Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(9):1958-1967. doi:10.7150/jca.30231 This issue Cite
Research Paper
Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas
1. Department of Pathology, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea
2. Comprehensive Gynecologic Cancer Center, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea
3. Institute for Clinical Research, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea
4. Department of Biomedical Science, Department of Pathology, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea
*These two authors contributed equally to this article.
Received 2018-9-27; Accepted 2019-2-25; Published 2019-5-12
Abstract
Background: Exosomes are extracellular microvesicles that are released by most cells and widely distributed in various body fluids. Malignant cells secrete large amounts of exosomes containing various molecular constituents reflecting the originating tumor. We investigated the difference in microRNA (miRNA) expression in serum exosomes from the patients with benign, borderline and malignant ovarian masses to assess the diagnostic relevance of serum exosomal miRNAs as biomarkers for preoperative diagnosis of ovarian carcinoma.
Methods: A total of 68 cases of ovarian masses were enrolled, comprising benign ovarian cysts (benign; n=10), borderline ovarian tumors (BOT, n=10), high-grade serous ovarian carcinomas (HGSOC, n=39) and non-HGSOCs (n=9). Exosomal RNA was extracted from the serum, and expression levels of seven miRNAs (miRNA-21, -93, -141, -145, -200a, -200b and -200c), which were reportedly dysregulated in serous ovarian cancer in previous studies, were quantified by real-time PCR, and compared between the four groups.
Results: MiR-93, -145, and -200c, showed significantly higher expression in serum exosomes of the cancer group (HGSOC and non-HGSOC) than of the non-cancer group (benign and BOT; all p<0.05). The remaining three miRs (miR-141, -200a, and -200b) were expressed at extremely low levels, and not appropriate as serological biomarkers. To test discrimination of cancer from non-cancer, the area under the receiver operating characteristic curves determined for cancer antigen 125 (CA125), miR-145, miR-200c, miR-21, and miR-93 were 0.801 (p<0.001), 0.910 (p<0.001), 0.802 (p<0.001), 0.585 (p=0.303), and 0.755 (p=0.002), respectively. MiR-145 showed superior sensitivity (91.6%), and miR-200c showed superior specificity (90.0%), compared with CA125.
Conclusion: Expression of exosomal miR-93, miR-145 and miR-200c was significantly elevated in the serum of ovarian cancer patients. Serum exosomal miR-145 in particular appeared to be the most promising biomarker for preoperative diagnosis of ovarian cancer.
Keywords: exosome, microRNA, ovarian cancer, serum, biomarker
Introduction
Ovarian cancer is the leading cause of death among gynecological malignancies in the United States, with a 5-year survival rate of 44% for all stages in all races. The lack of an effective early detection screening test and unfavorable anatomical location are associated with the majority of patients presenting with advanced stage at diagnosis, and consequent poor prognosis [1]. Preoperative differentiation between malignant and non-malignancy is very important to avoid unnecessary surgery, and to determine the optimal treatment plan. Even though several modalities, such as measurement of serum levels of cancer antigen 125 (CA125) and applying the risk of ovarian malignancy algorithm (ROMA), are currently used for preoperative discrimination of adnexal masses, none are sufficiently effective [2, 3]. Thus, there remains a need to develop useful biomarkers for preoperative diagnosis of ovarian carcinomas, especially for high-grade serous ovarian carcinoma (HGSOC), the most common and most lethal ovarian cancer.
MiRNAs (miRs) are a large family of small, non-coding RNAs of 21-25 nucleotides in length. miRs regulate gene expression post-transcriptionally by binding to the 3'-untranslated region of target messenger RNA (mRNA), resulting in mRNA degradation and/or translational repression[4]. Most biologic intracellular processes are under the control of miRs, which affect the activities of more than 50% of protein-coding genes in mammals. Dysregulation of miRs is associated with carcinogenic processes such as cell proliferation, metabolic perturbation, angiogenesis, invasion and metastasis in human cancers [5]. Deregulated expression of specific miRs is associated with various diseases, including solid and hematopoietic tumors. A number of miRs were increased or decreased in ovarian cancer tissues compared with normal tissues [6], including those identified in several high-throughput studies comparing miR expression profiles between normal tissue and serous carcinoma tissue (Table 1) [7-10]. Given that miRs are highly stable in a variety of body fluids, and that the composition of miRs in blood mirror the presence of various malignant diseases, we focused on the miRs most commonly reported to be altered in ovarian cancer tissues in the literature in order to investigate them as candidate serum biomarkers for detecting ovarian carcinoma. Among the many differentially expressed miRs reported in the high-throughput studies, we selected seven which were consistently dysregulated in serous carcinoma in at least three studies: six (miR-21, miR-93, miR-141, miR-200a, miR-200b, and miR-200c) were upregulated in serous carcinoma, and one (miR-145) was downregulated. In accordance with these studies, we have previously found that the expression level of miR-145 in HGSOC is significantly lower than in normal fallopian tube tissue [11].
To identify reliable serum biomarkers for ovarian cancer, we focused on the properties of exosomes; they are abundant in body fluids that can be less-invasively obtained, and reflect the characteristics of the originating cells. Exosomes are disk-shaped microvesicles with a diameter of 50-100 nm, which originate from intracellular microvesicular bodies. They are secreted by most types of proliferating cells and are found in various body fluids including blood, urine, saliva and ascites. Malignant cells secrete higher amounts of exosomes than normal cells, and tumor-derived exosomes contain various molecular constituents including proteins, DNA, and RNA that reflect the originating tumor cells [12-17].
In the present study, to identify novel biomarkers for discriminating ovarian carcinoma from a benign adnexal mass, we investigated the expression levels of seven candidate miRs in serum exosomes from patients with ovarian carcinomas and compared them with those of benign ovarian cysts (benign) and borderline ovarian tumors (BOT).
Materials and methods
Patients and samples
A total of 68 patients were enrolled in this study. Forty-eight patients with indeterminate ovarian masses who were scheduled to undergo surgery at CHA Bundang Medical Center were prospectively enrolled from October 2016 to April 2018, and peripheral bloods from the patients were sampled before the surgery. After final pathologic diagnosis of the ovarian masses, the 48 patients were divided into four disease groups: benign, BOT, HGSOC, and non-HGSOC. In addition, a total of 20 preoperatively collected serum samples from patients with HGSOC were kindly provided by the Keimyung University Dongsan Hospital Korea Regional Biobank and the Biobank of Inje University PAIK Hospital. Clinical data, including patient age at initial diagnosis, FIGO stage, presence of nodal metastasis, distant metastasis, local or systemic recurrence, were retrieved from electronic records and pathology reports. This study was approved by the institutional review board of CHA Bundang Medical Center. Informed consent was received from all patients.
The reference studies comparing the miRNA expression between serous carcinoma and normal tissue
| Study (year) | Method | Analysis platform | No. of serous carcinoma | Type of normal tissue | Upregulated miRNAs | Downregulated miRNAs |
|---|---|---|---|---|---|---|
| Nam et al. (2008) [7] | miRNA microarray | mirVana miRNA Labeling kit (Ambion) | 20 | ovarian tissue (unmatched) | miR-93, miR-23b, miR-20a, miR-27a, miR-16, miR-23a, miR-200a, miR-200b, miR-21, miR-200c, miR-141 | miR-145, miR-125b, miR-100, miR-99a, miR-26a, miR-10b, miR-143, miR-214, let-7b, miR-199a-AS, miR-29a, miR-125a |
| Li et al. (2014) [8] | miRNA microarray | mirVana miRNA Labeling kit (Applied Biosystems) | 8 | fallopian tube tissue (matched) | miR-452-5p, miR-409-3p, miR-224-5p, miR-382-5p, miR-4688, miR-4738-3p, miR-4535, miR-877-5p, miR-601, miR-202-3p, miR-370, miR-135b-5p, miR-3676-5p, miR-99b-3p, miR-1226-5p, miR-4476, miR-1185-2-3p, miR-663a, miR-4417, miR-4776-5p, miR-4741, miR-1202, miR-3960, miR-4634, miR-4687-3p, miR-3196, miR-4281, miR-1207-5p, miR-4539, miR-21-5p, miR-1225-5p, miR-939-5p, miR-1185-1-3p, miR-27a-3p, miR-483-5p, miR-575, miR-4739, miR-940, miR-642b-3p, miR-4530, miR-3663-3p, miR-134, miR-1290 | miR-34b-3p, miR-34c-5p, miR-34c-3p, miR-34b-5p, miR-129-1-3p, miR-450a-5p, miR-4423-3p, miR-542-3p, miR-449b-5p, miR-512-3p, miR-542-3p, miR-139-5p_v18.0, miR-503-5p, miR-375, miR-1180, miR-449a, miR-424-5p, miR-23b-5p, miR-129-2-3p, miR-126-5p, miR-125b-2-3p, miR-3607-3p, miR-135a-5p, miR-374c-5p, miR-328, miR-4324, miR-95, miR-99a-5p, miR-92b-3p, miR-139-3p, miR-505-5p, miR-145-3p, miR-548aa, miR-195-5p, miR-497-5p, miR-769-5p, miR-338-5p, miR-424-3p, miR-361-3p, miR-100-5p, miR-885-5p, miR-548d-5p, miR-744-5p, miR-4657, miR-140-3p, miR-625-5p, miR-339-3p, miR-423-3p, miR-4731-3p, miR-31-5p, miR-30a-5p, miR-598, let-7c, miR-145-5p, miR-140-5p, miR-29c-5p, miR-125b-5p, miR-423-5p, miR-1229-3p, miR-126-3p, miR-3653, miR-664a-3p, miR-101-3p |
| Vilming et al. (2014) [9] | miRNA microarray | GeneChip miRNA 2.0 Array (Affymetrix) | 12 (high grade only) | ovarian surface epithelium (unmatched) | miR-24-3p, miR-103a-3p, miR-126-3p, miR-885-5p, miR-106b-5p, miR-20a-5p, miR-17-5p, miR-106a-5p, miR-93-5p, miR-93-3p, miR-15b-5p, miR-200c-5p, miR-425-5p, miR-296-3p, miR-18a-5p, miR-378a-5p, miR-200b-5p, miR-183-5p, miR-200c-3p, miR-200b-3p, miR-182-5p, miR-200a-5p, miR-200a-3p, miR-141-3p | miR-202-3p, miR-383, miR-424-5p, miR-134, miR-542-5p, miR-424-3p, miR-483-5p, miR-381-3p, miR-4324, miR-376c-3p, miR-433, miR-493-3p, miR-379-5p, miR-509-3p, miR-509-3-5p, miR-127-5p, miR-299-3p, miR-887, miR-4269, miR-503-5p, miR-432-5p, miR-140-3p, miR-370, miR-214-3p, miR-154-5p, miR-29c-3p, miR-135a-3p, miR-127-3p, miR-514b-5p, miR-508-5p, miR-502-3p, miR-1271-5p, miR-99a-5p, miR-510, miR-501-3p, miR-500a-3p, miR-874, miR-101-3p, miR-532-5p, miR-362-5p, miR-532-3p, miR-204-5p, miR-188-5p, miR-143-3p, miR-328, miR-500a-5p, miR-99a-3p, miR-1291, miR-145-5p, miR-29b-2-5p, miR-509-5p, miR-29c-5p, miR-574-3p, miR-494 |
| Ibrahim et al. (2015) [10] | Real-time PCR | Cancer Focus microRNA PCR panel (Exiqon) | 22 | ovarian tissues (unmatched) | miR-200c, miR-141, miR-200b, miR-200a, miR-182, miR-31, miR-7, miR-203, miR-10a, miR-21, miR-18a, miR-93, miR-20b, miR-146a, miR-155, miR-15a, miR-106a, miR-210 | let-7a, miR-29a, miR-126, miR-29c, miR-132, miR-101, miR-26a, miR-7b, miR-143, miR-9, miR-7c, miR-214, miR-100, miR-125b, miR-202, miR-99a, miR-195, miR-145, miR-1, miR-133a |
Exosome isolation from serum
The whole blood samples were centrifuged at 3000 rpm for 15 min at room temperature. The resulting supernatant was designated serum, which was immediately apportioned into 500 μL aliquots and stored at -80°C. Frozen serum samples were thawed and then centrifuged at 2000 × g for 30 min to remove any cellular debris. The supernatant containing the cell-free serum was combined with 0.2 volumes of Total Exosome Isolation Reagent (Thermo Scientific, Worcester, MA). The samples were incubated at 4°C for 30 min and then centrifuged at 10000 × g for 10 min. The supernatant was discarded, and the exosomal pellet was resuspended in PBS.
Western blotting
To verify the isolation of exosomes from the serum, western blot analysis for CD9 and CD63, which are enriched in exosomes, was performed. Total proteins were extracted using lysis buffer (Pro-Prep, iNtRON Biotechnology, South Korea) and 20 μg of protein was separated by 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (GE Health care, Piscataway, NJ). After blocking with 5% skimmed milk for 1 hour at room temperature, membranes were incubated overnight at 4°C with primary antibody (anti-β-actin 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-CD9 1:1000 (Cell Signaling, Danvers, MA, USA) or anti-CD63 1:1000 (Abcam, Cambridge, UK)) followed by horseradish peroxidase-conjugated anti-mouse 1:1000 or anti-rabbit secondary antibody 1:1000 (Novus Biologicals, Littleton, CO, USA), and incubated for 1 hour at room temperature. After incubation, membranes were washed and proteins revealed by Western Blotting Luminol Reagent (Bio-Rad, Hercules, CA, USA).
RNA extraction from serum exosomes
Total RNA, including miRs, was extracted from the exosome pellet using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Briefly, 1.0 mL TRIzol reagent and 200 µL chloroform were added to the sample. After centrifugation at 12000 × g for 20 min at 4°C, the supernatant was incubated with 500 µL isopropanol at -20°C for 10 min. The mixture was centrifuged at 12000 × g for 10 min at 4°C to remove the supernatant and the RNA pellet was washed with 75% ethanol and then dissolved in 12 µL RNase-free water. The purity of isolated RNA was determined by OD260/280 using a Nanodrop ND-1000 spectro-photometer (Thermo Scientific, Worcester, MA).
Quantitative real-time PCR
The expression level of candidate miRs in serum exosomal RNA was evaluated by quantitative real-time PCR (qRT-PCR). Total RNA (2 ng) was reverse transcribed using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) and microRNA-specific stem-loop primers (part of the TaqMan microRNA Assay Kit; Applied Biosystems). The mixture was incubated at 16°C for 30 min, 42°C for 30 min and 85°C for 5min. QRT-PCR for miRNAs was conducted according to the TaqMan miR Assay protocol (Applied Biosystems Carlsbad, CA, USA) using a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). All PCR reactions were run in duplicate. The relative gene expression values for the target miRs were normalized to RNU48 and calculated using the 2-ΔCT method.
Statistical analysis
ANOVA and the Bonferroni post hoc t-test were used to compare the patient age between four disease groups. Kruskal-Wallis and Mann-Whitney U tests were used to compare the CA125 levels and the candidate miR expression levels between the four disease groups. Fisher's exact test was used to compare the FIGO stage between HGSOC and non-HGSOC groups. Receiver operating characteristic (ROC) curves were generated, and areas under the ROC curves (AUC) were calculated to evaluate the diagnostic performance of the markers. Diagnostic sensitivity, specificity and accuracy, in terms of ability of the biomarkers to assign the four known diagnoses to true positive (TP), true negative (TN), false negative (FN), and false positive (FP), were calculated as follows: sensitivity = TP/(TP+FN); specificity = TN/(TN + FP); and accuracy = (TN + TP)/(TN+TP+FN+FP). Statistical analysis was performed using SPSS software version 21.0 (SPSS, Chicago, IL). Statistical difference was considered significant when the p-value was <0.05.
Results
Clinical characteristics of the patients
The clinical characteristics of the patients are shown in Table 2. Sixty-eight patients were divided into four groups based on the pathologic diagnosis: benign (n=10), BOT (n=10), HGSOC (n=39), and non-HGSOC (n=9). The histologic types of non-HGSOC patients were clear cell carcinoma (n=3), mucinous carcinoma (n=4), endometrioid carcinoma (n=1), and low-grade serous carcinoma (n=1). The patients with HGSOC had the oldest mean age among the four groups (p=0.001). The preoperative serum CA125 level was significantly higher in the HGSOC group than the benign, BOT, and non-HGSOC groups (p<0.001). Based on a reference value of 35 U/mL that is generally accepted as the upper limit of the normal range, CA125 elevation above this cutoff was observed at a significantly higher frequency in the HGSOC group (97.4%) than the benign (14.3%), BOT (40.0%), and non-HGSOC (55.6%) groups (p<0.001). The FIGO stage at the time of diagnosis was significantly different between HGSOC and non-HGSOC patients; most non-HGSOC patients were stage I (77.8%), while the majority of HGSOC patients were stage III or IV (74.4%; p=0.004).
Identification of serum exosomes
To confirm that the pellets extracted using the exosome precipitation protocol were truly exosomes, western blot analysis for expression of CD9 and CD63, cell surface glycoproteins enriched in exosomes, was performed. CD9 and CD63 were readily detected in the pellets extracted from the sera of patients with ovarian cancer (HGSOC and non-HGSOC), indicating that exosomes had indeed been adequately purified using this methodology (Fig. S1).
Validation of expression of selected miRs in serum exosomes
Of the seven miRs selected for study by qRT-PCR of serum exosomes, miR-141, miR-200a and miR-200b did not show any detectable amplification up to 40 PCR cycles (or even beyond in most samples) regardless of patient group, suggesting that these miRs are expressed at extremely low levels in serum exosomes.
Clinicopathologic characteristics of the patients
| Parameters | Study groups | p | ||||
|---|---|---|---|---|---|---|
| Benign ovarian cyst (n=10) | BOT (n=10) | HGSOC (n=39) | non-HGSOC (n=9) | |||
| Age | Mean (Range) | 57.2 (35-70) | 41.6 (22-69) | 58.1 (35-83) | 43.7 (24-65) | <0.001 |
| Preoperative CA125 (U/mL) | Median (Range) | 9.1 (5.3-53.6) | 17.8(5.3-274.1) | 539.5(20.6-19805) | 41.6(10.8-465.6) | <0.001 |
| <35 U/mL | N (%) | 6(85.7) | 6 (60.0) | 1 (2.6) | 4 (44.4) | <0.001 |
| ≥35 U/mL | 1(14.3) | 4 (40.0) | 38 (97.4) | 5(55.6) | ||
| FIGO stage | I | NA | NA | 6(15.4%) | 7(77.8%) | 0.004 |
| II | NA | NA | 4(10.3%) | 0(0) | ||
| III | NA | NA | 11(28.2%) | 1(11.1%) | ||
| IV | NA | NA | 18(46.2%) | 1(11.1%) | ||
BOT, borderline ovarian tumor; HGSOC, high-grade serous ovarian carcinoma.
Expression levels of serum exosomal miR-145, 200c, 21, and 93 determined by qRT-PCR in patients with benign ovarian cyst (benign), borderline ovarian tumor (BOT), high-grade serous ovarian carcinoma (HGSOC), and non-HGSOC. The expression levels of miR-145 and miR-200c were significantly different between the HGSOC and non-cancer (benign or BOT) groups. By contrast, significant differences in miR-21 and miR-93, were found between non-HGSOC and non-cancer groups. The horizontal lines indicate median values.
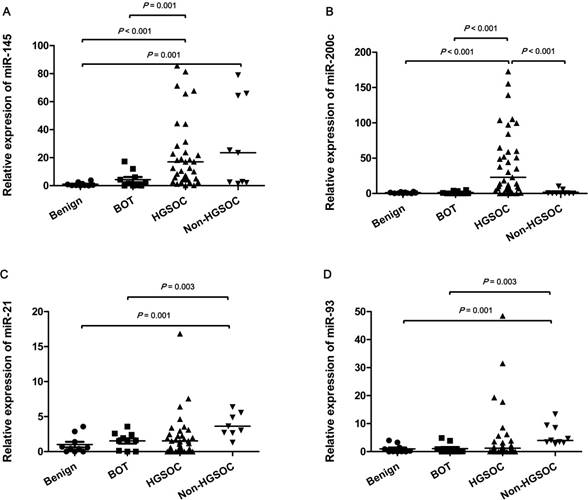
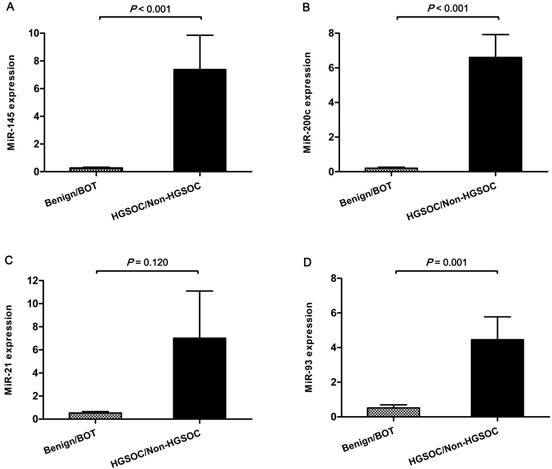
The differences in expression levels of miR-21, -93, -145 and -200c detected using qRT-PCR in serum exosomes from the benign, BOT, HGSOC and non-HGSOC groups are demonstrated in Fig 1. The mean expression level of miR-145 was significantly increased in the HGSOC group compared with the benign and BOT groups (47.7-fold and 9.9-fold; p<0.001 and p=0.001, respectively), and significantly increased in the non-HGSOC (29.6-fold) compared with the benign group (p=0.001). However, there was no significant difference in miR-145 level between the benign vs. BOT, BOT vs. non-HGSOC, and HGSOC vs. non-HGSOC (Fig. 1A). The mean expression level of miR-200c was significantly increased in the HGSOC group compared with the benign, BOT, and non-HGSOC groups (46.7-fold, 34.4-fold, and 25.5-fold; p<0.001, p<0.001, and p=0.001, respectively; Fig. 1B). The mean expression levels of miR-21 (26.4-fold and 17.4-fold; p=0.001 and p=0.003, respectively) and miR-93 (5.8-fold and 5.5-fold, respectively) were significantly increased in the non-HGSOC group compared with the benign and BOT groups, whereas their expression in the HGSOC group was not significantly altered (Fig. 1C and 1D). When the four groups were divided into simply non-cancer (benign and BOT, n=20) and cancer (HGSOC and non-HGSOC, n=48), the expression levels of miR-145, -200c, and -93 were significantly higher in the cancer group than in the non-cancer group (p<0.001, p<0.001, and p=0.001, respectively) (Fig. 2A, 2B and 2D). The expression level of miR-21 was slightly higher in the cancer than in non-cancer group; however, it was not statistically significant (p=0.120; Fig. 2C).
Performance of serum exosomal miRs as diagnostic markers compared with CA125
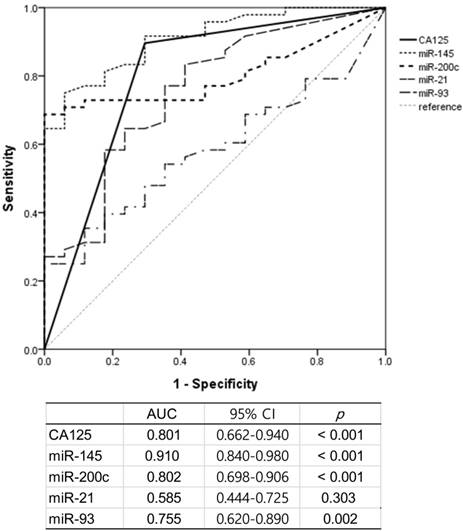
We performed ROC curve analysis to compare the performance of serum CA125 (>35 U/mL) and serum exosomal miRs for differential diagnosis of ovarian masses (benign/BOT vs. HGSOC/non-HGSOC). The AUC values (95% confidence interval; p-value) for CA125, miR-145, miR-200c, miR-21, and miR-93 were 0.801 (0.662-0.940; p<0.001), 0.910 (0.840-0.980; p<0.001), 0.802 (0.698-0.906; p<0.001), 0.585 (0.444-0.725; p=0.303), and 0.755 (0.620-0.890; p=0.002), respectively (Fig. 3). The AUC values for miR-145 and miR-200c were higher than that of CA125. The sensitivity, specificity, and accuracy of each miR and of CA125>35U/mL at predicting ovarian cancer are shown in Table 3. According to the AUC analysis, miR-145 was the best performing single marker for sensitivity (91.7%) and accuracy (86.8%). MiR-200c showed the highest specificity (90.0%) but a relatively low sensitivity (72.9%). The combination of CA125 and miR-145 had higher sensitivity (97.9%) and accuracy (86.8%). The combination of the three markers achieved perfect sensitivity (100%), but quite low specificity (55.0%).
Difference in expression of serum exosomal miRs between non-cancer (benign/BOT) and cancer (HGSOC/non-HGSOC) groups. Expression levels of miR-145, -200c, and -93 were significantly increased in the cancer group compared with the non-cancer group. Data represent mean ± standard error on the mean (SEM). Benign, benign ovarian cyst; BOT, borderline ovarian tumor; HGSOC, high-grade serous ovarian carcinoma.
Performance of serum CA125, serum exosomal miR-145, and miR-200c in predicting carcinoma
| Diagnostic markers | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Single marker | |||
| CA125+ | 89.5% | 70.6% | 84.6% |
| miR-145+ | 91.7% | 75.0% | 86.8% |
| miR-200c+ | 72.9% | 90.0% | 77.9% |
| Combination of two markers | |||
| CA125+ or miR-145+ | 97.9% | 60.0% | 86.8% |
| CA125+ or miR-200c+ | 93.8% | 70.0% | 86.8% |
| miR-145+ or miR-200c+ | 93.8% | 65.0% | 85.3% |
| Combination of three markers | |||
| CA125+ or miR-145+ or miR-200c+ | 100.0% | 55.0% | 86.8% |
Receiver operating characteristic (ROC) curve analysis for the prediction of carcinoma by CA125, serum exosomal miR-145, -200c, -21, and -93. AUC, area under the ROC curve; CI, confidence interval.
Correlation between serum exosomal miRNA expression and clinicopathologic features in patients with high-grade serous carcinoma
| Parameter | Number | miR-145 upregulation | p | miR-200c upregulation | p | miR-21 upregulation | p | miR-93 upregulation | p | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age, years | ||||||||||
| <56 | 19 | 6 (31.6%) | 0.389 | 13 (68.4%) | 0.915 | 11 (57.9%) | 0.621 | 14 (73.7%) | 0.557 | |
| ≥56 | 20 | 9 (45.0%) | 14 (70.0%) | 10 (50.0%) | 13 (65.0%) | |||||
| FIGO stage | ||||||||||
| I, II | 10 | 3 (30.0%) | 0.524 | 7 (70.0%) | 0.951 | 3 (30.0%) | 0.079 | 05 (50.0%) | 0.127 | |
| III, IV | 29 | 12 (41.1%) | 20 (69.0%) | 18 (62.1%) | 22 (75.9%) | |||||
| Nodal metastasis | ||||||||||
| Absent | 18 | 8 (44.4%) | 0.477 | 13 (72.2%) | 0.708 | 10 (55.6%) | 0.843 | 09 (50.0%) | 0.016 | |
| Present | 21 | 7 (33.3%) | 14 (66.7%) | 11 (52.4%) | 18 (85.7%) | |||||
| Distant metastasis | ||||||||||
| Absent | 21 | 5 (23.8%) | 0.042 | 12 (57.1%) | 0.077 | 08 (38.1%) | 0.033 | 15 (71.4%) | 0.748 | |
| Present | 18 | 10 (55.6%) | 15 (83.3%) | 13 (72.2%) | 12 (66.7%) | |||||
| Recurrence | ||||||||||
| Absent | 28 | 10 (35.7%) | 0.718 | 18 (64.3%) | 0.446 | 16 (57.1%) | 0.723 | 19 (67.9%) | 1 | |
| Present | 11 | 5 (45.5%) | 9 (81.8%) | 5 (45.5%) | 8 (72.7%) |
Correlation between serum exosomal miR expression and clinicopathologic factors in HGSOC patients
We analyzed whether the expressional elevation of the four miRs was associated with clinicopathologic features of the HGSOC patients (Table 4). Upregulation of miR-145, -200c, -21, and -93 detected by qRT-PCR was defined as normalized expression greater or equal to 3.6, 1.4, 0.6, and 0.3 times that of the endogenous control RNU48, respectively. Upregulation of miR-145 and miR-21 was observed at significantly higher frequency in the patients with distant metastasis than in those without (p<0.042 and 0.033, respectively). Upregulation of miR-93 was observed at significantly higher frequency in the patients with nodal metastasis than in those without (p=0.016).
Discussion
Ovarian carcinoma is the fifth most common cancer in women worldwide and remains the leading cause of mortality from female malignancy [18]. The major reason for the high mortality rate of ovarian carcinoma is late diagnosis due to obscure early symptoms and the lack of an early diagnostic marker. CA125 is the most widely used serum tumor marker in the gynecological field; however, it is not sufficiently effective to detect ovarian cancer early, being elevated above the normal serum level in only about 50% of patients with early-stage ovarian cancer [19]. In addition, elevation of CA125 is commonly observed in several benign gynecologic and non-gynecologic conditions, such as endometriosis, adenomyosis, pelvic inflammatory disease, and even pregnancy [20]. Therefore, a new reliable serum marker is necessary for the early diagnosis of ovarian carcinomas and for their differential diagnosis from a benign ovarian mass. In this regard, miRs, which appear in extracellular stable forms, represent promising candidate biomarkers for cancer.
There have been a great number of studies on the roles of miRs in cancer since their discovery about two decades ago. Alterations in miR expression have been detected in various human cancer tissues, including ovarian, in a number of studies [6]. Recently, several studies investigated the clinical relevance of blood circulating miRs as diagnostic and prognostic biomarkers for ovarian cancer [21]. Despite the ribonucleases in body fluids, the circulating miRs are reportedly highly stable because most are packaged in exosomes, or apoptotic bodies or are bound to argonaute 2 or lipoprotein complexes. Among these, exosomal miRs are thought to serve as biomarkers superior to other extracellular miRs because cancer cells secrete much larger amounts of exosomes than normal cells, therefore, measurement of exosomal RNA may reduce the background noise caused by RNAs shed from a variety of normal cells. We thus focused on the miRs contained in the serum exosomes rather than on free miRs circulating in serum. [15, 22-25].
As the first step towards identifying robust serum miR biomarkers, we selected the seven most commonly altered miRs in ovarian carcinoma tissues in published high-throughput miR profiling studies. We, then, examined their expression in serum exosomes from patients with benign and malignant ovarian tumors as candidate serum biomarkers. Of the seven miRs studied, expression levels of miR-145 and miR-200c were significantly higher in exosomes from HGSOC patients than in those from patients with benign or borderline tumors. The expression of miR-200c was no higher in non-HGSOCs than in non-cancerous tissue, whereas that of miR-145 was significantly higher in non-HGSOCs than in the benign control. The expression levels of miR-21 and miR-93 were significantly higher only in non-HGSOCs than in the other groups. In addition, we also found that upregulation of miR-145 and miR-21 was significantly associated with distant metastasis, representing their likely clinical implication. Interestingly, miR-145, identified in published reports as a marker downregulated in ovarian cancer tissue, was highly expressed in the serum exosomes of cancer patients. This unexpected finding suggests that miR expression in serum exosomes does not always mirror that of the originating tumor tissue. In accordance with our results, a previous study by Chan et al. demonstrated that among 20 miRs overexpressed in breast cancer tissue, only seven were overexpressed in both breast cancer tissues and sera, and the other 13 were dysregulated in breast cancer tissue and sera in opposite directions [26]. Dissimilar expression pattern of miRs between tumor tissue and blood have been reported in several subsequent studies [27-29]. Pigati et al. investigated the composition of extracellular exosomal miRs isolated from cell culture media, and intracellular miRs in MCF7 breast cancer cell line, and found that approximately 66% of extracellular exosomal miRs are closely reflected by the intracellular amount, while the remainder are either enriched in extracellular exosomes or retained in cells. These findings indicate that exosomal miRs are not simply unsorted products passively released by cells, and there may be as yet unknown mechanisms controlling the release of miRs into exosomes or the retention of miRs inside cells [30, 31].
The miR-145 gene is located on chromosome 5q32, and known as a tumor suppressor that is, as we and others have previously shown, downregulated in various human cancers including lung, prostate, colon, breast and ovarian cancer [1-3, 11]. Two previous studies reported that serum circulating miR-145 is also downregulated in patients with ovarian cancer, as in ovarian cancer tissues [4, 32]. However, in the present study, we found the opposite. This discrepancy could have resulted from the methodological difference; we analyzed the serum exosomal miR, not free circulating miR as they used. A recent study of thyroid cancer showed similar results to our study; miR-145 expression was significantly reduced in thyroid cancer tissue, but increased in serum exosomes, suggesting that miR-145 is actively secreted into exosomes by thyroid cancer cells [33]. Until this phenomenon can be explored, we suggest a “banishing theory” to explain it, wherein miR-145 is overexpressed in ovarian cancer cells as a response to inhibit cancer progression, but is immediately banished from the cancer cells in the form of an exosome. As a result, miR-145 would seemingly be upregulated in serum exosomes, while being downregulated in cancer tissue and serum.
MiR-200c is a member of the miR-200 family consisting of five members (miR-141, -200a, -200b, -200c, and -429) which plays crucial roles in cancer initiation and metastasis [33, 34]. Upregulation of miR-141, -200a, -200b, and -200c is a well-known miR signature of ovarian cancer tissue that has been identified in several studies [7-10]. We analyzed these four miR-200 family members but found that only miR-200c was highly expressed in serum exosomes, while the other three were present only at very low levels and inappropriate as serological markers.
In our study, of the chosen miRs and CA125 >35 U/mL, miR-145 was the best performing single marker at predicting carcinoma, with a sensitivity of 91.7%, while miR-200c showed the highest specificity (90.0%). When miR-145, miR-200c, and CA125 were combined, sensitivity reached 100%. Therefore, exosomal miR-145 or miR-200c might be promising biomarkers to discriminate ovarian cancer from benign lesions, and to overcome the limitation of CA125. To our knowledge, these biomarkers show the best performance among those detailed in the many previous studies of serum of ovarian cancer patients.
The level of serum CA125 was in the normal range (<35 U/mL) in five of the cancer patients in the present study (all of them were FIGO stage I or II), and elevated (≥35 U/mL) in five patients with benign and borderline disease. All five cancer patients with a normal CA125 level showed increased expression of miR-145 and/or miR-200c, indicating that serum exosomal miR-145 and -200c can compensate for the limited sensitivity of CA125 in preoperative prediction of ovarian carcinoma. In addition, four out of the five patients with a false positively elevated CA125 level showed a low expression level (below the cutoff value) of miR-200c, which suggests that miR-200c can compensate for the limited specificity of CA125.
To the best of our knowledge, this is the first study to demonstrate the potential role of serum exosomal miR-145 as a biomarker for ovarian cancer. Our results showed that upregulation of miR-145 and miR-200c were the most sensitive and specific markers for differential diagnosis of ovarian masses, performing better than CA125. A triple combination of miR-145 + miR-200c + CA125 was the most powerful marker with 100% sensitivity for ovarian cancer. A large-scaled study is now required to confirm the role of these miRs as biomarkers for ovarian cancer.
Supplementary Material
Supplementary figures.
Acknowledgements
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI16C1559).
Biospecimens and data used in this study were provided by the Biobank of Keimyung University, Dongsan Hospital Biobank and the biobank of Inje University PAIK Hospital (InjeBiobank), members of the Korea Biobank Network.
S.I. Park, a research nurse, supported the blood sampling of the patients enrolled at the CHA Bundang Medical Center.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
2. Karlsen MA, Sandhu N, Hogdall C, Christensen IJ, Nedergaard L, Lundvall L. et al. Evaluation of HE4, CA125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) as diagnostic tools of epithelial ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2012;127:379-83
3. Moore RG, Jabre-Raughley M, Brown AK, Robison KM, Miller MC, Allard WJ. et al. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol. 2010;203:228 e1-6
4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
5. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610
6. Zhang S, Lu Z, Unruh AK, Ivan C, Baggerly KA, Calin GA. et al. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res. 2015;13:393-401
7. Ibrahim FF, Jamal R, Syafruddin SE, Ab Mutalib NS, Saidin S, MdZin RR. et al. MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J Ovarian Res. 2015;8:56
8. Li Y, Yao L, Liu F, Hong J, Chen L, Zhang B. et al. Characterization of microRNA expression in serous ovarian carcinoma. Int J Mol Med. 2014;34:491-8
9. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH. et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690-5
10. Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC. et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. 2014;14:80
11. Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH. et al. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015;356:937-45
12. Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A. et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25:3703-7
13. Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A. et al. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66:9290-8
14. Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P. et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295-305
15. Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42-6
16. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21
17. Tang MK, Wong AS. Exosomes: Emerging biomarkers and targets for ovarian cancer. Cancer Lett. 2015;367:26-33
18. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30
19. Guo X, Li D, Chen M, Chen L, Zhang B, Wu T. et al. miRNA-145 inhibits VSMC proliferation by targeting CD40. Sci Rep. 2016;6:35302
20. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29
21. Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016;15:48
22. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679
23. Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M. et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159-67
24. Lance RS, Drake RR, Troyer DA. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Expert Rev Anticancer Ther. 2011;11:1341-3
25. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940-8
26. Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA. et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477-87
27. Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8:69934-44
28. Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng G. et al. Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thorac Cancer. 2016;7:348-54
29. Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P. et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098-106
30. Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D. et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515
31. Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as Biomarkers for Prostate Cancer. Front Genet. 2013;4:36
32. Sahu A, Jha PK, Prabhakar A, Singh HD, Gupta N, Chatterjee T. et al. MicroRNA-145 Impedes Thrombus Formation via Targeting Tissue Factor in Venous Thrombosis. EBioMedicine. 2017;26:175-86
33. Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472-98
34. Muralidhar GG, Barbolina MV. The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer. Int J Mol Sci. 2015;16:16833-47
Author contact
![]() Corresponding author: Hee-Jung An, MD PhD. Address: Department of Pathology, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea. Phone: +82-31-780-5439 Fax: +82-31-780-5476 Email: hjahnco.kr
Corresponding author: Hee-Jung An, MD PhD. Address: Department of Pathology, CHA Bundang Medical Center, CHA University, 59 Yatap-ro, Bundang-gu Seongnam-si Gyeonggi-do 13496, Republic of Korea. Phone: +82-31-780-5439 Fax: +82-31-780-5476 Email: hjahnco.kr

 Global reach, higher impact
Global reach, higher impact