Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(8):1825-1832. doi:10.7150/jca.29438 This issue Cite
Research Paper
Dysregulated Expression of Circular RNAs Serve as Prognostic and Clinicopathological Markers in Cancer
1. Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
2. Department of Endocrinology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
Received 2018-8-23; Accepted 2019-2-7; Published 2019-4-21
Abstract
Purpose: Circular RNAs (circRNAs) as prognostic biomarkers have spurred considerable interest in several types of tumors. In the present study, we aimed to elucidate the clinicopathological and prognostic values of circRNAs in human cancer.
Methods: We systematically searched PubMed Central (PMC), PubMed, Web of Science, EMBASE, Scopus, CBM and the Cochrane Library databases up to Nov 29, 2018. Eligible studies reporting on the association between circRNAs expression and clinicopathological and prognostic outcomes in cancer were incorporated. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess clinicopathological parameters, and hazard ratios (HRs) and 95% CIs to estimate overall survival (OS).
Results: Thirty-two studies involving 4529 patients were incorporated into our meta-analysis. Pooled results showed that high expression of oncogenic circRNAs was significantly associated with poor clinicopathological characteristics (tumor size: OR=1.29, 95%Cl: 1.10-1.51; TNM stage: OR=1.62, 95%Cl: 1.41-1.87; differentiation grade: OR=1.41, 95%Cl: 1.11-1.78; lymph node metastasis: OR=1.69; 95%Cl: 1.34-2.13; distant metastasis: OR=2.75; 95%Cl: 1.92-3.95) and a poor prognosis (OS: HR=2.75; 95%Cl: 2.34-3.15). Furthermore, we found that high expression of tumor-suppressor circRNAs was correlated with improved clinical characteristics (tumor size: OR=0.72; 95%Cl: 0.56-0.92; TNM stage: OR=0.77, 95%Cl: 0.68-0.88) and longer survival times (OS: HR=0.49; 95%Cl: 0.42-0.56). Subgroup analyses based on cancer types and circRNA types were also performed.
Conclusion: Our study indicates that circRNAs may serve as important biomarkers for clinicopathologic features and prognosis in human cancer.
Keywords: circRNA, cancer, prognosis, meta-analysis
Introduction
Circular RNA (circRNA) is a new class of endogenous non-coding RNA generated from the back-splicing by the canonical spliceosome [1]. Numerous circRNAs seem to be specifically expressed in a given cell type or developmental stage [2]. CircRNAs are characterized by a covalently closed loop structure with neither a 5' cap nor a 3' polyadenylated tail [3, 4]. Moreover, they are inherently resistant to exonucleolytic RNA decay. Taken their conserved and stable characteristics into account, circRNAs might be suitable as required novel biomarkers and therapeutic targets for human cancer [5-7]. Recent studies indicate that circRNAs might regulate transcription process and RNA splicing, function as efficient microRNA sponges, and can be translated into protein driven by N6-methyladenosine (m6A) modification [8, 9]. However, more underlying mechanisms and functions of circRNAs remain largely unknown. CircRNAs have been recently confirmed to have regulative functions in cell function, development of heart diseases, and pathogenesis of neurodegenerative diseases such as Alzheimer's disease [10]. Cancer is a major public health problem worldwide [11, 12]. The function of upregulated or downregulated circRNAs in various cancer types still require further investigation.
In this study, we performed a meta-analysis to summarize the clinicopathological and prognostic values of circRNAs in different types of cancer. Further prospective studies including more kinds of circRNAs in various tumors are warranted in the future.
Methods
Data search strategy
A computerized literature search was performed in the PubMed Central (PMC), PubMed, Web of Science, EMBASE, Scopus, CBM and the Cochrane Library databases up to Nov 29, 2018. A search strategy was developed based on the following terms: (“circRNA” or “circular RNA”) and (“cancer” or “carcinoma” or “tumor” or “tumour” or “neoplas*”). We additionally hand-searched the references of relevant articles and contacted investigators of certain studies when necessary. To be eligible for inclusion in the meta-analysis, a study must meet the following criteria: (1) case-control study or cohort study; (2) patients had a pathological diagnosis of cancer; (3) assessing the association between circRNA expression, clinicopathological features, and prognosis. Exclusion criteria were as follows: (1) literatures not pertinent to circRNA or cancer; or (2) similar studies from the same author as well as multiple duplicate data in the different works; or (3) animal experiments, case reports, correspondences, reviews, expert opinions, letters; or (4) no available data and the authors could not be contacted.
Data extraction and quality assessment
Two investigators (XH, ZCZ) evaluated the eligibility of all retrieved studies and extracted the relevant data independently. Extracted databases were then cross-checked between the two authors to rule out any discrepancy. Disagreement was resolved by consulting with a third investigator (ZWS). The following data of each collected studies were extracted independently: author, year of publication, circRNA type, cancer type, cases, detection method, role of circRNA and duration of follow-up. The study quality was assessed in accordance with the Newcastle-Ottawa Scale (NOS) (Supplementary Table S1). Eight items were extracted, and each item scored 1. The total scores ranged from 0 to 8. If the scores were≥7, then the study was considered high quality. Our investigation process was in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Statistical analysis
The statistical analysis was performed using STATA 14. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess clinicopathological parameters, and hazard ratios (HRs) and 95% CIs to estimate overall survival (OS). The between-study heterogeneity was evaluated by using the chi-square test and the I2 statistic. An I2 value of >50% of the I2 statistic was considered to indicate significant heterogeneity [13]. When a significant heterogeneity existed across the included studies, a random effects model was used for the analysis. Otherwise, the fixed effects model was used [14]. Subgroup analyses were performed to detect the source of heterogeneity. We further conducted sensitivity analyses to substantiate the stability of results and detect the potential source of heterogeneity. Publication bias was evaluated qualitatively by inspecting funnel plots and quantitatively through the Begg's and Egger's test. A two-tailed P-value<0.05 implies a statistically significant publication bias.
Results
Search results
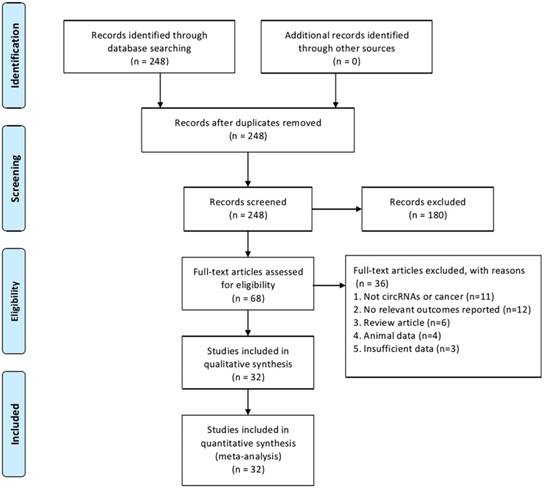
The study selection process is illustrated in Fig. 1. A total of 248 potential articles were identified from the databases search. Among these articles, 180 were excluded after abstract review, leaving 68 articles for the full-text review. In the review, 36 studies were excluded for the reasons as follows: eleven were eliminated because they were irrelevant to circRNA or cancer, twelve studies were of no relevant outcomes reported, six studies were of reviews, four studies involved non-human experiments, and three studies were excluded because of insufficient data for analysis. Finally, thirty-two studies with a total of 4529 patients that met the inclusion criteria were included in this meta-analysis.
Study selection and characteristics
Baseline characteristics of the included studies are presented in Table 1. The publication years of the eligible studies ranged from 2017 to 2018. Cancer types included gastric cancer (n=2), colorectal cancer (n=3), hepatocellular carcinoma (n=6), breast cancer (n=2), bladder cancer (n=5), lung cancer (n=4), osteosarcoma (n=5). The number of patients in each study ranged from 30 to 631. Additionally, the circRNA expression levels were measured by quantitative real time polymerase chain reaction (qRT-PCR). As indicated in Table 1, twenty-one circRNAs were recognized as tumor promoters and eleven were tumor suppressors. Moreover, the mean duration of follow-up ranged from 33 to 140 months. CircRNAs could serve as sponges to regulate gene expression via sequestering miRNAs. Therefore, we included corresponding miRNAs. All included studies screened out circRNAs from tumor tissues. According to the Newcastle-Ottawa Scale (NOS), the quality scores of the included trials ranged from 7 to 8, which indicated a high quality (Additional file 1).
Flowchart of the study selection process.
Meta-analysis for clinicopathological features
In the present study, we assessed the relationship between circRNAs expression and clinicopathological features of cancer patients (Table 2). High expression of oncogenic circRNAs was significantly associated with poor clinicopathological characteristics (tumor size: OR=1.29, 95%Cl: 1.10-1.51; TNM stage: OR=1.62, 95%Cl: 1.41-1.87; differentiation grade: OR=1.41, 95%Cl: 1.11-1.78; lymph node metastasis: OR=1.69; 95%Cl: 1.34-2.13; distant metastasis: OR=2.75; 95%Cl: 1.92-3.95). Furthermore, our study showed that high expression of tumor-suppressor circRNAs was correlated with improved clinical characteristics (tumor size: OR=0.72; 95%Cl: 0.56-0.92; TNM stage: OR=0.77, 95%Cl: 0.68-0.88). However, no significant relationship was observed between tumor-suppressor circRNAs overexpression and other clinical characteristics such as age, gender, differentiation grade, lymph node metastasis and distant metastasis.
Meta-analysis for overall survival
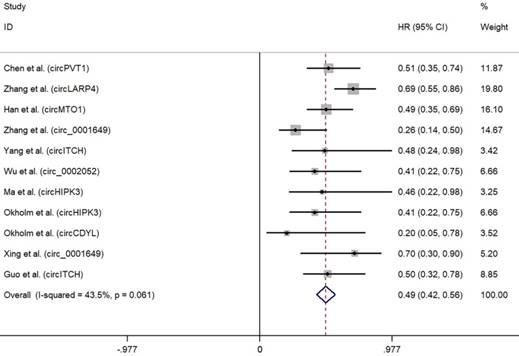
As depicted in Fig. 2, high expression of oncogenic circRNAs was significantly associated with a poor prognosis (OS: HR=2.75; 95%Cl: 2.34-3.15; p<0.001), and the fixed-effect model was adopted in terms of no significant heterogeneity among the studies (I²=0.5%, p=0.452). Furthermore, high expression of tumor-suppressor circRNAs was correlated with longer survival times (OS: HR=0.49; 95%Cl: 0.42-0.56; p<0.001). No significant heterogeneity among the studies (I²=43.5%, p=0.061) was found and the fixed-effect model was adopted (Fig. 3).
Subgroup analysis in terms of various cancer types
We further conducted subgroup analysis by factors of cancer types to explore the source of heterogeneity (Table 3). High expression of circRNAs was correlated with longer survival times in gastric cancer (OS: HR=0.62; 95%Cl: 0.50-0.74), hepatocellular carcinoma (OS: HR=0.44; 95%Cl: 0.33-0.55), bladder cancer (OS: HR=0.49; 95%Cl: 0.33-0.65) and osteosarcoma (OS: HR=0.49; 95%Cl: 0.28-0.71). However, high expression of circRNAs was correlated with poor survival in colorectal cancer (OS: HR=2.52; 95%Cl: 1.61-3.43), breast cancer (OS: HR=3.47; 95%Cl: 1.95-5.00) and lung cancer (OS: HR=2.91; 95%Cl: 1.92-3.91). Relatively significant heterogeneities were observed in hepatocellular carcinoma (I²= 86.9%), lung cancer (I²= 71.0%) and osteosarcoma (I²= 70.3%).
Subgroup analysis in terms of various circRNAs types
When subgrouped by circRNAs types (Table 4), our study found that high expression of circRNAs was correlated with longer survival times in circPVT1 (OS: HR=0.54; 95%Cl: 0.35-0.74), circHIPK3 (OS: HR=0.50; 95%Cl: 0.29-0.72), circ_0001649 (OS: HR= 0.35; 95%Cl: 0.20-0.51) and circ-ITCH (OS: HR=0.49; 95%Cl: 0.30-0.69). However, high expression of circRNAs was correlated with poor survival in circRNA Cdr1as (OS: HR=2.77; 95%Cl: 1.70-3.83) and circ_0067934 (OS: HR=3.66; 95%Cl: 2.15-5.16). No significant heterogeneities were observed in circRNA Cdr1as (I²=46.3%), circ- ITCH (I²=0.0%) and circ_0067934 (I²=0.0%).
Main characteristics of the studies included in this meta-analysis.
| Study | Year | CircRNA | Cancer type | mRNA | Sample | CircRNA expression | Detection method | Expression status | Follow -up (months) | Cita- tion | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | ||||||||||
| Zhou et al. | 2018 | circ_0008717 | Osteosarcoma | miR-203 | Tumor tissue | 45 | 45 | qRT-PCR | Up-regulated | 80 | [15] |
| Zhu et al. | 2018 | circPVT1 | Osteosarcoma | NA | Tumor tissue | 30 | 50 | qRT-PCR | Up-regulated | 62 | [16] |
| Zhang et al. | 2017 | circUBAP2 | Osteosarcoma | miR-143 | Tumor tissue | 42 | 50 | qRT-PCR | Up-regulated | 60 | [17] |
| Hsiao et al. | 2017 | circCCDC66 | Colorectal cancer | miR-33b, miR-93 | Tumor tissue | 131 | 98 | qRT-PCR | Up-regulated | 58 | [18] |
| Weng et al. | 2018 | ciRS-7 | Colorectal cancer | miR-7 | Tumor tissue | 89 | 76 | qRT-PCR | Up-regulated | 83 | [19] |
| He et al. | 2017 | circGFRA1 | Breast cancer | miR-34a | Tumor tissue | 109 | 103 | qRT-PCR | Up-regulated | 140 | [20] |
| Jiang et al. | 2017 | circCdr1as | Cholangiocarcinoma | NA | Tumor tissue | 24 | 30 | qRT-PCR | Up-regulated | 45 | [21] |
| Zhong et al. | 2017 | circMYLK | Bladder cancer | miR-29a | Tumor tissue | 16 | 16 | qRT-PCR | Up-regulated | 33 | [22] |
| Liu et al. | 2018 | circ_103809 | Lung cancer | miR-4302 | Tumor tissue | 22 | 22 | qRT-PCR | Up-regulated | 76 | [23] |
| Yao et al. | 2017 | circ_100876 | Lung cancer | NA | Tumor tissue | 48 | 52 | qRT-PCR | Up-regulated | 40 | [24] |
| Zhao et al. | 2017 | circFADS2 | Lung cancer | miR-498 | Tumor tissue | 20 | 23 | qRT-PCR | Up-regulated | 60 | [25] |
| Luan et al. | 2018 | circ_0084043 | Melanoma | miR-153-3p | Tumor tissue | 15 | 15 | qRT-PCR | Up-regulated | 60 | [26] |
| Wei et al. | 2018 | circZFR | Papillary thyroid cancer | miR-1261 | Tumor tissue | 41 | 41 | qRT-PCR | Up-regulated | 55 | [27] |
| Zhang et al. | 2018 | circ_0023404 | Cervical cancer | miR-136 | Tumor tissue | 27 | 26 | qRT-PCR | Up-regulated | 78 | [28] |
| Verduci et al. | 2017 | circPVT1 | Head and neck squamous cell carcinoma | miR-497-5p | Tumor tissue | 71 | 35 | qRT-PCR | Up-regulated | 70 | [29] |
| Xu et al. | 2017 | circCdr1as | Hepatocellular carcinoma | miR-7 | Tumor tissue | 48 | 47 | qRT-PCR | Up-regulated | 62 | [30] |
| Zeng et al. | 2017 | circHIPK3 | Colorectal cancer | miR-7 | Tumor tissue | 89 | 89 | qRT-PCR | Up-regulated | 90 | [31] |
| Li et al. | 2017 | circHIPK3 | Bladder cancer | miR-558 | Tumor tissue | 45 | 179 | qRT-PCR | Up-regulated | 112 | [32] |
| Meng et al. | 2018 | circ_10720 | Hepatocellular carcinoma | NA | Tumor tissue | 32 | 65 | qRT-PCR | Up-regulated | 118 | [33] |
| Wu et al. | 2018 | circIRAK3 | Breast cancer | miR-3607 | Tumor tissue | 60 | 62 | qRT-PCR | Up-regulated | 120 | [34] |
| Wang et al. | 2018 | circ_0067934 | Lung cancer | NA | Tumor tissue | 79 | 80 | qRT-PCR | Up-regulated | 60 | [35] |
| Zhu et al. | 2018 | circ_0067934 | Hepatocellular carcinoma | miR-1324 | Tumor tissue | 25 | 25 | qRT-PCR | Up-regulated | 60 | [36] |
| Chen et al. | 2017 | circPVT1 | Gastric cancer | miR-125 | Tumor tissue | 107 | 80 | qRT-PCR | Down-regulated | 85 | [37] |
| Zhang et al. | 2017 | circLARP4 | Gastric cancer | miR-424-5p | Tumor tissue | 220 | 411 | qRT-PCR | Down-regulated | 110 | [38] |
| Han et al. | 2017 | circMTO1 | Hepatocellular carcinoma | miR-9 | Tumor tissue | 116 | 116 | qRT-PCR | Down-regulated | 80 | [39] |
| Zhang et al. | 2018 | circ_0001649 | Hepatocellular carcinoma | NA | Tumor tissue | 35 | 42 | qRT-PCR | Down-regulated | 44 | [40] |
| Yang et al. | 2018 | circITCH | Bladder cancer | miR-17, miR-224 | Tumor tissue | 25 | 45 | qRT-PCR | Down-regulated | 60 | [41] |
| Wu et al. | 2018 | circ_0002052 | Osteosarcoma | miR-1205 | Tumor tissue | 54 | 54 | qRT-PCR | Down-regulated | 50 | [42] |
| Ma et al. | 2018 | circHIPK3 | Osteosarcoma | NA | Tumor tissue | 37 | 45 | qRT-PCR | Down-regulated | 60 | [43] |
| Okholm et al. | 2017 | circHIPK3 | Bladder cancer | NA | Tumor tissue | 228 | 229 | qRT-PCR | Down-regulated | 75 | [44] |
| Okholm et al. | 2017 | circCDYL | Bladder cancer | NA | Tumor tissue | 228 | 229 | qRT-PCR | Down-regulated | 75 | [44] |
| Xing et al. | 2018 | circ_0001649 | Retinoblastoma | NA | Tumor tissue | 30 | 30 | qRT-PCR | Down-regulated | 60 | [45] |
| Guo et al. | 2017 | circITCH | Hepatocellular carcinoma | NA | Tumor tissue | 100 | 188 | qRT-PCR | Down-regulated | 83 | [46] |
Abbreviations: qRT-PCR, quantitative real time polymerase chain reaction; NA, not available.
Clinical characteristics of circRNAs in cancer.
| Tumor promoter | Tumor suppressor | |||||
|---|---|---|---|---|---|---|
| OR | 95% Cl | P | OR | 95% Cl | P | |
| Age | 0.794 | 0.592-1.065 | 0.124 | 1.008 | 0.804-1.263 | 0.946 |
| Gender (M/W) | 1.264 | 0.879-1.817 | 0.207 | 1.020 | 0.896-1.161 | 0.763 |
| Tumor size | 1.291 | 1.104-1.510 | 0.001 | 0.717 | 0.560-0.917 | 0.008 |
| TNM stage (III+IV/I+II) | 1.621 | 1.407-1.868 | 0.000 | 0.773 | 0.683-0.875 | 0.000 |
| Differentiation grade | 1.406 | 1.112-1.778 | 0.004 | 0.889 | 0.760-1.040 | 0.141 |
| Lymph node metastasis (Y/N) | 1.687 | 1.337-2.129 | 0.000 | 0.993 | 0.889-1.110 | 0.906 |
| Distant metastasis (Y/N) | 2.753 | 1.919-3.949 | 0.000 | 0.608 | 0.360-1.027 | 0.063 |
Abbreviations: M, men; W, women; Y, yes; N, no; OR, odds ratio; CI, confidence interval. The results are in bold if P < 0.05.
Subgroup analysis of circRNAs in various cancer types.
| Subgroup analysis | Studies (n) | CircRNA | HR | 95% CI | p-value | Heterogeneity | ||
|---|---|---|---|---|---|---|---|---|
| I2 (%) | PQ | Model | ||||||
| Gastric cancer | Chen et al. (2017) | circPVT1 | 0.508 | 0.347-0.745 | ||||
| Zhang et al. (2017) | circLARP4 | 0.689 | 0.552 -0.860 | |||||
| Total | 0.621 | 0.500-0.743 | 0.000 | 49.6% | 0.159 | Fixed | ||
| Colorectal cancer | Hsiao et al. (2017) | circCCDC66 | 2.266 | 1.265-4.061 | ||||
| Weng et al. (2018) | ciRS-7 | 2.441 | 1.298-4.594 | |||||
| Zeng et al. (2017) | circHIPK3 | 3.047 | 1.525 -5.147 | |||||
| Total | 2.518 | 1.608 -3.429 | 0.000 | 0.0% | 0.809 | Fixed | ||
| Hepatocellular carcinoma | Han et al. (2017) | circMTO1 | 0.491 | 0.349 -0.691 | ||||
| Zhang et al. (2018) | circ_0001649 | 0.265 | 0.141-0.498 | |||||
| Meng et al. (2018) | circ_10720 | 4.300 | 1.495-6.984 | |||||
| Xu et al. (2017) | circCdr1as | 3.621 | 2.108-5.325 | |||||
| Guo et al. (2017) | circITCH | 0.512 | 0.320-0.781 | |||||
| Zhu et al. (2018) | circ_0067934 | 3.605 | 1.816-5.546 | |||||
| Total | 0.441 | 0.333-0.549 | 0.000 | 86.9% | 0.000 | Random | ||
| Breast cancer | He et al. (2017) | circGFRA1 | 3.790 | 2.011-7.142 | ||||
| Wu et al. (2018) | circIRAK3 | 3.328 | 1.208-5.234 | |||||
| Total | 3.474 | 1.947-5.000 | 0.000 | 0.0% | 0.764 | Fixed | ||
| Bladder cancer | Yang et al. (2018) | circITCH | 0.480 | 0.236-0.976 | ||||
| Zhong et al. (2017) | circMYLK | 2.595 | 1.010-6.668 | |||||
| Okholm et al. (2017) | circHIPK3 | 0.406 | 0.220-0.750 | |||||
| Okholm et al. (2017) | circCDYL | 0.533 | 0.325-0.780 | |||||
| Li et al. (2017) | circHIPK3 | 4.325 | 2.800-6.907 | |||||
| Total | 0.490 | 0.332-0.654 | 0.000 | 71.0% | 0.008 | Random | ||
| Lung cancer | Liu et al. (2018) | circ_103809 | 2.494 | 1.036-6.005 | ||||
| Yao et al. (2017) | circ_100876 | 2.731 | 1.709-4.363 | |||||
| Zhao et al. (2017) | circFADS2 | 3.232 | 1.495-6.984 | |||||
| Wang et al. (2018) | circ_0067934 | 3.774 | 1.498-6.670 | |||||
| Total | 2.913 | 1.919-3.907 | 0.000 | 0.0% | 0.883 | Fixed | ||
| Osteosarcoma | Wu et al. (2018) | circ_0002052 | 0.406 | 0.220-0.750 | ||||
| Ma et al. (2018) | circHIPK3 | 0.461 | 0.218-0.977 | |||||
| Zhou et al. (2018) | circ-0008717 | 2.729 | 1.100-6.773 | |||||
| Zhu et al. (2018) | circPVT1 | 3.306 | 1.663-6.570 | |||||
| Zhang et al. (2017) | circUBAP2 | 2.364 | 1.275-4.382 | |||||
| Total | 0.496 | 0.282-0.710 | 0.000 | 70.3% | 0.009 | Random | ||
Abbreviations: HR, hazard ratio; CI, confidence interval.
Subgroup analysis in terms of various circRNAs types.
| Subgroup analysis | Studies (n) | Cancer type | HR | 95% CI | p-value | Heterogeneity | ||
|---|---|---|---|---|---|---|---|---|
| I2 (%) | PQ | Model | ||||||
| circPVT1 | Chen et al. (2017) | Gastric cancer | 0.508 | 0.347-0.745 | ||||
| Zhu et al. (2018) | Osteosarcoma | 3.306 | 1.663-6.570 | |||||
| Verduci et al. (2017) | Head and neck squamous cell carcinoma | 2.120 | 1.213-4.950 | |||||
| Total | 0.544 | 0.347-0.741 | 0.000 | 73.8 | 0.022 | Random | ||
| circHIPK3 | Zeng et al. (2017) | Colorectal cancer | 3.012 | 1.534-5.052 | ||||
| Okholm et al. (2017) | Bladder cancer | 0.406 | 0.220-0.750 | |||||
| Li et al. (2017) | Bladder cancer | 4.011 | 2.856-6.901 | |||||
| Ma et al. (2018) | Osteosarcoma | 0.461 | 0.218-0.977 | |||||
| Total | 0.502 | 0.287-0.716 | 0.000 | 84.7 | 0.000 | Random | ||
| circRNA Cdr1as | Xu et al. (2017) | Hepatocellular carcinoma | 3.612 | 2.109-5.315 | ||||
| Jiang et al. (2017) | Cholangiocarcinoma | 2.108 | 1.120-3.968 | |||||
| Total | 2.767 | 1.704-3.831 | 0.000 | 46.3 | 0.172 | Fixed | ||
| circ_0001649 | Zhang et al. (2018) | Hepatocellular carcinoma | 0.265 | 0.141-0.498 | ||||
| Xing et al. (2018) | Retinoblastoma | 0.611 | 0.335-0.901 | |||||
| Total | 0.353 | 0.199-0.506 | 0.000 | 71.8 | 0.060 | Random | ||
| circ-ITCH | Guo et al. (2017) | Hepatocellular carcinoma | 0.500 | 0.320-0.780 | ||||
| Yang et al. (2018) | Bladder cancer | 0.480 | 0.236-0.976 | |||||
| Total | 0.494 | 0.299-0.690 | 0.000 | 0.0 | 0.927 | Fixed | ||
| circ_0067934 | Zhu et al. (2018) | Hepatocellular carcinoma | 3.635 | 1.821-5.508 | ||||
| Wang et al. (2018) | Lung cancer | 3.774 | 1.498-6.670 | |||||
| Total | 3.659 | 2.154-5.164 | 0.000 | 0.0 | 0.915 | Fixed | ||
Abbreviations: HR, hazard ratio; CI, confidence interval.
Publication bias and sensitivity analysis
The funnel plot did not indicate any evidence of publication bias in this analysis (Figure S2). No evidence of publication bias was observed from Begg's funnel plot (P=0.369) (Figure S3) and Egger's test (P=0.082) (Figure S4). To sum up, the possibility of publication bias could be excluded. The sensitivity analysis showed that the results of the meta-analysis did not change when studies were omitted one by one (Figure S5).
Discussion
The present study revealed a significant association between high expression of circRNAs and clinicopathological and prognostic significance in human cancer. Thirty-two studies involving 4529 patients were incorporated into our meta-analysis. Since the expression of circRNAs were upregulated or downregulated in different cancers, we decided to recognize twenty-one circRNAs as tumor promoters and eleven as tumor suppressors and analysis them respectively. Pooled results showed that high expression of oncogenic circRNAs was significantly associated with poor clinicopathological characteristics including tumor size, TNM stage, differentiation grade, lymph node metastasis and distant metastasis. A significant association between oncogenic circRNAs and a poor prognosis was also detected in our study. Furthermore, we found that high expression of tumor-suppressor circRNAs was correlated with longer survival times and improved clinical characteristics such as tumor size and TNM stage.
Forest plots for OS according to the type of oncogenic circRNAs in cancer.

Forest plots for OS according to the type of tumor suppressor circRNAs in cancer.
Relatively significant heterogeneities were observed in our study. To explore the source of heterogeneity, we performed sensitivity analysis and found that none of those studies altered the pooled OR significantly, indicating that other unknown factors might be the cause. Furthermore, we predicted that disease type may account for the heterogeneity and the stratified analyses were then performed. Subgroup analysis focused mainly on seven cancer types, including gastric cancer [37, 38], colorectal cancer [18, 19, 31], hepatocellular carcinoma [30, 33, 36, 39, 40, 46], bladder cancer [22, 32, 41, 44], breast cancer [20, 34], lung cancer [23-25, 35], osteosarcoma [15-17, 42, 43]. Because of only one article included for other cancer types, we failed to perform further meta-analysis. Relatively significant heterogeneities were observed in three cancer types including hepatocellular carcinoma, lung cancer and osteosarcoma (I²>50%). Small sample size and limited article included may account for the significant heterogeneity. Neither the Egger test nor the Begg's funnel plot showed significant publication bias for the association between circRNAs expression and clinicopathological and prognostic significances. Even though the results are reliable, additional relevant studies are warranted to further confirm the findings of this meta-analysis.
Four previous meta-analysis by Wang et al. [5], Ding et al. [47], Li et al. [48], and Chen et al. [49] were also performed to detect the association between circRNAs and cancer. As for Li et al., they included 10 articles about circRNAs as diagnostic biomarkers for cancer. In the study of Wang et al., they highlighted the diagnostic value of circRNAs for human cancers especially in HCC diagnosis with 17 publications. Chen et al. just focused on circRNAs as potential biomarkers for the diagnosis of digestive system malignancy. Li et al., Wang et al., and Chen et al. failed to discuss anything about the prognostic and clinicopathological significances of circRNAs. Moreover, limited studies and sample sizes were included in their studies, which decreased the reliability of conclusions. Ding et al. assessed the expression of circRNAs as a promising biomarker in the diagnosis and prognosis of cancers. However, only 11 articles were included in the prognostic meta-analysis. In our study, a computerized literature search was performed and thirty-two studies involving 4529 patients were included. Moreover, we assessed both prognostic and clinicopathological significance of circRNAs expression in cancer patients. A further subgroup analysis in different cancer types were also performed. Nevertheless, large-scale and better-designed trials are warranted to further identify the clinicopathological and prognostic significance of circRNAs expression in cancer.
Limitations
Despite the promising data, some limitations still should be acknowledged. Firstly, because of limited number of studies, we failed to perform subgroup analysis in terms of different kinds of circRNAs. More circRNAs types and other aspects of cancer including chemotherapeutic susceptibility and relapse should be explored. Secondly, functional studies are needed to clarify the underlying mechanisms of circRNAs in the tumorigenesis. Thirdly, the extensive clinical application of circRNA requires further study. Moreover, the number of subjects in the included studies are relatively small, which might result in a lack of statistical power and prevent a meaningful analysis of the results. With the updating of gene chip and microarray platform technology and an explosion of circRNAs research in cancer, a significant extension of our finding and re-analysis including more patients, could be accomplished in near future. Finally, when not reported in original articles, HRs were extrapolated from the Kaplan-Meier curves or calculated from the provided data within the papers according to the method of Parmar et al. [50], which could introduce potential source of bias. However, this practice has not been shown to yield results significantly different from direct methods of HR estimation.
Conclusions
The present meta-analysis suggests a significant association between high expression of circRNAs and clinicopathological and prognostic significance in human cancer. Additionally, circRNAs may be promising biomarkers and therapeutic targets for cancer. Nevertheless, large-scale studies using standardized approaches are warranted to provide a new insight into the prognostic value of circRNAs.
Supplementary Material
Supplementary figures and tables.
Abbreviations
CircRNA, Circular RNA; OR, Odds ratio; CI, Confidence interval; HR, Hazard ratios; OS, Overall survival; NOS, Newcastle-Ottawa Scale; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses; qRT-PCR, quantitative real time polymerase chain reaction.
Acknowledgements
This work is supported by the National Key Research and Development Program of China (2016YFC1100100), the Major Research Plan of National Natural Science Foundation of China (No. 91649204), and the Natural Science Foundation of Hubei Province, China (No. 2018CFB118).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH. et al. Exon circularization requires canonical splice signals. Cell reports. 2015;10:103-11
2. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141-57
3. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA biology. 2015;12:381-8
4. Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. Embo J. 2013;32:923-5
5. Wang M, Yang YX, Xu J, Bai W, Ren XL, Wu HJ. CircRNAs as biomarkers of cancer: a meta-analysis. Bmc Cancer. 2018:18
6. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Molecular cancer. 2017;16:94
7. Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in hepatocellular carcinoma patients: A meta-analysis. Cancer medicine. 2019;00:1-9
8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
9. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell research. 2017;27:626-41
10. Lukiw W, Zhao YH, Rogaev E, Bhattacharjee S. A Circular RNA (circRNA) ciRS-7 in Alzheimer's disease (AD) targets miRNA-7 trafficking and promotes deficits in the expression of the ubiquitin conjugase (UBE2A) and the epidermal growth factor receptor (EGFR). Faseb J. 2016:30
11. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2018. Ca-Cancer J Clin. 2018;68:7-30
12. Huang X, Zhang W, Shao Z. Association between long non-coding RNA polymorphisms and cancer risk: a meta-analysis. Bioscience reports. 2018:38
13. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629-34
14. Huang X, Zhang W, Zhang Z, Shi D, Wu F, Zhong B. et al. Prognostic Value of Programmed Cell Death 1 Ligand-1 (PD-L1) or PD-1 Expression in Patients with Osteosarcoma: A Meta-Analysis. Journal of Cancer. 2018;9:2525-31
15. Zhou X, Natino D, Qin Z, Wang D, Tian Z, Cai X. et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget. 2018;9:22288-300
16. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. International journal of biological sciences. 2018;14:321-30
17. Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W. et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687-97
18. Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS. et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017;77:2339-50
19. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T. et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:3918-28
20. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. Journal of experimental & clinical cancer research: CR. 2017;36:145
21. Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui YF. et al. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. European review for medical and pharmacological sciences. 2018;22:365-71
22. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L. et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer letters. 2017;403:305-17
23. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochemical and biophysical research communications. 2018;500:846-51
24. Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ. et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathology, research and practice. 2017;213:453-6
25. Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Bioscience reports. 2018:38
26. Luan W, Shi Y, Zhou Z, Xia Y, Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochemical and biophysical research communications. 2018;502:22-9
27. Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochemical and biophysical research communications. 2018;503:56-61
28. Zhang J, Zhao X, Zhang J, Zheng X, Li F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochemical and biophysical research communications. 2018;501:428-33
29. Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T. et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017:18
30. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2017;143:17-27
31. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell death & disease. 2018;9:417
32. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C. et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. 2017; 18: 1646-59.
33. Meng J, Chen S. Twist1 regulates Vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. 2018; 78: 4150-62.
34. Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J. et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer letters. 2018;430:179-92
35. Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. European review for medical and pharmacological sciences. 2018;22:3053-60
36. Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D. et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochemical and biophysical research communications. 2018;497:626-32
37. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer letters. 2017;388:208-19
38. Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Molecular cancer. 2017;16:151
39. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (Baltimore, Md). 2017;66:1151-64
40. Zhang X, Qiu S, Luo P, Zhou H, Jing W, Liang C. et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer biomarkers: section A of Disease markers. 2018
41. Yang C, Yuan W, Yang X, Li P, Wang J, Han J. et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Molecular cancer. 2018;17:19
42. Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochemical and biophysical research communications. 2018;502:465-71
43. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. Journal of Cancer. 2018;9:1856-62
44. Okholm TLH, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J. et al. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. 2017; 2: 36.
45. Xing L, Zhang L, Feng Y, Cui Z, Ding L. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;105:326-33
46. Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S. et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169-77
47. Ding HX, Lv Z, Yuan Y, Xu Q. The expression of circRNAs as a promising biomarker in the diagnosis and prognosis of human cancers: a systematic review and meta-analysis. Oncotarget. 2018;9:11824-36
48. Li Y, Zeng X, He J, Gui Y, Zhao S, Chen H. et al. Circular RNA as a biomarker for cancer: A systematic meta-analysis. Oncol Lett. 2018;16:4078-84
49. Chen Z, Zhang L, Han G, Zuo X, Zhang Y, Zhu Q. et al. A Meta-Analysis of the Diagnostic Accuracy of Circular RNAs in Digestive System Malignancy. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:962-72
50. Parmar MKB, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815-34
Author contact
![]() Corresponding author: Zengwu Shao M.D., Ph.D., Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China. E-mail: szwprocom
Corresponding author: Zengwu Shao M.D., Ph.D., Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China. E-mail: szwprocom

 Global reach, higher impact
Global reach, higher impact