3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(8):1814-1824. doi:10.7150/jca.29535 This issue Cite
Research Paper
Challenges of Providing Concordant Interpretation of Somatic Variants in Non-Small Cell Lung Cancer: A Multicenter Study
1. National Center for Clinical Laboratories, Beijing Hospital, National Center of Gerontology, Beijing, People's Republic of China;
2. Graduate School, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, People's Republic of China;
3. Beijing Engineering Research Center of Laboratory Medicine, Beijing Hospital, Beijing, People's Republic of China
Received 2018-8-28; Accepted 2019-2-21; Published 2019-4-21
Abstract
Background: Success of multiple-gene mutation tests by next-generation sequencing (NGS), associated with molecular targeting therapies for cancers, depending on the accuracy and consistency of interpreting variants. Here, we summarized reports from clinical laboratories for cases with non-small cell lung cancer (NSCLC) and discussed conflicting interpretations of somatic variants.
Methods: Three mimetic DNA samples, containing six somatic mutations, were prepared based on three clinical case reports of NSCLC. Clinical reports and genetic testing questionnaires were collected from 67 laboratories enrolled in this investigation.
Results: Thirty-four laboratories with correct variant results identified two variants, based on FDA approval of targeted drugs for the same tumor, consistently, with strong clinical significance, whereas the other variants were classified with conflicting interpretations. Discordant interpretations were reported for ERBB2 with three different classifications, including strong clinical significance (53.0%, 18/34), potential clinical significance (38.2%, 13/34), and unknown significance (8.8%, 3/34). In the variant therapeutic drug recommendation section, 32.4% of the laboratories (11/34) did not recommend all the available therapeutic drugs designated by the National Comprehensive Cancer Network (NCCN). In the remaining group of 33 laboratories with incorrect variant results, less correct classifications were acquired for the variants with strong clinical significance.
Conclusions: Owing to numerous reasons, the interpretation of variants differed greatly, which might in turn lead to the inappropriate clinical care of patients with NSCLC. By analyzing the limitations of different databases used by laboratories, we integrated various types of databases with different levels of evidence to form a comprehensive and detailed variant interpretation pipeline, aiming to standardize the variant classification and provide accurate and sufficient therapeutic drug recommendation to clinicians for minimal-inappropriate therapeutic options.
Keywords: clinical significance, next-generation sequencing, non-small cell lung cancer, somatic variant
Introduction
Tumor genotyping to identify actionable oncogenic driver mutations for targeted therapy has become increasingly important in the management of cancers [1]. In non-small cell lung cancer (NSCLC), several somatic genomic targets are recommended in the National Comprehensive Cancer Network (NCCN) NSCLC guidelines for expanding targeted treatment options, such as for EGFR or ERBB2 mutations, or ALK, ROS1, or RET fusions [2]. Rapid progress in molecular targeting therapies for NSCLC has boosted the development of multiple-gene mutation tests via next-generation sequencing (NGS), which can simultaneously screen all mutations relevant to the drugs currently available. At the same time, variants with other genetic alterations that are engaged in ongoing clinical trials or are of uncertain functional significance are also frequent, thus increasing challenges in interpreting variants [3].
With hundreds of somatic variants observed, oncologists are trying to identify the clinical significance and the recommended drugs, based on the interpretation from different laboratories. Thus, the success of sequencing-based individual-targeted therapies depends on the accuracy and consistency of variant interpretations, which include variation classification, treatment recommendation, and prognostic or diagnostic implications. A somatic variant can be considered a biomarker, and the variants are classified into different tiers based on their clinical impacts [4]. Variation classification, based on the strength of evidence, is considered by the physicians in the decision-making process, and some clinical laboratories only report the variants of a high-level category with enough evidence of clinical impact [5]. However, accurate variant interpretation is challenging due to numerous influencing factors. First, different standardized systems for the classification of somatic variants have been set up, such as the SVC [6], PHIAL [7], and BWH/DFCI methods [8], as well as a recent consensus classification system established by the Association for Molecular Pathology (AMP), American Society of Clinical Oncology (ASCO), and College of American Pathologists (CAP) [4]. Second, each public database or web-based tool has its own criteria for the collection and explanation of the clinical evidence and targeted therapy, which leads to different interpretations and various therapeutic options [9]. In total, the databases of somatic variant information could be divided into two groups [10]: variant catalogs, such as the Catalogue of Somatic Mutations In Cancer (COSMIC) [11], or variant interpretive databases, such as ClinVar and My Cancer Genome, which deliver information on the effect of tumor variants on sensitivity to targeted therapeutics provided by expert contributors [10,12]. Third, the laboratories have different decision support frameworks with various combinations of databases and different rules for annotations [5].
Till date, NGS has been widely used in routine clinical care for patients with NSCLC. However, concordance for interpretation of somatic variants has not been achieved, especially for mutations not listed in FDA-approved indicators. Here, we summarized the clinical reports of NSCLC cases from 67 laboratories with the aims of describing the conflicting interpretation of somatic variants and explaining the possible reasons underlying the same. Our study indicated that the interpretation of the same mutation may differ greatly across laboratories, hence leading to incorrect clinical care of the patients with NSCLC.
Materials and Methods
Study Design
We selected three clinical case reports of NSCLC, retrieved from PubMed, which were associated with confirmed targeted therapies (Table S1) [13-15]. Three corresponding mimetic DNA samples, containing six reported variants, were prepared [16]. We defined the six variants as follows: type 1, variants and genes with associations based on FDA-approved drugs for the same tumor [EML4 exon 13-ALK exon 20 and EGFR c.2235_2249delGGAATTAAGAGAAGC (p.Glu746_Ala750 del) ]; type 2, variants and genes with clinically proven associations based on NCCN guidelines [EGFR c.2155G>T (p.Gly719Cys)], EGFR c.2303G>T (p.Ser768Ile), and ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys)]; and type 3, genes with clinically proven associations based on NCCN guidelines, but variants with no clinically proven associations [KRAS c.145G>A (p.Glu49Lys)].
Samples were assigned to different laboratories, and detailed clinical information, identical to the case reports published, were also provided. These laboratories were required to submit their clinical reports and complete questionnaires on the somatic genomic alterations, the cancer-specific databases used, and the classification and interpretation of the detected variants. Somatic mutations were to be classified as per the following four tiers: variants of strong clinical significance (tier I), variants of potential clinical significance (tier II), variants of unknown significance (tier III), and variants that are benign or likely benign (tier IV). For each variant, classifications and drug recommendations, reported by the laboratories, were compared to determine concordance or discordance. Each variant was classified, and the drugs recommended by public databases were submitted by clinical laboratories to present the variability of evidence from public databases. For variants with more than two classifications, the submitted interpretation from each laboratory was compared to our reassessment result with the AMP-ASCO-CAP criteria, using the same public database combination, to determine whether some other factors caused the conflicting classifications. For statistical comparisons, the Mann-Whitney test and Fisher's exact probability test were used. All statistical tests were two-sided, and a P value of less than 0.05 was considered statistically significant.
Participant Enrollment
Initially, 101 participants were enrolled in this analysis; nine participants did not submit their results. Of the 41 laboratories reporting correct results and 51 laboratories reporting incorrect results, the ones that did not report all the expected variants (since the expected variant fell outside the specific detectable range) were excluded from the analysis. Consequently, data from 34 laboratories with correct variant results were analyzed in this study (Figure 1A). To present the interpretation of laboratories in the real-world setting, the variant classification and therapeutic recommendation from the remaining 33 laboratories with incorrect variant results were also summarized.
Results
Databases Used by the 34 Laboratories with Correct Variant Results
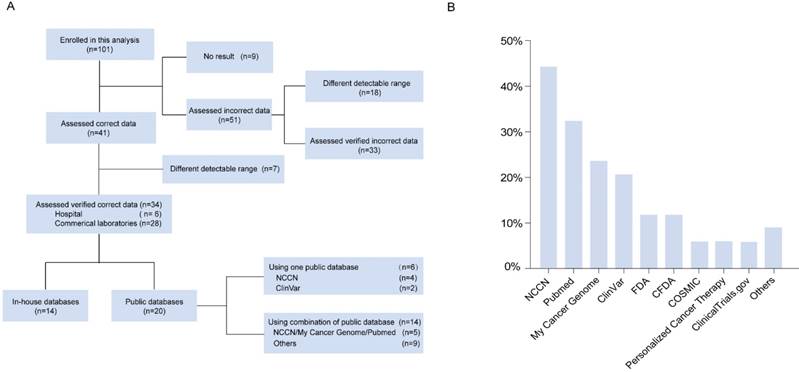
Overall, 34 participants were considered eligible for this analysis, including six hospital or clinical laboratories (17.6%) and 28 commercial laboratories (82.4%). Of the 34 laboratories included in this analysis, 14 (41.2%) used established in-house databases, while 20 (58.8%) used either a single public database or a combination of different types of public databases in their routine work (Figure 1A), including NCCN, My Cancer Genome, ClinVar, the Food and Drug Administration (FDA), the China Food and Drug Administration (CFDA), COSMIC, Personalized Cancer Therapy, ClinicalTrials.gov, and PubMed (Figure 1B). NCCN (version 1.2017), a professional guideline, was the most widely used public resource among laboratories (15/34, 44.1%). In public cancer- specific databases, My Cancer Genome (23.5%, 8/34) was the most frequently used, followed by ClinVar (20.6%, 7/34). The FDA and the CFDA, were the authorized institutions for recommending drugs towards personalized cancer therapy, which were used as reference by five laboratories (25.0%) (Figure 1B). Among the 20 laboratories that used public databases, 14 (70.0%) combined at least two public databases as their resource. The most common combination was of NCCN and My Cancer Genome, which was used by five laboratories (25.0%) (Figure 1A). In addition, PubMed was used as a literature database by 11 (32.4%, 11/34) laboratories. For the in-house databases used by 14 laboratories, in our study, combinations of some public databases and the detailed information for each in-house database were not provided by the laboratories.
Consistency in Somatic Variant Classifications across 34 Laboratories
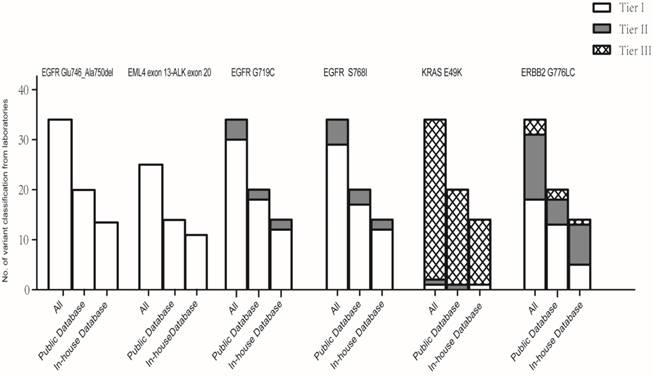
We compared the variant classifications submitted by each laboratory; no statistical difference was observed between the classifications using public and in-house databases for each variant (Figure 2). EML4 exon 13-ALK exon 20 and EGFR c.2235_2249del GGAATTAAGAGAAGC (p.Glu746_Ala750del) were consistently classified as variants of strong clinical significance (Table S2-3); other variants were classified discordantly. Of the 34 laboratories, tier I was reported for EGFR c.2155G>T (p.Gly719Cys) and EGFR c.2303G>T (p.Ser768Ile) by 30 laboratories (88.2%) and 29 laboratories (85.3%), respectively, while other laboratories classified the two variants as of potential clinical significance (tier II). Of the 20 laboratories that used a combination of public databases, two (10.0%) reported tier II for EGFR c.2155G>T (p. Gly719Cys) and three (15.0%) reported tier II for EGFR c.2303G>T (p.Ser768Ile).
The classification for ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys) was greatly discordant across the laboratories, with three different interprettations, including 18 of strong clinical significance (53.0%), 13 of potential clinical significance (38.2%), and three of unknown significance (8.8%). The laboratories using combined public databases reported five tier II (25.0%) and two tier III (10.0%) classifications for ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys). Although the laboratories also presented three kinds of classifications for KRAS c.145G>A (p.Glu49Lys), most (94.2%) assessed it as a variant with unknown significance.
Classification of Somatic Variants Based on Each Database
We then compared the categorization of each mutation with the AMP-ASCO-CAP somatic variant classification criteria based on the evidence from each public database (Table 1). These databases were last accessed on April 30, 2017. EML4 exon 13-ALK exon 20 and EGFR c.2235_2249delGGAATTAAGAGAAGC (p.Glu746_Ala750del) held definitely strong clinical significance and were classified into tier I consistently by the six databases, despite different levels of evidence. FDA, NCCN, and Personalized Cancer Therapy databases provided evidence of level A, ranking matched therapies by evidence of clinical benefits. My Cancer Genome, ClinVar, and COSMIC did not incorporate clinical practice guideline recommendations or rank the relevant targeted agents by the regulatory approval status [17]. My Cancer Genome links variants to specific targeted therapies after an expert review, COSMIC is curated by groups of experts and lists relevant information from the literature, and ClinVar interprets the variants as pathogenic or benign and appropriately ranks a few related drugs. We regarded the information from the three databases as well-powered studies with expert consensus and thus listed the evidence into level B.
The variants EGFR c.2155G>T (p. Gly719Cys) and EGFR c.2303G>T (p.Ser768Ile) are common activating mutations, thought to be associated with the sensitivity to EGFR tyrosine kinase inhibitors (TKIs), as suggested by NCCN. Undoubtedly, NCCN provided level A evidence, whereas My Cancer Genome, ClinVar, and COSMIC provided level B evidence and categorized both variants into tier I. Personalized Cancer Therapy presented EGFR c.2303G>T (p.Ser768Ile) as an actionable functional alteration; however, the therapeutic significance was conflicting, based on clinical studies. Thus, the evidence from this database was listed into level C (tier II).
A schematic illustration of enrolled laboratories and databases used. (A) Flow diagram of the laboratories enrolled in the analysis. (B) Distribution of databases used by the 34 laboratories.
Classification of each variant by the laboratories. The classification of each mutation was compared between two groups of laboratories using public databases and in-house databases with the Mann-Whitney test. No statistical differences were observed between the classifications by the two groups of laboratories using public and in-house databases for each mutation. *P values were 1.000, 1.000, 0.707, 0.955, 0.764, and 0.164 for EGFR c.2235_2249delGGAATTAAGAGAAGC (p.Glu746_Ala750 del), EML4 exon 13-ALK exon 20, EGFR c.2155G>T (p.Gly719Cys), EGFR c.2303G>T (p.Ser768Ile), KRAS c.145G>A(p.Glu49Lys), and ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys), respectively.
Categorization of Each Mutation with AMP-ASCO-CAP Somatic Variant Classification Criteria Based on the Evidences from Public Databases
| Level of evidence/ Categorizationa | |||||||
|---|---|---|---|---|---|---|---|
| FDA/ CFDA, Professional guidelines | Variant interpretive databases | Variant catalogs | |||||
Somatic Variants | FDA/CFDA | NCCN | My Cancer Genome | Personalized Cancer Therapy | ClinVar | COSMIC | Totalb |
| EGFR c.2235_2249delGGAATTAAGAGAAGC (p.Glu746_Ala750del) | Level 1A /Tier I | Level 1A /Tier I | Level B /Tier I | Level 1A /Tier I | Level B /Tier I | Level B /Tier I | Level 1A /Tier I |
| EML4 exon13-ALK exon20 | Level 1A /Tier I | Level 1A /Tier I | Level B /Tier I | Level 1A /Tier I | Level B /Tier I | Level B /Tier I | Level 1A /Tier I |
| EGFR c.2155G>T (p. Gly719Cys) | NA | Level 2A /Tier I | Level B /Tier I | Level B /Tier I | Level B /Tier I | Level B /Tier I | Level 2A /Tier I |
| EGFR c.2303G>T (p.Ser768Ile) | NA | Level 2A /Tier I | Level B /Tier I | Level 2C /Tier II | Level B /Tier I | Level B /Tier I | Level 2A /Tier I |
| KRAS c.145G>A (p.Glu49Lys) | NA | NA | NA | NA | NA | Tier III | Tier III |
| ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys) | NA | Level 2A /Tier I | Level C / Tier II | Level D / Tier II | Level D / Tier II | Tier III | Level 2A / Tier I |
aStandards of level of evidence and categorization for somatic variants were from Reference4. NA: not available.
bTotal: The classification of mutations with AMP-ASCO-CAP somatic variant classification criteria based on the evidence from all the database public listed.
Patients with NSCLC and ERBB2 exon 20 insertion have been demonstrated to be sensitive to trastuzumab and afatinib therapies, but show no confirmed response to other ERBB2-targeted drugs [18]. NCCN recommended trastuzumab and afatinib for patients with NSCLC and ERBB2 mutations. Personalized Cancer Therapy presented evidence as level D for ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys). My Cancer Genome provided interpretations for ERBB2 exon 20 insertions without the designation for a specific variant, which presented the evidence of preclinical studies or case reports (level C) and categorized the variants into tier II. ClinVar interpreted the variant as likely pathogenic, representing the evidence as level D, which would classify the ERBB2 variant into tier II. COSMIC incorporated the ERBB2 variant and listed in one study (tier III) [13].
Although KRAS mutations are also mentioned in NCCN, their clinical significances are unclear. KRAS c.145G>A (p.Glu49Lys) could not be found in any of the databases, except COSMIC, and therefore, it is functionally unknown. COSMIC only demonstrated a somatic variant, which classified the variant into tier III.
Reassessment of Variants with Conflicting Interpretations
For the four variants with conflicting interpretations, the submitted interpretations of each laboratory was compared to that from our reassessment with the AMP-ASCO-CAP criteria using the same public database combination (Table S4-7). The concordance rates were only 60.0% (12/20) for ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys). Besides, the other three variants had a higher concordance with 90.0% (18/20) for EGFR c.2155G>T (p.Gly719Cys), 85.0% (17/20) for EGFR c.2303G>T (p.Ser768Ile), and 95.0% (19/20) for KRAS c.145G>A (p.Glu49Lys).
Consistency in Therapeutic Drug Recommendations across 34 Laboratories
The recommendations for therapeutic drugs could be divided into three categories according to the different levels of evidence: (1) drugs approved by the FDA or CFDA in selected tumor types (companion diagnostic), (2) drugs described in guidelines, and (3) drugs with predictive associations based on the results from preclinical studies, clinical trials, case reports, and drugs currently being investigated in clinical trials.
The laboratories recommended the FDA-approved drugs with corresponding indications (the same variant and tumor type) concordantly, which is considered to be “standard of care.” All the laboratories recommended the FDA-approved drugs for ALK fusion and an EGFR deletion in exon 19. Notably, icotinib, approved by the CFDA in China, was recommended by more than 50% of the laboratories for all the three EGFR mutations. Brigatinib, recently examined by the FDA for patients with NSCLC and ALK fusion (approved on April 10, 2017, after our study) was provided as the therapeutic choice by 28.0% (7/25) of the laboratories.
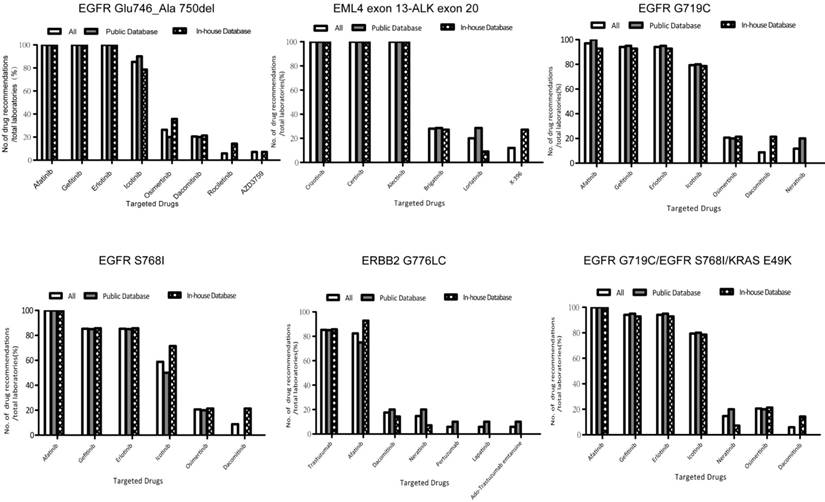
The therapeutic drugs afatinib, gefitinib, and erlotinib were recommended consistently for G719X and S768I by 91.2% (31/34) and 85.3% (29/34) of the laboratories, respectively. Trastuzumab and afatinib were suggested simultaneously for the ERBB2 mutation by 79.4% (27/34) of the laboratories as per the NCCN guideline for NSCLC. In total, 32.4% (11/34) of the laboratories did not follow the guideline (Table S2-7). We noticed that, 11.1% (2/18) of the laboratories that classified the ERBB2 mutation as tier I, 23.4% (3/13) that regarded the ERBB2 mutation as tier II, and all the three laboratories that classified ERBB2 mutations as tier III, missed at least one of the two drugs.
Some drugs, confirmed to be efficacious based on evidence from clinical studies were recommended by partial laboratories (Figure 3). Detailed information in this regard is listed in Table S2-5, and Table S7. For example, dacomitinib was demonstrated to be sensitive to a subgroup of EGFR exon 19 deletions in a clinical trial (phase II) in My Cancer Genome and Personalized Cancer Therapy [19]. Laboratory 80 recommended drugs being assessed in clinical trials, such as AZD3759 and ensartinib, and stated the availability of trials in clinical reports.
In addition, databases with different levels of evidence may give diverse therapeutic drug recommendations (Table 2). For example, afatinib, gefitinib and erlotinib were recommended by the FDA and NCCN for EGFR c.2235_2249del GGAATTAAGAGAAGC (p.Glu746_Ala750del). My Cancer Genome and Personalized Cancer Therapy not only covered these therapeutic treatments, but referred to osimertinib, daconitinib and rociletinib for this variant; AZD3759 was also mentioned in ClinicalTrials.gov (Table 2).
Consistency in the Interpretation across the 33 Laboratories with Incorrect Variant Results
The five variants of EGFR deletion in exon 19, ALK fusion, EGFR G719X, EGFR S768I, and ERBB2 G776LC were classified as variants of strong clinical significance by only 57.6%, 82.4%, 84.9%, 72.7%, and 9.1% of the laboratories, respectively, which are much less than those in the group of 34 laboratories with correct variant results (Table S8-9). Of the 91 false-positive results reported in the three cases, 53.8% (49/91) were interpreted as tier III or tier IV, and no therapeutic recommendation was provided by the laboratories (Table S10), and 40.7% (37/91) were not interpreted at all.
Discussion
For any practicing oncologist, the current challenge is not to identify genomic alterations, but rather how to best utilize the emerging information to select the optimally approved or investigational therapy in multiplex tumor genomic testing. Thus, we suggest that the laboratories should interpret the variants from two aspects, including clinical validity and clinical actionability for somatic variants in tumor-profiling NGS tests [20].
Therapeutic drugs recommended by the laboratories for each variant. The recommended therapeutic drugs for each mutation were compared between the two groups of laboratories using public databases and in-house databases via the Fisher's exact probability test. No statistical differences were observed between the therapeutic drugs recommended by the two groups of laboratories using public and in-house databases for each mutation. *P values were 0.645, 0.493, 0.370, 0.559, and 0.645 for EGFR c.2235_2249delGGAATTAA GAGAAGC (p.Glu746_Ala750 del), EML4 exon 13-ALK exon 20, EGFR c.2155G>T (p.Gly719Cys), EGFR c.2303G>T (p.Ser768Ile), KRAS c.145G>A(p.Glu49Lys), and ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys), respectively.
Category and Evidence of the Drugs Recommended by the Laboratories
| Variant | Drug recommended | Category | FDA or CFDA | Professional guideline | Other evidencesa |
|---|---|---|---|---|---|
| EGFR c.2235_2249del GGAATTAAGAGAAGC (p.Glu746_Ala750del) | Afatinib | Category 1 | FDA | NCCN | My Cancer Genome, Personalized Cancer Therapy, COSMIC |
| Gefitinib | Category 1 | FDA | NCCN | My Cancer Genome, Personalized Cancer Therapy, ClinVar, COSMIC | |
| Erlotinib | Category 1 | FDA | NCCN | My Cancer Genome, Personalized Cancer Therapy, COSMIC | |
| Icotinib | Category 1 | CFDA | None | My Cancer Genome, Personalized Cancer Therapy | |
| Osimertinib | Category 3 | FDA approved for T790M positive NSCLC patients | None | My Cancer Genome, Personalized Cancer Therapy | |
| Dacomitinib | Category 3 | None | None | My Cancer Genome, Personalized Cancer Therapy | |
| Rociletinib | Category 3 | None | None | My Cancer Genome, Personalized Cancer Therapy | |
| AZD3759 | Category 3 | None | None | PMID: 27928026, ClinicalTrials.gov (NCT02228369) | |
| EML4 exon 13-ALK exon 20 | Crizotinib | Category 1 | FDA | NCCN | My Cancer Genome, Personalized Cancer Therapy |
| Ceritinib | Category 1 | FDA | NCCN, | My Cancer Genome, Personalized Cancer Therapy | |
| Alectinib | Category 1 | FDA | NCCN | Personalized Cancer Therapy, None | |
| Brigatinib | Category 2 | FDA approved (April.29th,2017 after our study) | NCCN | My Cancer Genome, Personalized Cancer Therapy | |
| Lorlatinib | Category 3 | None | None | Personalized Cancer Therapy, PMID: 26698910, ClinicalTrials.gov (NCT01970865) | |
| Ensartinib | Category 3 | None | None | Personalized Cancer Therapy, ClinicalTrials.gov (NCT01625234) | |
| EGFR c.2155G>T (p. Gly719Cys) | Afatinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | COSMIC |
| Gefitinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome, Personalized Cancer Therapy, ClinVar, COSMIC | |
| Erlotinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome | |
| Icotinib | Category 1 | CFDA approved for EGFR sensitive mutations | None | None | |
| Osimertinib | Category 3 | FDA approved for T790M positive NSCLC patients | None | ClinicalTrials.gov (NCT02759835) | |
| Dacomitinib | Category 3 | None | None | PMID: 25456362, ClinicalTrials.gov (NCT01774721) | |
| Neratinib | Category 3 | None | None | My Cancer Genome | |
| EGFR c.2303G>T (p.Ser768Ile) | Afatinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome, Personalized Cancer Therapy, COSMIC |
| Gefitinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome, Personalized Cancer Therapy, COSMIC | |
| Erlotinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome, Personalized Cancer Therapy, COSMIC | |
| Icotinib | Category 1 | CFDA approved for EGFR sensitive mutations | None | None | |
| Osimertinib | Category 3 | FDA approved for T790M positive NSCLC patients | None | ClinicalTrials.gov (NCT02759835) | |
| Dacomitinib | Category 3 | None | None | My Cancer Genome, PMID: 25456362, ClinicalTrials.gov (NCT01774721) | |
| ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys) | Trastuzumab | Category 2 | FDA approved for ERBB2 amplification breast cancer patients | NCCN | My Cancer Genome, ClinVar |
| Afatinib | Category 2 | FDA approved for EGFR exon 19 deletions and L858R positive NSCLC patients | NCCN | My Cancer Genome, Personalized Cancer Therapy | |
| Dacomitinib | Category 3 | None | None | PMID: 25899785, ClinicalTrials.gov (NCT0114286) | |
| Neratinib | Category 3 | None | None | My Cancer Genome, | |
| Pertuzumab | Category 3 | FDA approved for ERBB2 amplification breast cancer patients | None | ClinicalTrials.gov (NCT00063154), | |
| Lapatinib | Category 3 | FDA approved for ERBB2 amplification breast cancer patients | None | ClinicalTrials.gov (NCT01306045) | |
| Trastuzumab emtansine | Category 3 | FDA approved for ERBB2 amplification breast cancer patients | None | ClinicalTrials.gov (NCT02675829, NCT02289833) |
a Evidences from COSMIC, PubMed and ClinicalTrials.gov are listed when there are no confirmed evidence from FDA or CFDA, professional guideline and interpretive databases.
First, the laboratories should provide exactly the same interpretation and therapeutic recommendation for the variants, with definitive evidence. For targeted therapy, clinical validity refers to a test where a somatic variant predicts the relations of response or resistance to a targeted drug. FDA/CFDA-approved therapies and professional guidelines provide the most accurate evidence to prove the clinical validity [21]. Therefore, these variants should be classified as variants of strong clinical significance. Results providing incorrect interpretation for clinical validity of the variants and missing the targeted therapy from companion diagnostics and professional guidelines would be a wrong approach. In our study, all the five variants (type 1 and type 2) are biomarkers with strong clinical significance. The laboratories presented concordant classifications for type 1 variants. The categorization of ERBB2 mutation was the most controversial among the laboratory reports. Of the 34 ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys) reports, 13 (38.2%) claimed the variant to be of potential clinical significance, and three (8.8%) claimed it to be of unknown significance. EGFR c.2155G>T (p.Gly719Cys) and EGFR c.2303G>T (p.Ser768Ile) were also interpreted differently. Of note, one laboratory reported KRAS c.145G>A (p.Glu49Lys) as a variant with strong clinical significance. Major conflicts may occur due to two reasons. First, some laboratories did not include FDA, CFDA, and professional guidelines when interpreting the variants. Second, differences in understanding of the same level of evidence may also have led to conflicting classifications. Forty percent (8/20) of our classifications, according to each combination, disagreed with the results from the laboratories. For ERBB2 c.2326delGinsTTAT (p.Gly776delinsLeuCys), the laboratories using NCCN should have interpret the variant as tier I theoretically; however, only 60.0% (9/15) of these laboratories regarded the mutation as a variant of strong clinical significance. We speculated that these laboratories reported conflicting interpretations of the ERBB2 mutation due to different considerations of NCCN. Similarly, for KRAS c.145G>A (p.Glu49Lys), no clinical evidence for the clinical implication of this variant has been shown in previous studies or databases. The laboratory might present wrong interpretations of tier I, according to the significance of KRAS, instead of the variant. Since levels of evidence may vary between specific variants of the same gene, decisions regarding individual patients should be based on the variant instead of the gene [22]. In the Cancer Genome Atlas (TCGA) project, the somatic cancer variants were annotated in four different layers, including tumor types, genes, variants, and drugs [23]. We suggest that laboratories annotate the variants based on variant and tumor type when using consensus and variant interpretive databases (Figure 4). Although the 33 laboratories with incorrect variant results presented similar discordance in their variant interpretation, these laboratories provided less correct variant classification results owing to the poor testing results, which further emphasizes the importance to improve their accuracy in laboratories for variant detection by NGS.
Pipeline for variant interpretation in clinical laboratories.
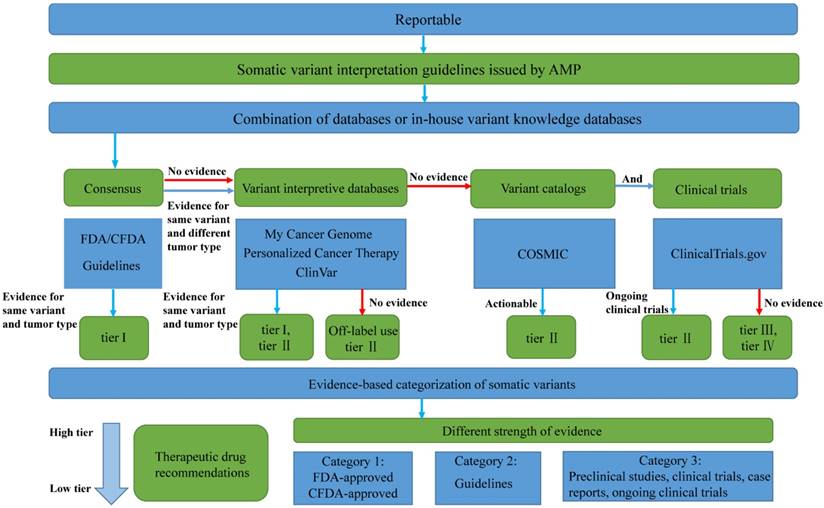
Second, the laboratories should provide the comprehensive and correct interpretation and therapeutic recommendation, based on correct evidence for the variants with only limited evidence at present. The off-label use of an FDA/CFDA-approved drug, large multicenter studies, clinical trials, small clinical studies, and case reports provides different levels of evidence to suggest possible clinical actionability [20,24]. It would not be wrong for laboratories to only review the evidence in FDA/CFDA-approved therapies and professional guidelines, and neglect other information. However, in this approach, patients might miss the opportunity of further investigational agents. In our study, we showed that laboratories may use various combinations of public databases or in-house databases, which might be created from multiple sources by linking public variant knowledge databases. Different types of public databases may result in conflicting variant classification results, with different levels of evidences, each type of database providing limited and different information depending on their database characteristics. For example, ERBB2 mutation was mentioned in NCCN guideline and was classified as tier I with a level 2A evidence, whereas the evidence presented by My Cancer Genome, Personalized Cancer Therapy, and ClinVar categorized it into tier II, and COSMIC categorized it into tier III. Therefore, we suggest that the combination of databases should contain at least four layers of resources (Figure 4): (1) FDA, CFDA, and professional guidelines; (2) variant interpretive databases and PubMed implementing evidence-based expert consensus for curating genes and variants, which summarizes varying levels of evidence from well-powered studies, clinical trials, small studies, and case reports; (3) variant catalogs covering the rare variants of variant interpretive databases; and (4) databases containing information on on-going clinical trials.
Compared to previous targeted therapeutics for the three clinical cases reported, the choices of drugs has increased greatly owing to the rapid development of new drugs and clinical studies. Multiple drug recommendations by various laboratories varied, probably due to four reasons. First, the laboratories classifying the variant as low tier may recommend fewer targeted drugs than those giving high-tier evaluations for that variant. For example, the three laboratories classifying ERBB2 mutations as variants of unknown significance did not suggest any drug for the patient. It is not acceptable for the results to miss the targeted therapy supported by companion diagnostics and professional guidelines. Second, different therapeutic drugs may be recommended for the variants, depending on the databases used (Table 2). All the targeted therapy recommendations based on correct evidence are preferable, but the laboratories should adequately illustrate the level and source of evidence when recommending the targeted therapy, since this information would be used for decision-making in the clinical context. Third, some laboratories only reported the drugs with strong evidence, instead of all the drugs in the databases, for clinical decisions. For example, four laboratories using a combination of the NCCN and My Cancer Genome databases only reported the four TKI inhibitors for EGFR G719C, as described in the NCCN or CFDA indicators, whereas neratinib is also recommended by My Cancer Genome. The approach of the four laboratories did not provide enough information about investigational therapy to physicians. Fourth, some laboratories recommended drugs with no confirmed evidence from their database used. For example, two laboratories (lab 72 and lab 86) recommending pertuzumab, lapatinib, and trastuzumab emtansine, used ClinVar, which did not address the clinical implications. Four laboratories (lab 20, lab 30, lab 35, and lab 37) using NCCN, recommended osimertinib, a third-generation TKI for EGFR G719C and S768I mutations. Osimertinib was approved by the FDA for patients with NSCLC that were positive for T790M (Table 2). However, NCCN (version 1.2017) did not recommend osimertinib as a therapeutic option for EGFR G719C and S768I mutations. It is undesirable for laboratories to provide targeted therapy without any source of evidence. We will also emphasize on the rapid progress of targeted therapies, since osimertinib was recently reported to have satisfactory efficacy in the first-line treatment of EGFR mutation-positive advanced NSCLC and has been included in NCCN (version 3. 2018) as the recommendation drug for first-line therapy of patients with NSCLC and sensitizing EGFR mutations [25, 26]. However, prior to our study, no consensus evidence was shown regarding the efficacy of osimertinib against other EGFR mutations, except T790M. Similarly, trastuzumab emtansine has been recommended by NCCN (version 3. 2018), based on the results from clinical trial NCT02675829, which showed trastuzumab emtansine treatment to be more effective than previous therapies [27]. Therefore, it is essential for laboratories to provide interpretation based on updated evidence.
According to the limitations highlighted in the interpretation of clinical laboratories, we summarized a pipeline for variant interpretation (Figure 4), which might be helpful to solve the challenges in variant classification and drug recommendation. Compared to the previously established interpretation pipelines [28], our variant interpretation pipeline provided the corresponding databases with different levels of clinical evidence to interpret variant classification in AMP-ASCO-CAP guideline, and became a clearly and visualized algorithm to bring more possibilities to realize the consistency in variant interpretation. It was worth noticing that databases included in our pipeline represented all types of databases, rather than limit to those exemplified databases that submitted by participating laboratories. Our study merely uncovered the principle types of databases that should be included in the variant interpretation process and did not determine the specific databases. A major limitation of this study is the small number of variants included. The results might be more representative, if the variability in interpretation could be collected from more cancer variants, associated with insufficient evidence, to lead to a clear consensus. However, for the six variants included in this study, many targeted therapies are also based on insufficient evidence, since the number of available targeted therapeutic agents approved by the FDA/CFDA and recommended by professional guidelines is very small. In our practice of precision medicine, the problem of insufficient evidence exists in the classification of the variants, as well as the recommendation of targeted therapies.
It should be emphasized here, that the variant interpretation of laboratories is different from the treatment recommendation offered by physicians, which is made in clinical settings. The physicians will make decisions according to clinical symptoms, complications, therapy history, affordability, and clinical report from laboratories. It is not easy for oncologists to track all the literature about the relevance of targeted therapies with variants, due to the rapid progress in precision oncology. Therefore, accurate reports by laboratories, translated from genomic data, are extremely helpful for oncologists to select an optimal therapy for the benefit of the patients.
In conclusion, multiplex tumor genetic testing depends not only on precise variant detection based on massively parallel sequencing, but also on the accurate interpretation of variants. Inconsistent variant interpretation occurs frequently due to numerous reasons. Our study emphasized the importance of making a comprehensive and detailed variant interpretation pipeline to standardize variant classification and recommend therapeutic drugs to minimize inappropriate therapeutic options.
Abbreviations
NGS: next-generation sequencing; NSCLC: non-small cell lung cancer; NCCN: National Comprehensive Cancer Network; AMP: the Association for Molecular Pathology; ASCO: American Society of Clinical Oncology; CAP: College of American Pathologists; COSMIC: the Catalogue of Somatic Mutations In Cancer; FDA: the Food and Drug Administration; CFDA: the China Food and Drug Administration; TCGA: The Cancer Genome Atlas; EGFR: epidermal growth factor receptor; EML4: echinoderm microtubule associated protein like 4 gene; ALK: anaplastic lymphoma receptor tyrosine kinase gene; KRAS: Kirsten rat sarcoma 2 viral oncogene homolog; ERBB2: erb-b2 receptor tyrosine kinase 2; ROS1: ROS proto-oncogene 1; RET: ret proto-oncogene.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported by a grant from the Fund for Dongcheng Excellent Talents (Rui Zhang), Beijing Hospital Nova Project grant BJ-2018-136 (Rui Zhang) and National Natural Science Foundation of China grant 81772273 (Jinming Li).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kamps R, Brandao RD, Bosch BJ. et al. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18:E308
2. Ettinger DS, Wood DE, Aisner DL. et al. Non-small cell lung cancer, version 5.2017, nccn clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:504-35
3. Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80
4. Li MM, Datto M, Duncavage EJ. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, american society of clinical oncology, and college of american pathologists. J Mol Diagn. 2017;19:4-23
5. Dienstmann R, Dong F, Borger D. et al. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol Oncol. 2014;8:859-73
6. Sukhai MA, Craddock KJ, Thomas M. et al. A classification system for clinical relevance of somatic variants identified in molecular profiling of cancer. Genet Med. 2016;18:128-36
7. Van Allen EM, Wagle N, Stojanov P. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682-8
8. Hoskinson DC, Dubuc AM, Mason-Suares H. The current state of clinical interpretation of sequence variants. Curr Opin Genet Dev. 2017;42:33-9
9. Patel JM, Knopf J, Reiner E. et al. Mutation based treatment recommendations from next generation sequencing data: A comparison of web tools. Oncotarget. 2016;7:22064-76
10. Ritter DI, Roychowdhury S, Roy A. et al. Somatic cancer variant curation and harmonization through consensus minimum variant level data. Genome Med. 2016;8:117
11. Forbes SA, Beare D, Boutselakis H. et al. Cosmic: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777-83
12. Landrum MJ, Lee JM, Benson M. et al. Clinvar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862-8
13. Cappuzzo F, Bemis L, Varella-Garcia M. HER2 mutation and response to trastuzumab therapy in non-small-cell lung cancer. N Engl J Med. 2006;354:2619-21
14. Tanizaki J, Banno E, Togashi Y. et al. Case report: Durable response to afatinib in a patient with lung cancer harboring two uncommon mutations of EGFR and a KRAS mutation. Lung Cancer. 2016;101:11-5
15. Xu CW, Cai XY, Shao Y. et al. A case of lung adenocarcinoma with a concurrent EGFR mutation and ALK rearrangement: A case report and literature review. Mol Med Rep. 2015;12:4370-5
16. Zhang R, Ding J, Han Y. et al. The reliable assurance of detecting somatic mutations in cancer-related genes by next-generation sequencing: the results of external quality assessment in China. Oncotarget. 2016;7:58500-15
17. Prawira A, Pugh TJ, Stockley TL. et al. Data resources for the identification and interpretation of actionable mutations by clinicians. Ann Oncol. 2017;28:946-57
18. Mazieres J, Peters S, Lepage B. et al. Lung cancer that harbors an her2 mutation: Epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31:1997-2003
19. Ramalingam SS, Blackhall F, Krzakowski M. et al. Randomized phase II study of dacomitinib (pf-00299804), an irreversible pan-human epidermal growth factor receptor inhibitor, versus erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2012;30:3337-44
20. Sukhai MA, Craddock KJ, Thomas M. et al. A classification system for clinical relevance of somatic variants identified in molecular profiling of cancer. Genet Med. 2016;18:128-36
21. U.S. Food and Drug Administration, CDRH'S approach to tumor profiling next generation sequencing tests. Accessed on 14 May 2018. Available online: https://. www.fda.gov/downloads/medicaldevices/productsandmedicalprocedures/invitrodiagnostics/ucm584603.pdf
22. Meric-Bernstam F, Johnson A, Holla V. et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015:107
23. Dienstmann R, Jang IS, Bot B. et al. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015;5:118-23
24. Kulkarni S, Pfeifer J. Clinical Genomics. 1st ed. Elsevier Inc: Academic Press. 2014
25. Soria JC, Ohe Y, Vansteenkiste J. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113-25
26. National Comprehensive Cancer Network. (NCCN) Clinical Practice Guidelines in Oncology. Non-Small Cell Lung Cancer, Version 3. 2018. Accessed on 14 May 2018. Available online: https://. www.nccn.org/professionals/physician_gls/default.aspx
27. Li BT, Shen R, Buonocore D. et al. Ado-trastuzumab emtansine in patients with HER2 mutant lung cancers: Results from a phase II basket trial. J Clin Oncol. 2017;35:15_suppl 8510
28. Dienstmann R, Dong F, Borger D. et al. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol Oncol. 2014;8:859-73
Author contact
![]() Corresponding authors: Rui Zhang, ruizhangorg.cn and Jinming Li, jmliorg.cn, National Center for Clinical Laboratories, Beijing Hospital, No.1 Dahua Road, Dongdan, Beijing, 100730, People's Republic of China. Tel: +86-10-58115053; Fax: +86-10-65212064.
Corresponding authors: Rui Zhang, ruizhangorg.cn and Jinming Li, jmliorg.cn, National Center for Clinical Laboratories, Beijing Hospital, No.1 Dahua Road, Dongdan, Beijing, 100730, People's Republic of China. Tel: +86-10-58115053; Fax: +86-10-65212064.

 Global reach, higher impact
Global reach, higher impact