time imaging of tumors and metastases using tumor-targeting light-emitting Escherichia coli. Molecular imaging and biology. 2008;10(1):54-61
191. Riedel CU, Monk IR, Casey PG, Morrissey D, O'Sullivan GC, Tangney M. et al. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Applied and environmental microbiology. 2007;73(9):3091-4
192. Felfoul O, Mohammadi M, Taherkhani S, De Lanauze D, Xu YZ, Loghin D. et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nature nanotechnology. 2016;11(11):941
193. Martel S. Targeting active cancer cells with smart bullets. Ther Deliv. 2017;8(5):301-12
194. Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, Luckett W. et al. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proceedings of the National Academy of Sciences. 2004;101(38):13832-7
195. Na HS, Kim HJ, Lee H-C, Hong Y, Rhee JH, Choy HE. Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine. 2006;24(12):2027-34
196. Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10(11):785
Author contact
![]() Corresponding author: zyuan27ca; Phone: (519) 953-6641; Fax: (519) 457-3997
Corresponding author: zyuan27ca; Phone: (519) 953-6641; Fax: (519) 457-3997

 Global reach, higher impact
Global reach, higher impact