3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(8):1879-1889. doi:10.7150/jca.30477 This issue Cite
Research Paper
The Fibroblast TIAM2 Promotes Lung Cancer Cell Invasion and Metastasis
1. Institute of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha 410078, People's Republic of China.
2. Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China.
3. School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai 200240, People's Republic of China.
4. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha, Hunan 410008, People's Republic of China.
Received 2018-10-7; Accepted 2019-2-22; Published 2019-4-21
Abstract
Purpose: TIAM2 (T-cell lymphoma invasion and metastasis 2), a RAC1 guanine nucleotide exchange factor, plays crucial roles in human cancer cells. Its homolog, TIAM1, has been reported to promote the migration and invasion of cancer cells through regulating the functions of cancer associated fibroblasts (CAFs). However, the functions of TIAM2 in CAFs have not been investigated. In this study, we explored how fibroblast TIAM2 influences the migration and invasion of lung cancer cells.
Methods: We cultured primary lung CAFs and adjacent normal lung fibroblasts (NFs) from 12 non-small cell lung cancer (NSCLC) patients. RT-PCR and western blot were used to compare TIAM2 levels between CAFs and NFs. Two co-culture systems were designed, in which cancer cells were directly co-cultured with fibroblasts and indirectly co-cultured with conditional medium (CM) from fibroblasts. Subsequently, the wound healing and transwell tests were conducted to assess the migration and invasion ability of fibroblasts and co-cultured cancer cells. Finally, cytokine antibody arrays were used to screen differentially secreted cytokines in the CM.
Results: The expression levels of TIAM2 were significantly higher in CAFs than NFs, and TIAM2-silenced fibroblasts showed decreased migration and invasion ability. In the direct co-culture system, the migration and invasion of cancer cells were retarded when co-culturing with TIAM2-silenced fibroblasts, and the expression levels of EMT-related genes also changed in cancer cells. Decreased migration and invasion of cancer cells were also observed when culturing with the CM from TIAM2-silenced fibroblasts. In addition, the cytokine antibody arrays revealed that Osteoprotegerin (OPG) was significantly decreased in the CM of TIAM2-silenced fibroblasts. This result suggested that OPG might be one of the main cytokines contributing to the migration and invasion of cancer cells in co-culture systems.
Conclusion: Our results suggest that fibroblast TIAM2 promotes the invasion and migration of lung cancer cell, and OPG might be one of the main cytokines contributing to this pro-cancer process.
Keywords: TIAM2, lung cancer, cancer-associated fibroblasts, migration and invasion, osteoprotegerin
Introduction
Tumor microenvironment is strongly associated with tumor progression and metastasis [1, 2]. Previous studies have reported that cancer associated fibroblasts (CAFs) are abundant in cancer microenvironment and play a crucial role in cancer cell migration and invasion [3, 4]. For example, CAFs can secrete soluble factors into the extracellular matrix and, thus, influence the invasion of cancer cells [5-8]. Therefore, it is necessary to elucidate the mechanisms of how CAFs affect the migration and invasion ability of cancer cells. Meanwhile, researchers have been concentrated on finding various biomarkers of CAFs which are expect to be potential targets for cancer prevention and treatment [9-13].
TIAM2 is a biomarker which has been reported to promote the proliferation and invasion of cancer cells. It has been found that the expression of TIAM2 promoted epithelial-to-mesenchymal transition (EMT), resulting in liver cancer metastasis [14-16]. Zhao et al. reported that TIAM2 promotes invasion and motility of lung cancer cells through activating Rac1 and EMT process [16]. Besides, TIAM2 is also a Rac1 specific guanine nucleotide exchange factor (GEF) and is initially identified as the homolog of TIAM1 [17]. In contrast, TIAM1 has been proved to play a crucial role in migration and invasion of both cancer cells [18, 19] and fibroblasts [20-22]. Nevertheless, the expression of TIAM2 in cancer associated fibroblasts has not been investigated, and the influence of fibroblasts TIAM2 on the migration and invasion of CAFs is still unraveled.
Although previous studies have revealed that TIAM2 exerts its pro-migration and pro-invasion function through activating Rac1 [23-26], the mechanism of how fibroblast TIAM2 affects cancer cells in the microenvironment is elusive. In contrast, TIAM1 has been found to promote cancer cell invasion and metastasis through CAFs [20, 21]. The experiments of Xu et al. indicated that fibroblast TIAM1 may play a role in modulating the tumor microenvironment and, thus affecting cancer cell invasion and metastasis [20]. It was also found that fibroblast TIAM1 could regulate breast cancer invasion and metastasis through TIAM1-osteopontin pathway [21]. Additionally, TIAM1-regulated Osteopontin in senescent fibroblasts was also found to promote the migration and invasion of associated epithelial cells [22]. However, whether fibroblast TIAM2 could affect the migration and invasion of cancer cells through similar mechanisms has not been characterized.
In the current study, we investigated the expression of TIAM2 on CAFs and its influence on the migration and invasion of cancer cells. To determine how cancer cells were affected by fibroblast TIAM2, we silenced the expression of TIAM2 in fibroblasts, and assessed the migration and invasion ability of cancer cells in the direct and indirect co-culture system. Specifically, the conditional medium (CM) of fibroblasts was used to culture cancer cells in the second system. Furthermore, cytokine antibody arrays were used to screen differentially secreted cytokines that might contribute to the migration and invasion of cancer cells.
Materials and methods
Reagents and antibodies
Anti-β-actin and GAPDH antibodies were purchased from Sigma-Aldrich (St Louis, MO, USA). Anti-α-SMA and Vimentin antibodies were purchased from Abcam (Cambridge, UK). Anti-TIAM2 and E-cadherin antibodies were purchased from Santa Cruz (CA, USA). Anti-N-cadherin antibody was purchased from Cell Signaling Technology (MA, USA). RNA interference was performed using Lipofectamine RNAiMAX Reagent (Invitrogen, Thermo Fisher Scientific, USA). Three siRNAs of TIAM2 (siT-1, siT-2, siT-3) were designed and synthesized by Ribobio (Guangzhou, China). The target sequences were: 5'-GAGGCTCTTTCCTTTATTA-3' (siT-1), 5'-CCGTCATACTGGTTTATAA-3' (siT-2) and 5'-GGGAACTGATCCATACGAA-3' (siT-3). A scramble siRNA was purchased from Santa Cruz (CA, USA) and used as control siRNA (siNC).
Cell line culture
Human lung adenocarcinoma cell line A549 and human lung fibroblast cell line MRC5 were bought from Cell bank of Chinese Academy of Sciences (Shanghai, China). Human umbilical vein endothelial cell line HUVEC was bought from Genechem (Shanghai Genechem Co., Ltd. China). A549 and HUVEC were cultured in RPMI-1640 (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% FBS. MRC-5 was cultured in αMEM (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% FBS. All the cell lines were incubated at constant conditions 37 °C and 5% CO2.
Isolation and culture of CAFs and NFs
Human lung cancer-associated fibroblasts (CAFs) and paired normal lung fibroblasts (NFs) were isolated from surgically resected lungs from NSCLC (non-small cell lung cancer) patients (Table 1) as previously reported [27, 28]. Briefly, approximately 5mm3 lung cancer and normal lung tissues were minced into 1mm3 pieces and planted into cell culture flasks respectively. DMEM/F12 (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% FBS and 1% antibotics (penicillin and streptomycin) were added to the flasks. After 3 to 7 days, the cells outgrew from the tissue pieces. When the cells reached 80% confluency, they were harvested and re-planted at a density of 1 × 104 cells/cm2. Cells were incubated at 37 °C in 5% CO2 atmosphere. The fibroblasts were separated from contaminating epithelial and immune cells by differential trypsinization. Finally, cells from passages 3-7 were used for all experiments.
Patient demographics, tumor stage and histology
| Sample | Age | Sex | Stage | Histology |
|---|---|---|---|---|
| Patient1 | 51 | Female | T3N0M0 IIB | Adenocarcinoma |
| Patient2 | 48 | Male | T2bN0M0 IIA | Squamous cell carcinoma |
| Patient3 | 70 | Female | T1N2M0 IIIA | Adenocarcinoma |
| Patient4 | 66 | Male | T2bN0M0 IIA | Adenocarcinoma |
| Patient5 | 62 | Male | T1bN0M0 IA2 | Adenocarcinoma |
| Patient6 | 54 | Male | T2N2M0 IIIA | Adenocarcinoma |
| Patient7 | 48 | Male | T3N1M0 IIIA | Squamous cell carcinoma |
| Patient8 | 56 | Male | T2bN0M0 IIA | Adenocarcinoma |
| Patient9 | 65 | Male | T3N0M0 IIB | Squamous cell carcinoma |
| Patient10 | 54 | Female | T4N1M0 IIIA | Adenocarcinoma |
| Patient11 | 52 | Female | T2N0M0 IIA | Adenocarcinoma |
| Patient12 | 76 | Male | T2N2M0 IIIA | Squamous cell carcinoma |
Immunofluorescence microscopy
Cells were cultured in confocal dishes, fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.2% TritonX-100 for 10 min. Blocking was performed in 5% BSA for 30 min at room temperature. Subsequently, cells were incubated with primary antibodies overnight at 4 °C. Cells were then washed with PBS and incubated with Alexa Fluor secondary antibodies at room temperature for 1 hour. After washed with PBS, cells were mounted with DAPI staining solution. Primary antibodies targeting αSMA and vimentin were purchased from Abcam (Cambridge, UK). Besides, Alexa Fluor secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Finally, the fluorescence was observed by confocal laser-scanning microscope system DeltaVision (GE Healthcare, UK), and images were processed with relevant software.
Flow cytometry and cell sorting
Human lung adenocarcinoma cell line A549 was harvested from flasks and rinsed with PBS for three times. Then, it was stained with cell labeling reagent Calcein-AM (AAT Bioquest, USA) at 37 °C in 5% CO2 atmosphere for 30 minutes. We washed the cell line A549 three times with PBS and directly co-cultured it with lung fibroblast cell line MRC5, respectively. After three days, cancer cells were sorted using a FACSAria II (BD Biosciences).
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) following the manufacturer's instructions. RNA was reverse-transcribed into cDNA using the Primescript RT reagent kit with gDNA Eraser (Takara Bio Inc., Japan). The PCR system was performed using SYBR Premix Dimer Eraser kit (Takara Bio Inc). At the end of the PCR cycles, melting curve analyses were conducted to confirm the specificity of the PCR products. The GAPDH primer sequences were 5'-GGAGCGAGATCCCTCCAAAAT-3' (forward) and 5'-GGCTGTTGTCATACTTCTCATGG-3' (reverse). The TIAM2 primer sequences were 5' -TACCACCTGACGGAAGCACTA-3' (forward) and 5'-ACACGGTCCCATAATCCTCATA-3' (reverse).
Western blot
The cells were lysed in RIPA buffer with 1% PMSF. The proteins were separated on 10% SDS-polyacrylamide gels and were transferred to PVDF membranes (Millipore). The membranes were blocked in 5% fat-free milk in TBST and then incubated with primary antibodies overnight at 4 °C. After washed three times with TBST, membranes were incubated with secondary antibodies at room temperature for 1 hour and were washed three times again. Protein bands were visualized using an enhanced chemiluminescence reagent (GE Healthcare, UK) and quantified by densitometry using ChemiDoc XRS+ image analyzer (Bio-Rad, USA).
Migration and invasion assay
In order to perform the migration assay, we conducted the wound healing and transwell experiments. In the wound healing assay, cells were seeded in 6-well plates. When cells grew to 100% confluency, the monolayers were scratched by a 1000-μL pipette tip and washed with PBS. The photographs were taken every 24 hours and the migration rates were calculated. Specifically, we designed three groups of wound healing experiments. (1) Human lung fibroblast cell line MRC5 was transfected with TIAM2-siRNA/NC-siRNA after 100% confluency. (2) MRC5 was transfected with TIAM2-siRNA/NC- siRNA and co-cultured with cancer cell line A549. (3) The conditional medium produced by TIAM2- siRNA/NC-siRNA treated MRC5 was used to culture cancer cell line A549. In addition, transwell experiments were also performed using 24-well transwell chambers with 8-μm pore size (Corning). Cells were harvested and rinsed with PBS. Then, cells were resuspended in serum-free RPMI- 1640 and seeded in the upper chamber. The lower chamber contained 600 μL of complete growth medium (RPMI-1640 supplemented with 10% FBS). In the invasion assay, the upper chambers were coated with Matrigel. After the incubation at 37 °C for 24 hours, non-invading cells on the upper membrane were removed with a cotton swab. Cells invaded to the lower surface of the membrane were stained with 0.1% crystal violet. The stained cells were photographed and quantified by counting the number of cells.
Conditional medium and cytokine antibody array
Fibroblasts were seeded in 6-well plates and transfected by TIAM2-siRNA or NC-siRNA. When cells reached to 100% confluency, the medium was replaced with serum-free RPMI-1640. After 48 hours, the cell-culture media were collected and centrifuged at 10000 g for 15 min at 4 °C. The condition medium was used for various cell experiments and the cytokine antibody array, according to the standard instruction. The cytokines with significantly different expression levels were screened out by the array.
Statistical Analysis
All the experiments data in our study were processed using graphpad and expressed as the mean ± SEM. Statistical significance was evaluated by student t test or paired t test, and p < 0.05 were considered as statistically significant.
Results
CAFs expressed higher levels of TIAM2 than NFs
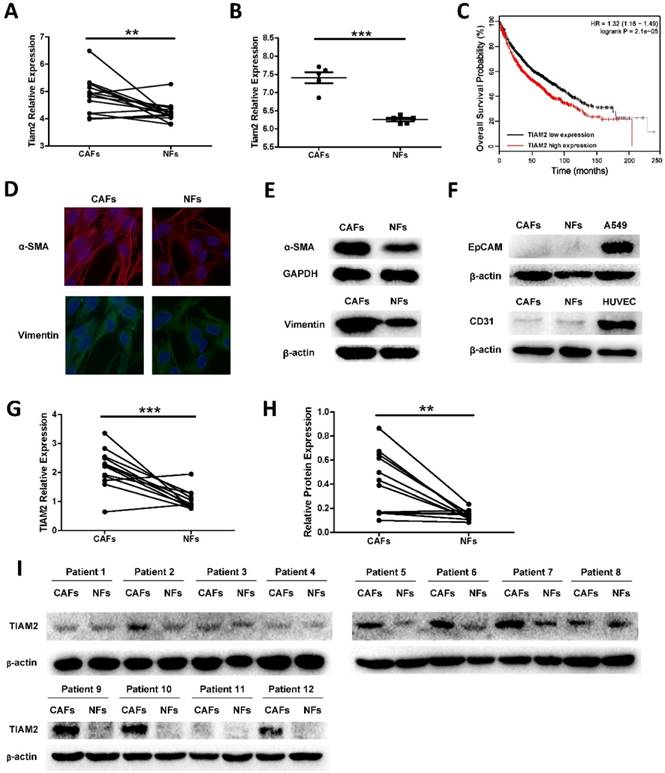
TIAM2 has been proved to promote cancer cell invasion and migration in many researches. However, the functions of TIAM2 in CAFs were not investigated. To determine whether TIAM2 is a differentially expressed gene between CAFs and NFs, we searched available GEO microarray data which compare CAFs and NFs, and we focused on the expression difference of TIAM2. As shown in Figure 1A, we used the data from Series GSE22874, and the microarray gene expression analysis of 15 pairs CAFs and NFs from NSCLC patients showed that TIAM2 expression levels were significantly higher in CAFs than NFs (p = 0.0058). In addition, we analyzed the data from Series GSE48397, and found that TIAM2 expression levels were higher in mouse lung CAFs than NFs (p < 0.0001, Figure 1B). More importantly, Kaplan-Meier survival analysis of data from the online tool Kmplot [29] revealed that the expression of TIAM2 is adversely related to the outcome of lung cancer patients (Figure 1C).
In our research, twelve paired cancer-associated fibroblasts (CAFs) and normal fibroblasts (NFs) were isolated from lung cancer tissues and adjacent lung tissues as previously mentioned. Cell authentication and purity were checked by specific immune staining with Vimentin, a mesenchymal marker and α-smooth muscle actin (α-SMA), a marker of myofibroblasts (Figure 1D). We confirmed this result by performing western blot experiments (Figure 1E). Consistent with previous reports [3], CAFs expressed higher levels of fibroblast markers compared with NFs. In order to exclude the existence of other cell types, the epithelial marker Ep-cam and the endothelial marker CD31 were also examined. As shown in Figure 1F, primary cell CAFs and NFs expressed no EpCAM compared with the lung cancer cell line A549, and no CD31 was expressed compared with the endothelial cell line HUVEC. These results collaboratively confirmed the purity of CAFs and NFs cultured in our experiments. After the verification and purification process, we examined TIAM2 expression levels of these CAFs and NFs using RT-qPCR (Figure 1G, p = 0.0006), and the results were consistent with the data from GEO database. Besides, identical results were obtained when we examined the protein levels of TIAM2 by western blot (Figure 1H-I, p = 0.0029). These results cohesively confirmed that lung CAFs express higher levels of TIAM2 than NFs.
Silencing TIAM2 attenuated fibroblast migration and invasion
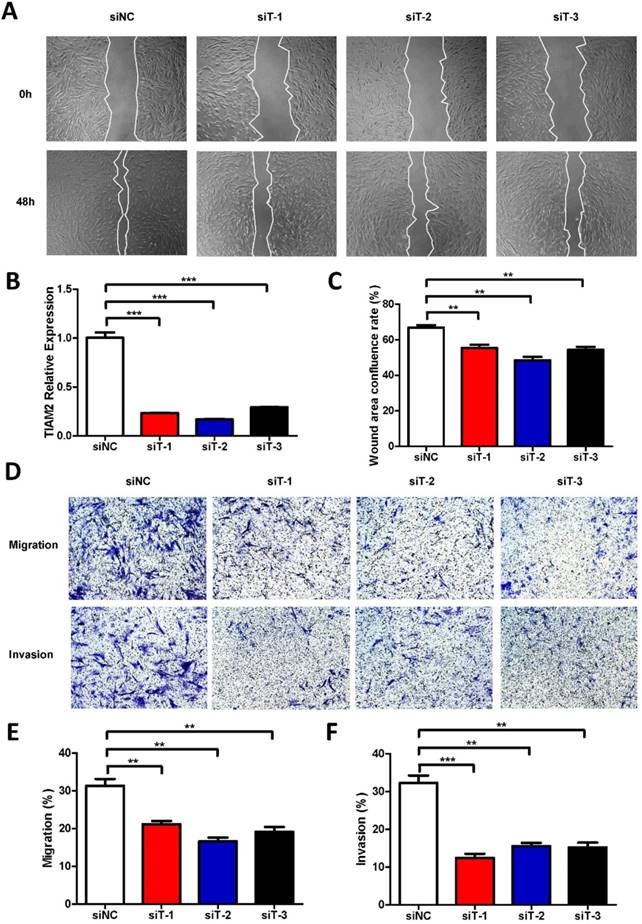
In previous researches, TIAM2 showed the ability of promoting the migration and invasion of lung cancer cells, and CAFs also play similar roles in the cancer microenvironment. Moreover, fibroblast TIAM1, the homolog of TIAM2, has also been proved as a modulator of tumor cell invasion and metastasis. Therefore, we inferred that TIAM2 might exert similar functions as TIAM1. Firstly, we performed the wound healing experiments with respect to the human lung fibroblast cell line MRC5 to investigate the relationship between TIAM2 and fibroblasts (Figure 2A). We silenced the expression of TIAM2 in MRC5 using three siRNAs (siT-1, siT-2, siT-3). As shown in Figure 2B, the result indicated that the expression of TIAM2 was successfully downregulated after performing the silencing. The wound healing confluence rates of the control group siNC and three TIAM2-siRNAs were 66.85% ± 1.35%, 55.40% ± 1.80%, 48.41% ± 1.92% and 54.37% ± 1.64%, respectively (Figure 2C, p1 = 0.007, p2 = 0.0014, p3 = 0.0042). These statistics showed that the migration of cells was delayed after silencing TIAM2 compared with the control group. In addition, Figure 2D showed decreased number of passed cells in the transwell tests. Specifically, the migration rates of siNC and three TIAM2-siRNAs were 31.34% ± 1.81%, 21.17% ± 0.84%, 16.63% ± 0.97% and 19.14% ± 1.25%, respectively (Figure 2E, p1 = 0.007, p2 = 0.002, p3 = 0.0052). The invasion rates of siNC and three TIAM2-siRNAs were 32.26% ± 1.97%, 12.42% ± 1.11%, 15.53% ± 0.88% and 15.25% ± 1.23% (Figure 2F, p1 = 0.0009, p2 = 0.0015, p3 = 0.0018). According to these results, we demonstrated that TIAM2 promoted the migration and invasion of lung fibroblasts.
TIAM2 expression levels in lung CAFs and NFs. We analyzed the expression difference using data from GEO database and cultured primary lung CAFs and NFs. (A) TIAM2 expression levels difference among 15 pairs of human primary fibroblasts in GEO database (GSE22874). (B) TIAM2 expression levels difference of 5 pairs mouse primary fibroblasts in GEO database (GSE48397). (C) Kaplan-Meier survival curves in lung cancer patients showing overall survival (OS) with high (red) and low (black) expression of TIAM2. Hazard ratio (HR) and p value are indicated. (D) Immunofluorescence images of CAFs and NFs showing α-SMA and Vimentin positive staining. (E) Western blot showing the expression of Vimentin and α-SMA in purified CAFs and NFs cells. (F) Western blot showing CAFs and NFs express no EpCAM and CD31 compared with the cancer cell line A549 and the endothelial cell line HUVEC. (G) RNA expression levels of TIAM2 (RT-PCR) of human primary fibroblasts cultured in our experiment. (H-I) Protein levels of TIAM2 in human primary fibroblasts by western blot.
Silencing TIAM2 fibroblasts retarded cancer cell invasion and migration in directly co-cultured model
Given that TIAM2-siRNA slowed down the migration and invasion of fibroblasts, we further examined whether TIAM2-silenced fibroblasts could affect the migration and invasion of cancer cells. Therefore, we co-cultured TIAM2-silenced fibroblasts with A549 cells. As shown in Figure 3A, we conducted the wound healing tests to show the difference after performing the silencing. The wound healing confluence rates of siNC and three TIAM2-siRNAs were 68.31% ± 1.60%, 37.73% ± 1.79%, 38.46% ± 2.24% and 45.86% ± 0.43% (Figure 3B, p1 = 0.0002, p2 = 0.0004, p3 = 0.0002). These results revealed that TIAM2-silenced fibroblasts delayed the migration of cancer cells. In order to highlight the variation of the migration and invasion ability of cancer cells, we sorted calcium-AM labeled cancer cells by flow cytometry as the schematic shown in Figure 3C. Figure 3D showed the process of separating calcium-AM labeled cancer cells from fibroblasts by flow cytometry. After that, we performed the transwell experiments using these sorted cancer cells (Figure 3E). The migration rates of siNC and three TIAM2-siRNAs were 53.63% ± 1.52%, 31.81% ± 0.97%, 22.59% ± 1.35% and 34.75% ± 2.30%, respectively (Figure 3F, p1 = 0.0003, p2 = 0.0001, p3 = 0.0024). The invasion rates of siNC and three TIAM2-siRNAs were 65.98% ± 2.27%, 31.31% ± 1.36%, 31.85% ± 1.15% and 42.05% ± 1.28% (Figure 3G, p1 = 0.0002, p2 = 0.0002, p3 = 0.0008). These results indicated that the migration and invasion of cancer cells were inhibited after co-culturing with TIAM2-silenced fibroblasts.
Silencing TIAM2 delayed the migration and invasion of MRC5. (A) The wound healing experiments after silencing TIAM2 in MRC5. (B) Decreased number of cells after silencing TIAM2 in the transwell tests. (C) TIAM2 expression difference after the silencing. (D) Averages of wound area confluence rate. (E-G) Averages of migration rate and invasion rate.
TIAM2-silenced fibroblasts directly co-cultured with cancer cells. (A) The wound healing experiments of co-cultured cancer cells and TIAM2-silenced fibroblasts. (B) Averages of wound area confluence rate. (C) Scheme showing the method of co-culturing TIAM2-silenced fibroblasts with cancer cells. (D) Cancer cell sorting using flow cytometry from co-cultured cells. (E) Transwell experiments using sorted cancer cells from flow cytometry. (F, G) Averages of migration rate and invasion rate. (H) The expression of E-cadherin, N-cadherin and Vimentin in sorted cancer cells by western blot.
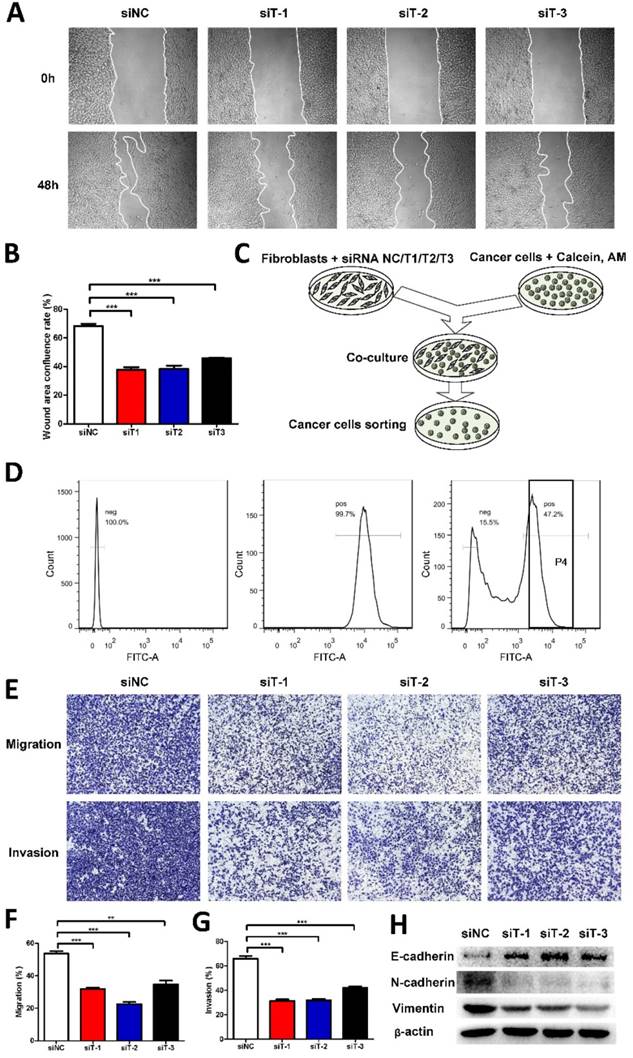
EMT process has been reported to be crucial to cell migration and invasion [3, 30]. In addition, the epithelium marker E-cadherin and mesenchymal marker Vimentin are widely used as representative EMT markers. As a result, we examined the expression of E-cadherin, N-cadherin and Vimentin of sorted cancer cells by western blot. As compared in Figure 3H, we can see that the expression of E-cadherin was increased after the knockdown of TIAM2 in fibroblasts, while the N-cadherin and Vimentin protein levels were decreased. These results suggested that TIAM2-silenced fibroblasts might retard the migration and invasion of co-cultured cancer cells through the EMT process.
Cancer cell invasion and migration retarded by conditional medium of TIAM2-silenced fibroblasts
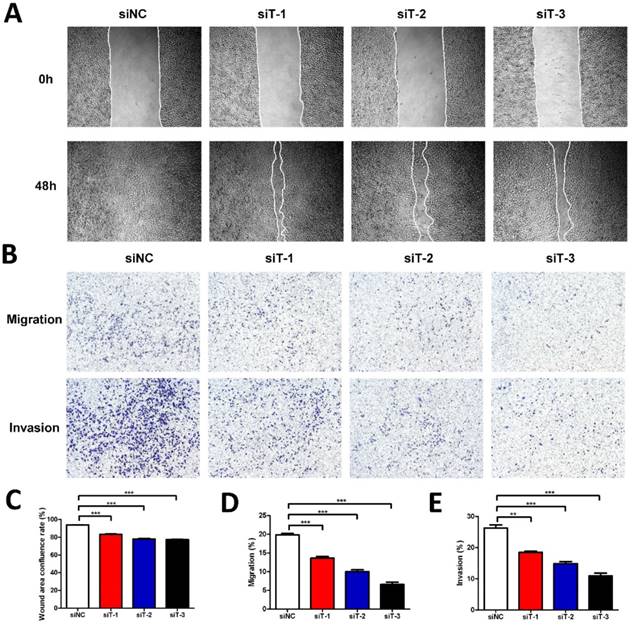
Considering that the secretion of fibroblasts can influence cancer cells, to further investigate the mechanism how TIAM2-silenced fibroblasts affect the cancer cells in detail, we used the conditional medium (CM) of TIAM2-silenced fibroblasts to culture A549 cell line. Similarly, we also conducted the wound healing experiments (Figure 4A). As the statistics given in Figure 4C, the wound healing confluence rates of siNC and three TIAM2-siRNAs were 93.78% ± 0.26%, 83.09% ±0.57%, 78.00% ±0.62% and 77.43% ± 0.20% (p1, p2, p3 < 0.0001). These results revealed that the CM of TIAM2-silenced fibroblasts delayed the migration of cancer cells. Moreover, as the transwell experiments shown in Figure 4B, the migration rates of siNC and three TIAM2-siRNAs were 19.84% ±0.39%, 13.63% ±0.46%, 10.03% ±0.48% and 6.57% ±0.64% (Figure 4D, p1 = 0.0005, p2 < 0.0001, p3 < 0.0001), and the invasion rates of siNC and three TIAM2-siRNAs were 26.26% ± 1.00%, 18.46% ± 0.34%,14.89% ± 0.64% and 10.95% ± 0.90% (Figure 4E, p1 = 0.0018, p2 = 0.0007, p3 = 0.0003). These results also confirmed the decreased migration and invasion ability of cancer cells in the CM produced by TIAM2-silenced fibroblasts.
Cancer cells cultured with conditional medium from TIAM2-silenced fibroblasts. (A) The wound healing experiments of A549 cultured with conditional medium. (B) Transwell experiments of A549 cultured with conditional medium. (C-E) Averages of wound area confluence rate, migration rate and invasion rate.
Osteoprotegerin significantly decreased in the conditional medium of TIAM2-silenced fibroblasts
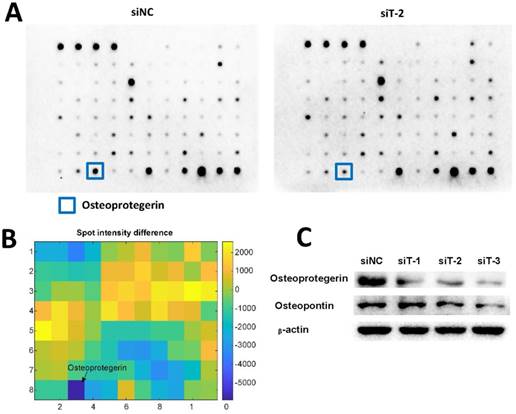
Cytokines have been reported to play important roles in regulating cancer cell migration and invasion [31-33]. Therefore, we used Abcam human cytokine antibody array to screen the differential secretion profiles of cytokines in the CM from siNC-silenced MRC5 and TIAM2-silenced MRC5. Given that the interfering performance of siT-2 was better than siT-1 and siT-3, we used the CM from siT-2-silenced MRC5. As shown in Figure 5A-B, Osteoprotegerin was the most significantly decreased cytokine in siT-2-CM compared with siNC-CM. In addition, the decreased expression levels of Osteoprotegerin in TIAM2-silenced fibroblasts were confirmed by western blot (Figure 5C).
Considering the expression of Osteopontin was significantly increased in TIAM1-silenced fibroblasts as previously reported [21, 22] and TIAM1 is the homolog of TIAM2 [17], similarly, we examined the expression of Osteopontin in TIAM2-silenced fibroblasts. As shown in Figure 5C, there was no significant Osteopontin expression difference between the siT-1-silenced MRC5 and siNC-silenced MRC5. In contrast, the expression of Osteopontin in siT-2 and siT-3 silenced MRC5 were slightly lower than siNC-silenced MRC5. These results collaboratively indicated that the knockdown of TIAM2 in fibroblasts induced the variation of relevant cytokines secreted by fibroblasts, which might inhibit the migration and invasion of cancer cells.
Differentially secreted cytokines in conditional medium. (A) The cytokine profiles of conditional medium from siNC silenced MRC5 and siT-2 silenced MRC5 were screened using the Abcam human cytokine antibody array. The signal of the most significantly decreased cytokine Osteoprotegerin was marked. (B) Analysis of the spot intensity difference between siNC-CM and siT-2-CM. (C) Verification of Osteoprotegerin and Osteopontin expression using western blot.
Discussion
CAFs have been proved to promote the migration and invasion of cancer cells [3, 4], and we also noted that the TIAM-family could promote the proliferation and invasion of cancer cells in previous researches [14, 15]. Specifically, the functions of TIAM1 with respect to fibroblasts and cancer cells have been clarified [19, 21] and TIAM2 has also been proved to directly increase the migration and invasion ability of cancer cells [14-15, 34]. Nevertheless, the functions of TIAM2 in CAFs were largely unknown. In the current study, we established primary cell CAFs and NFs from lung cancer patients, and found that human lung CAFs expressed significantly higher levels of TIAM2 than NFs. In addition, TIAM2 might induce the CAFs to secrete several types of cytokines which promote the migration and invasion of cancer cells in co-culture models. These findings indicated the roles of TIAM2 in lung cancer microenvironment.
Given that CAFs could promote the migration and invasion of cancer cells, we inferred that TIAM2 might affect cancer cells through CAFs. Since the roles of TIAM2 in cancer associated fibroblasts remained unclear, we first compared the expression levels of TIAM2 between CAFs and NFs, and our experiments showed that human lung CAFs expressed higher levels of TIAM2 than NFs (Figure 1). This was consistent with the research of Chen et al., which found that the liver tumor tissues expressed higher level of TIAM2 than matched non-tumor tissues [14].
Other studies showed that TIAM2 promoted the proliferation and invasion of cancer cells [14-16]. For example, Chen et al. reported that TIAM2 could promote the proliferation and invasion of liver cancer cells [14]. Zhao et al. also found that TIAM2 enhanced non-small cell lung cancer cell invasion and motility [16]. Moreover, TIAM2 was also reported to regulate cellular motility by activating Rac1-mediated focal adhesion disassembly, which was required for cell migration [23]. To further investigate the mechanism of how TIAM2 affected cancer cells in the tumor microenvironment, TIAM2-silenced fibroblasts were directly and indirectly co-cultured with cancer cells. This experiment design was inspired by the research of Yoshida et al., in which two kinds of co-culture systems were applied [35]. Their results indicated that the direct co-culture system could simulate the microenvironment of CAFs to the greatest extent while the indirect approach could only reflect the effect of conditional medium. In our study, we at first directly co-cultured the MRC5 with A549 (Figure 3C) and the results suggested that TIAM2 might exerted the function of pro-migration and pro-invasion to cancer cells partly through fibroblasts. These findings went a step further than previous researches which only found the relationship between TIAM2 and cancer cells [14-16]. Compared with TIAM1, which is involved in both anti-cancer and pro-cancer mechanisms [18, 20], TIAM2 only exerted pro-cancer function.
Meanwhile, we also conducted the indirect co-culture to determine whether TIAM2 affected cancer cells by secreting cytokines in the conditional medium. Our findings showed that the CM produced by TIAM2-silenced fibroblasts could retard the migration and invasion of cancer cells. Besides, we found that cytokine Osteoprotegerin (OPG) significantly decreased in the CM of TIAM2-silenced fibroblasts (Figure 5B). Cytokine OPG was also known as the tumor necrosis factor receptor superfamily member 11B (TNFRSF11B), which was first discovered to play a role in the regulation of bone density [36]. Previous studies also showed that OPG promoted tumor cell growth and facilitated metastasis of breast, prostate and lung cancer [37-39]. For example, Zahia et al. showed that OPG regulated the migration of cancer cells through SDF-1/CXCR4 axis and promoted cancer development [40]. Russmueller et al. revealed the upregulation of OPG expression was related to bone invasion in oral cancer [41]. Our findings suggested that fibroblast TIAM2 influenced the expression of OPG, and OPG might be one of the main cytokines contributing to the pro-cancer process. This phenomenon suggested that OPG might be a potential therapeutic target in tumor treatment. In contrast, TIAM1-deficient fibroblasts have been proved to induce increased cancer cell migration and this process was dependent on fibroblast secretion of Osteopontin [21, 22]. However, our results showed that Osteopontin was slightly downregulated in TIAM2-deficient fibroblasts.
In this study, we demonstrate that CAFs express higher level of TIAM2 than NFs, and the expression of TIAM2 in CAFs promotes cancer cell migration and invasion. Moreover, cytokine OPG might be one of the main contributors to this effect. Our findings also indicate that more researches should be performed to elucidate other mechanisms of how CAFs influence the migration and invasion of cancer cells.
Abbreviations
TIAM2: T-cell lymphoma invasion and metastasis 2; TIAM1: T-cell lymphoma invasion and metastasis 1; GEF: guanine nucleotide exchange factor; CAFs: cancer-associated fibroblasts; NFs: normal fibroblasts; NSCLC: non-small cell lung cancer; OS: overall survival; HR: hazard ratio; α-SMA: alpha smooth muscle actin; EMT: epithelial-to-mesenchymal transition; CM: conditional medium; OPG: Osteoprotegerin.
Acknowledgements
We sincerely appreciate helpful comments of reviewers.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFC1306900, 2016YFC0905002), National Natural Science Foundation of China (81573508, 81874327), and the Strategy-Oriented Special Project of Central South University in China (ZLXD2017003).
Human rights statement and informed consent
All of the tissues were from patients who had written informed consents, in compliance with the code of ethics of the World Medical Association (Declaration of Helsinki) at the time of the surgery. The protocol was approved by the Ethics Committee of Xiangya School of Medicine, Central South University (registration number of CTXY-110008-3).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423-37
2. Cao Y, DePinho RA, Ernst M, Vousden K. Cancer research: past, present and future. Nat Rev Cancer. 2011;11(10):749-54
3. Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582-98
4. LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech. 2018;11(4):dmm029447
5. Calon A, Tauriello DV, Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin. Cancer Biol. 2014;25:15-22
6. Bai YP, Shang K, Chen H, Ding F, Wang Z, Liang C, Xu Y, Sun MH, Li YY. FGF-1/-3/FGFR4 signaling in cancer-associated fibroblasts promotes tumor progression in colon cancer through Erk and MMP-7. Cancer Sci. 2015;106:1278-1287
7. Liu J, Chen S, Wang W, Ning BF, Chen F, Shen W, Ding J, Chen W, Xie WF, Zhang X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Lett. 2016;379(1):49-59
8. Min A, Zhu C, Wang J, Peng S, Shuai C, Gao S, Tang Z, Su T. Focal adhesion kinase knockdown in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis via downregulating MCP-1/CCL2 expression. J Biochem Mol Toxicol. 2015;29(2):70-6
9. Navab R, Strumpf D, Bandarchi B, Zhu CQ, Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L, Barczyk M, Panchal D, To C, Yun JJ, Der S, Shepherd FA, Jurisica I, Tsao MS. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2011;108(17):7160-5
10. Vicent S, Sayles LC, Vaka D, Khatri P, Gevaert O, Chen R, Zheng Y, Gillespie AK, Clarke N, Xu Y, Shrager J, Hoang CD, Plevritis S, Butte AJ, Sweet-Cordero EA. Cross-species functional analysis of cancer-associated fibroblasts identifies a critical role for CLCF1 and IL-6 in non-small cell lung cancer in vivo. Cancer Res. 2012;72(22):5744-56
11. Su S, Chen J, Yao H, Liu J, Yu S, Lao L, Wang M, Luo M, Xing Y, Chen F, Huang D, Zhao J, Yang L, Liao D, Su F, Li M, Liu Q, Song E. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172(4):841-856.e16
12. Paulsson J, Micke P. Prognostic relevance of cancer-associated fibroblasts in human cancer. Semin Cancer Biol. 2014;25:61-8
13. Wang J, Min A, Gao S, Tang Z. Genetic regulation and potentially therapeutic application of cancer-associated fibroblasts in oral cancer. J Oral Pathol Med. 2014;43(5):323-34
14. Chen JS, Su IJ, Leu YW, Young KC, Sun HS. Expression of T-cell lymphoma invasion and metastasis 2 (TIAM2) promotes proliferation and invasion of liver cancer. Int J Cancer. 2012;130(6):1302-13
15. Yen WH, Ke WS, Hung JJ, Chen TM, Chen JS, Sun HS. Sp1-mediated ectopic expression of T-cell lymphoma invasion and metastasis 2 in hepatocellular carcinoma. Cancer Med. 2016;5(3):465-77
16. Zhao ZY, Han CG, Liu JT, Wang CL, Wang Y, Cheng LY. TIAM2 enhances non-small cell lung cancer cell invasion and motility. Asian Pac J Cancer Prev. 2013;14(11):6305-9
17. Chiu CY, Leng S, Martin KA, Kim E, Gorman S, Duhl DM. Cloning and characterization of T-cell lymphoma invasion and metastasis 2 (TIAM2), a novel guanine nucleotide exchange factor related to TIAM1. Genomics. 1999;61(1):66-73
18. Boissier P, Huynh-Do U. The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling. Cell Signal. 2014;26(3):483-91
19. Vaughan L, Tan CT, Chapman A, Nonaka D, Mack NA, Smith D, Booton R, Hurlstone AF, Malliri A. HUWE1 ubiquitylates and degrades the RAC activator TIAM1 promoting cell-cell adhesion disassembly, migration, and invasion. Cell Rep. 2015;10(1):88-102
20. Xu K, Rajagopal S, Klebba I, Dong S, Ji Y, Liu J, Kuperwasser C, Garlick JA, Naber SP, Buchsbaum RJ. The role of fibroblast Tiam1 in tumor cell invasion and metastasis. Oncogene. 2010;29(50):6533-42
21. Xu K, Tian X, Oh SY, Movassaghi M, Naber SP, Kuperwasser C, Buchsbaum RJ. The fibroblast Tiam1-osteopontin pathway modulates breast cancer invasion and metastasis. Breast Cancer Res. 2016;18(1):14
22. Liu J, Xu K, Chase M, Ji Y, Logan JK, Buchsbaum RJ. Tiam1-regulated osteopontin in senescent fibroblasts contributes to the migration and invasion of associated epithelial cells. J Cell Sci. 2012;125(Pt 2):376-86
23. Rooney C, White G, Nazgiewicz A, Woodcock SA, Anderson KI, Ballestrem C, Malliri A. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010;11(4):292-8
24. Woroniuk A, Porter A, White G, Newman DT, Diamantopoulou Z, Waring T, Rooney C, Strathdee D, Marston DJ, Hahn KM, Sansom OJ, Zech T, Malliri A. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat Commun. 2018;9(1):2124
25. Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8:23
26. Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510-23
27. Hashimoto H, Suda Y, Miyashita T, Ochiai A, Tsuboi M, Masutomi K, Kiyono T, Ishii G. A novel method to generate single-cell-derived cancer-associated fibroblast clones. J Cancer Res Clin Oncol. 2017;143(8):1409-1419
28. Ishii G, Hashimoto H, Asada K, Ito T, Hoshino A, Fujii S, Kojima M, Kuwata T, Harigaya K, Nagai K, Ushijima T, Ochiai A. Fibroblasts associated with cancer cells keep enhanced migration activity after separation from cancer cells: a novel character of tumor educated fibroblasts. Int J Oncol. 2010;37(2):317-25
29. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241
30. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21-45
31. Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br J Cancer. 2014;110:724-732
32. Dai Z, Wu J, Chen F, Cheng Q, Zhang M, Wang Y, Guo Y, Song T. CXCL5 promotes the proliferation and migration of glioma cells in autocrine- and paracrine-dependent manners. Oncol Rep. 2016;36(6):3303-3310
33. Mo M, Zhou M, Wang L, Qi L, Zhou K, Liu LF, Chen Z, Zu XB. CCL21/CCR7 enhances the proliferation, migration, and invasion of human bladder cancer T24 cells. PLoS One. 2015;10(3):e0119506
34. Krøigård AB, Larsen MJ, Lænkholm AV, Knoop AS, Jensen JD, Bak M, Mollenhauer J, Thomassen M, Kruse TA. Identification of metastasis driver genes by massive parallel sequencing of successive steps of breast cancer progression. PLoS One. 2018;13(1):e0189887
35. Yoshida T, Ishii G, Goto K, Neri S, Hashimoto H, Yoh K, Niho S, Umemura S, Matsumoto S, Ohmatsu H, Iida S, Niimi A, Nagai K, Ohe Y, Ochiai A. Podoplanin-positive cancer-associated fibroblasts in the tumor microenvironment induce primary resistance to EGFR-TKIs in lung adenocarcinoma with EGFR mutation. Clin Cancer Res. 2015;21(3):642-51
36. Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, Shimamoto G, DeRose M, Elliott R, Colombero A, Tan HL, Trail G, Sullivan J, Davy E, Bucay N, Renshaw-Gegg L, Hughes TM, Hill D, Pattison W, Campbell P, Sander S, Van G, Tarpley J, Derby P, Lee R, Boyle WJ. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89(2):309-19
37. Weichhaus M, Chung ST, Connelly L. Osteoprotegerin in breast cancer: beyond bone remodeling. Mol Cancer. 2015;14:117
38. Baud'huin M, Duplomb L, Teletchea S, Lamoureux F, Ruiz-Velasco C, Maillasson M, Redini F, Heymann MF, Heymann D. Osteoprotegerin: multiple partners for multiple functions. Cytokine Growth Factor Rev. 2013;24(5):401-9
39. Chung ST, Geerts D, Roseman K, Renaud A, Connelly L. Osteoprotegerin mediates tumor-promoting effects of Interleukin-1beta in breast cancer cells. Mol Cancer. 2017;16(1):27
40. Benslimane-Ahmim Z, Pereira J, Lokajczyk A, Dizier B, Galy-Fauroux I, Fischer AM, Heymann D, Boisson-Vidal C. Osteoprotegerin regulates cancer cell migration through SDF-1/CXCR4 axis and promotes tumour development by increasing neovascularization. Cancer Lett. 2017;395:11-19
41. Russmueller G, Moser D, Würger T, Wrba F, Christopoulos P, Kostakis G, Seemann R, Stadler V, Wimmer G, Kornek G, Psyrri A, Filipits M, Perisanidis C. Upregulation of osteoprotegerin expression correlates with bone invasion and predicts poor clinical outcome in oral cancer. Oral Oncol. 2015;51(3):247-53
Author contact
![]() Corresponding author: Zhaoqian Liu, Institute of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha 410078, People's Republic of China; Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China. Tel: +86 731 89753845, Fax: +86 731 82354476, E-mail: liuzhaoqian63com.
Corresponding author: Zhaoqian Liu, Institute of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha 410078, People's Republic of China; Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China. Tel: +86 731 89753845, Fax: +86 731 82354476, E-mail: liuzhaoqian63com.

 Global reach, higher impact
Global reach, higher impact