3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(9):2025-2034. doi:10.7150/jca.29457 This issue Cite
Research Paper
Long non-coding RNA UASR1 promotes proliferation and migration of breast cancer cells through the AKT/mTOR pathway
1. Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University. 283 Tongzipo Road, Changsha 410013, Hunan, China.
2. College of Bioscience and Biotechnology, Hunan Agricultural University, Furong District, Changsha 410128, China.
3. Department of Medical Microbiology, Immunology & Cell Biology, Southern Illinois University School of Medicine. 913 N. Rutledge Street, Springfield, IL 62794.
4. Department of Pharmaceutical Engineering, School of Chemical Engineering, Xiangtan University, Xiangtan, 411105, China.
5. Biotechnology Department, College of Science, Baghdad University, Baghdad, Iraq, 10071.
6. Laboratory of Hepatobiliary and Pancreatic Surgery, Affiliated Hospital of Guilin Medical University, 15 Lequn Road, Guilin 541001, Guangxi, China.
Received 2018-8-24; Accepted 2019-3-28; Published 2019-5-12
Abstract
Long non-coding RNAs (lncRNAs) are non-coding RNAs longer than 200 nucleotides that function as regulatory factors in many human diseases, including cancer. However, majority of lncRNAs remain to be characterized. In this study, we characterized a novel lncRNA transcript, named UNC5B antisense RNA1 (UASR1). UASR1 is 647bp in length consisting of two exons. This lncRNA is an antisense of intron 1 of unc-5 netrin receptor B (UNC5B) gene. In breast cancer tissues, UASR1 was upregulated. Ectopic expression of UASR1 promoted proliferation and clonogenic growth of breast cancer cells MCF7 and MDA-MB-231. The migration of these cells also increased as demonstrated by wound healing and transwell assays. In contrast, silencing of UASR1 suppressed cell proliferation and migration. Further studies showed that UASR1 activated AKT and AKT-mediated mTOR signaling pathway to stimulate cell proliferation and growth. In these cells, active pAKT, pTSC2, p4EBP1 and pp70S6K were increased. Taken together, our data suggest that UASR1 plays an oncogenic role in breast cancer cells through activation of the AKT/mTOR signaling pathway, being a novel RNA oncogene.
Keywords: Long non-coding RNA, UASR1, oncogenic lncRNA, AKT/mTOR, Breast cancer.
Introduction
Only 1.5% of nucleic acids in human genome are used for protein encoding, but other 98.5% of the genome does not encode proteins. Some of them are for non-coding RNA transcripts longer than 200 nucleotides, called long non-coding RNAs (lncRNAs) [1, 2]. To date, it is well known that lncRNAs participate in chromatin modifications and transcriptional and translational regulation of protein-encoding genes or regulate protein function and activity through specific binding [3-5].
In tumor tissues, lncRNAs are markedly deregulated and pervasively involved in development and progression of cancers [6-8]. In breast cancer, lncRNAs are abnormally expressed and demonstrate differential expression in subtypes of tumors [9, 10]. Some lncRNAs affect the growth and development of breast cancer by regulating cell proliferation and apoptosis, and others are involved in invasion and metastasis and drug resistance of breast cancer [6]. For instance, H19 is the first lncRNA transcribed by RNA polymerase II and is expressed mainly in human embryonic cells. H19 gene is reactivated and up-regulated during tissue regeneration or tumorigenesis [11, 12]. Oncogene E2F1 is an important transcription factor, promoting cell growth and proliferation, and c-Myc is an oncogene activated in multiple tumors; both E2F1 and c-Myc are stimulating factors of H19 expression, and the H19 lncRNA functions as a downstream effector of c-Myc and E2F1 in tumor development and progression [11]. H19 may play as an oncogene or suppressor depending on tumor types [13].
HOX antisense intergenic RNA (HOTAIR) is overexpressed in multiple malignant tumors, such as breast cancer, liver cancer and colon cancer [14]. In breast cancer, HOTAIR promotes transformation of normal epithelial cells by competitively binding to the tumor suppressor BRCA1 [15, 16]. HOTAIR is upregulated in primary and metastatic breast cancer when compared to the normal breast epithelium, and the expression of HOTAIR is associated with distant metastasis and poor prognosis of patients [17]. Therefore, oncogenic HOTAIR is an independent prognostic factor in breast cancer.
Breast cancer is a leading cause of cancer death in women, and in the United States, over 266,000 new invasive breast cancer cases plus 63,000 new carcinoma in situ (CIS) are diagnosed annually, leading to more than 40,000 cancer death [18]. LncRNAs function as tumor suppressors or oncogenes in the development and progression of breast cancer [19, 20]. Several lncRNAs have been well characterized in breast cancer, such as HOTAIR and BORG [21, 22], but lncRNAs that are involved in breast cancer and their pathogenic role in disease progression remain to be fully understood. In this study, we characterized a novel oncogenic lncRNA, named UNC5B antisense RNA 1 (UASR1). This UASR1 lncRNA, encoded by a gene located at 10q22.1 chromosome, consists of two exons with 647bp in length (Gene ID: 728978). Our data demonstrated that UASR1 promoted growth and migration of breast cancer cells through activation of the AKT/mTOR signaling pathway, functioning as an oncogene in breast cancer.
Materials and Methods
Human sample procurement. Frozen breast cancer (n=30 pairs) and matched normal adjacent tissues were procured following approved IRB protocol from the Tissue Bank of Simmons Cancer Institute at the Southern Illinois University, Springfield, IL. These frozen specimens were used for RNA extraction and real-time RT-PCR analyses of UASR1 expression.
Cell culture. Human breast cancer cells (MCF7 and MDA-MB‑231) were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO) and 1% penicillin/streptomycin at 37˚C, 5% CO2. The basal medium for HEK 293T cells was the same as that for MCF7 cells, but 2.0 mM L-glutamine was added. All cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA).
RNA preparation and quantitative RT-PCR. Total RNA was extracted using TRIzol reagent (Invitrogen, CA). RNA was treated with RNase-free DNase I and the first strand cDNA was synthesized from 1.0μg of total RNA with oligo dT primers and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase according to the manufacturer's protocol (New England Biolab, Beverly, MA). To detect the UASR1 transcript, SYBR Green mix was used with UASR1 primers: forward, 5'-GCGGATCGCAGACCCTAAG-3' and reverse, 5'-GAGAACACTTTGCGGAAGGC-3'. The internal control was β-actin. ΔΔCt values were used to determine relative expression levels as fold changes [23].
Plasmid construction. For ectopic expression of UASR1, its cDNA was amplified by RT-PCR and then subcloned into the pCDH-CMV-EF1-copGFP vector (System Biosciences, Mountain View, CA). High Fidelity Phusion DNA polymerase (Thermo Fisher Scientific, FL) was used for PCR amplification, and conventional sequencing was conducted to confirm the DNA sequence. PCR primers for UASR1 subcloning were: forward, 5'-CCCTCCTCAAACACACATCC-3' and reverse, 5'-TTAAGGAAATTAAAAATACC.
Three shRNA constructs were designed to knock down UASR1 expression. shRNA-1 is forward 5'-CACCTGCTTAATACACATT-3' and reverse 5'-AATGTGTATTAAGCAGGTG-3'; shRNA-2 is forward 5'-CCTCAGGGAAATGCATGGA-3' and reverse 5'-TCCATGCATTTCCCTGAGG-3'; and shRNA-3 is forward 5'-CCGCAAAGTGTTCTCTCCTT-3' and reverse 5'-AAGGAGAGAACACTTTGCGG-3'.
Cell transfection and infection. For packaging of lentiviruses, HEK 293T cells were seeded in 10cm dishes. Cells were transfected with UASR1 expression or shRNA plasmids plus packaging plasmids REV, VSVG and GAG for 24h. Then the medium was changed and cells were continued to culture for 48 -72h to collect viral particles [24]. For infection, cells were spread at 60% of confluence and exposed to lentiviruses (MOI=1) with 1:2000 diluted Polybrene (Millipore, Billerica, MA).
Cell proliferation. For cell proliferation assays, cells were split into 96-well plates at 2000-3000 cells/per well. Viable cells were measured by tetrazolium salt, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays over 5 days [25]. For cell counting assays, cells were split into 12-plates. At the indicated time points, cells were collected and subjected to trypan blue staining. Viable cells were counted in a Vi-cell counter (Beckman Coulter, Brea, CA). For rapamycin treatment, cells were fed with fresh medium containing rapamycin (20 nM) for 24 hours and then subjected to counting.
Colony formation assays. Cells were harvested by trypsin and seeded in 6‑well plates (100 cells/well). The culture medium was refreshed every three days. Once colonies formed were visible, cells were rinsed twice with phosphate buffered saline (PBS) followed by fixation with 4% paraformaldehyde and staining with Giemsa for 5 min. The number of colonies was photographed and counted.
Wound healing assays. Cells were seeded in 6-well plates at 90% confluence and cultured in medium supplemented with 2% FBS. Scratches were made using a 200μl tip. Migration distance was estimated with photographs taken at each time point [26].
Transwell migration assays. Transwell migration assays were performed using 8.0μm pore inserts (BD Biosciences, San Jose, CA). A total of 2-5x104 cells were suspended in 500μl serum-free medium and loaded into upper wells; lower chambers were filled with 750μl complete medium with 10% FBS. Migration chambers were incubated in a 5% CO2 incubator at 37˚C for 36-48h. Cells were then stained and counted for 5 random microscope fields [27].
Western blotting. Cells were lysed in a cell lysis buffer (Roche, IN) with a protease inhibitor cocktail followed by centrifugation at 14,000 rpm for 5 min to collect soluble proteins in supernatant. Cell lysates were separated by 10~12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto 0.22μm PVDF membranes (Millipore, CA) [28]. The membranes were incubated with an appropriate primary antibody with gentle shaking at 4˚C overnight. After incubation with a goat anti-rabbit or anti-mouse secondary antibody at 37˚C for 1h. Protein bands were visualized using an enhanced chemiluminescence.
Results
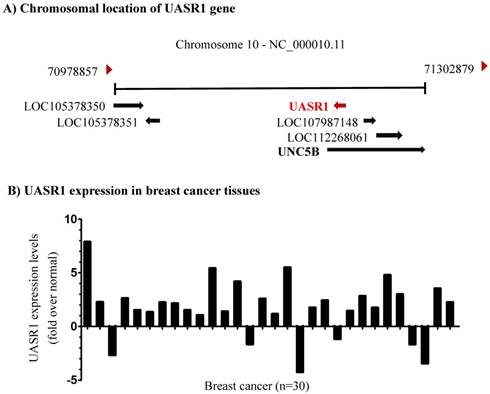
Identification of a novel long non-coding RNA transcript UASR1. Unc-5 netrin receptor B (UNC5B) is a ligand-dependent receptor. In absence of netrins, the intracellular death domain is cleaved by caspase 3 and then combined with death associated protein kinase (DAPK) to induce cell apoptosis; however, in the presence of netrins, the death domain is not cleaved, and apoptosis is inhibited [29]. The lncRNA LOC28978 (Gene ID: 728978) is named UNC5B antisense RNA 1 (UASR1) and we investigated its function in this study. DNA sequence analyses indicated that UASR1 is located in intron 1 of UNC5B gene (Fig. 1A). The intron 1 of UNC5B gene is composed of 66,758bp and UASR1 is located at 5,164 to 4,160bp as an antisense. UASR1 RNA is 647bp in length, consisting of two exons; the intron of UASR1 gene is 358bp in length (Supplementary Figure S1).
Chromosomal location and expression of UASR1 in breast cancer tissues. (A) UASR1 chromosome localization. Data were edited from the web page https://www.ncbi.nlm.nih.gov/gene/?term=728978. (B) UASR1 expression in breast cancer tissues (n=30 pairs). Total RNA extracted from tissues was converted into cDNA for templates. UASR1 RNA levels were determined by quantitative real-time RT-PCR using the SYBR Green technology. ΔΔCt values were used to determine relative expression levels. Data are expressed as fold over the UASR1 level in the matched adjacent normal tissues.
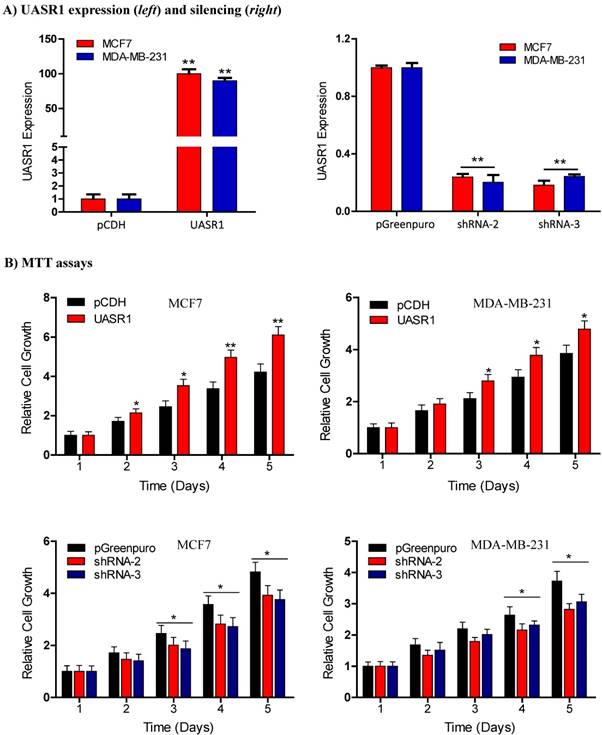
Breast cancer cell growth promoted by UASR1. (A) Expression or shRNA-mediated silencing of UASR1 in MCF7 and MDA-MB-231 cells. Cells were infected with lentivirus pCDH (vector control), UASR1, pGreenpuro (vector control), shRNA-2 or shRNA-3 for 96 hours. UASR1 mRNA levels were detected by SYBR Green quantitative real-time RT-PCR with β-actin mRNA as an internal control. **P<0.01 compared to vector control. (B) MTT assays. Viable cells were measured by a tetrazolium salt, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method as described in Material and Methods. Upper panel: UASR1 expression; lower panel: UASR1 silencing. Data indicate mean ± SD from three independent experiments. *P<0.05 and **P<0.01 compared to vector controls.
UASR1 is up-regulates in breast cancer tissues. To investigate the biological function of UASR1, we first evaluated the expression of UASR1 in breast cancer tissues. As shown in Fig. 1B, in 30 paired breast cancer and adjacent normal tissues, UASR1 expression was increased in 24 (80%) breast cancer tissues compared to the adjacent normal breast tissues, in which UASR1 was increased by >2.0 fold in 15 (50%) breast cancer cases. These data imply that UASR1 may be a new oncogenic RNA transcript in breast cancer.
UASR1 promotes growth and proliferation of breast cancer cells. To estimate the biological function of UASR1 in breast cancer cells, we used the lentivirus system to deliver UASR1 cDNA or shRNA for targeted expression or silencing in breast cancer cells MCF7 and MDA-MB-231. Empty vectors were used as controls, and RT-qPCR was used to verify the expression of UASR1 in these cell models (Fig. 2A). We tested three UASR1 shRNAs, and our data showed that shRNA-2 and shRNA-3 effectively knocked down UASR1 expression to 20~30% in MCF7 and MDA-MB-231 cells. The shRNA-1 did not work efficiently (data not shown).
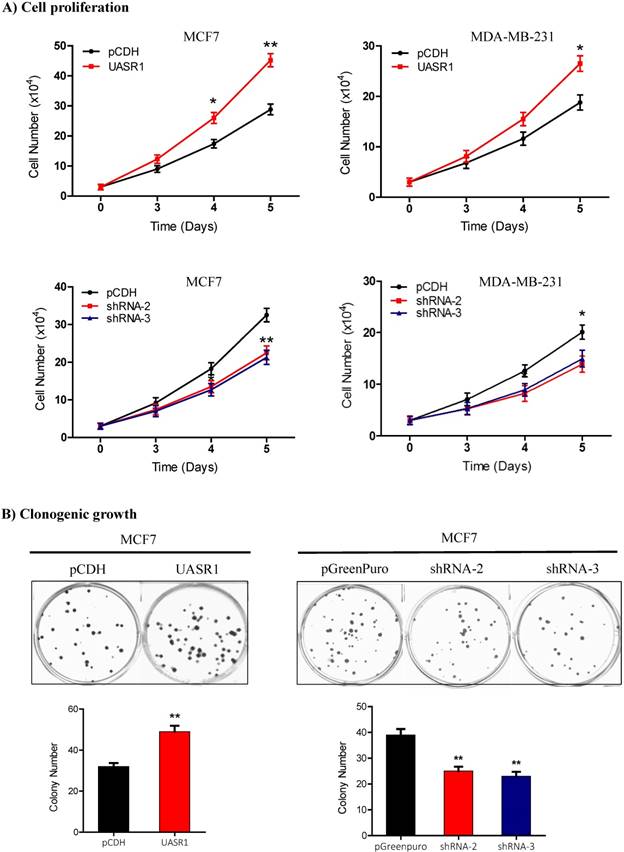
We then observed the effect of UASR1 expression on growth and proliferation of breast cancer cells. Results showed that UASR1 expression increased the growth and proliferation of MCF7 and MDA-MB-231 cells compared to the empty vector control (Fig. 2B, upper panel). In contrast, silencing of UASR1 inhibited the cell growth and proliferation in MCF7 and MDA-MB-231 cells (Fig. 2B, lower panel). The stimulation of UASR1 on proliferation of MCF7 and MDA-MB-231 cells was further confirmed by cell counting assays (Fig. 3A).
Clonogenic growth is a key feature of cancer cells. We further estimated the effect of UASR1 on clonogenicity of breast cancer cells. As shown in Fig. 3B, UASR1 expression promoted colony formation in MCF7 cells compared to the vector control. In contrast, UASR1 silencing inhibited the colonization of MCF7 cells. Together these data suggest that UASR1 promotes the cell growth and proliferation in breast cancer.
Proliferation and clonogenic growth of breast cancer cells promoted by UASR1. (A) Cell proliferation by viable cell counting. Cells (3x104/well) were spread into 12-well plates. At the indicated time points, cells were collected by trypsinization and subjected to trypan blue staining. Viable cells were counted using a Vi-cell counter. Upper panel: UASR1 expression; lower panel: UASR1 silencing. *P<0.05 and **P<0.01 compared to controls. (B) Colony formation assays. Cells (100 cells/well) were seeded in 6-well plates. After 2 weeks, visible colonies were fixed with 4% paraformaldehyde and stained with Giemsa for 5 min to photograph and count colony number. Data indicate mean ± SD from three independent experiments. *P<0.05 and **P<0.01 compared to controls.
Wound healing and migration of breast cancer cells enhanced by UASR1. UASR1 cDNA or shRNAs were delivered by lentiviral vectors into MCF7 and MDA-MB-231 cells, and would healing and migration assays were conducted as described in Materials and Methods. (A) Ectopic UASR1 expression. Upper panel: wound healing; lower panel: migration. Bar charts indicate the cell number migrated. Data indicate mean ± SD from three independent experiments. **P<0.01 compared to controls. (B) UASR1 silencing in MCF7 cells. Left panel: wound healing; right panel: migration. Bar charts indicate cell number migrated. Data indicate mean ± SD from three independent experiments. **P<0.01 compared to controls.
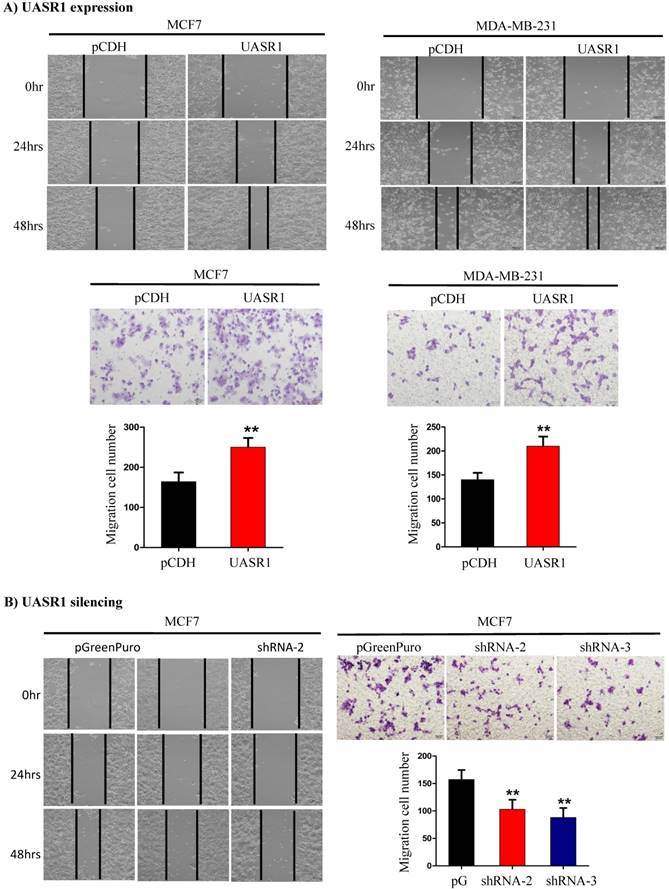
UASR1 promotes migration of breast cancer cells. We further tested the effect of UASR1 on migration of breast cancer cells by wound healing and transwell migration assays. As shown in Fig. 4A, targeted expression of UASR1 promoted wound healing and cell migration through micro-holes in transwells in MCF7 and MDA-MB-231 cells compared to the vector controls. In contrast, silencing of UASR1 inhibited the wound healing and migration of breast cancer cells (Fig. 4B and Supplementary Fig. S2A & B). These data suggest that UASR1 promotes wound healing and migration of breast cancer cells.
UASR1 promotes cell proliferation and migration through activation of the AKT/mTOR signaling pathway. After we determined the role of UASR1 in growth and proliferation of breast cancer, we moved forward to investigate the underlying mechanism of action. UASR1 is an antisense of UNC5B (intron 1), and thus we first estimated the effect of targeted expression of UASR1 on cellular UNC5B protein levels and neither ectopic expression nor silencing of UASR1 altered UNC5B protein levels (data not shown), indicating that UASR1 does not function via targeting UNC5B.
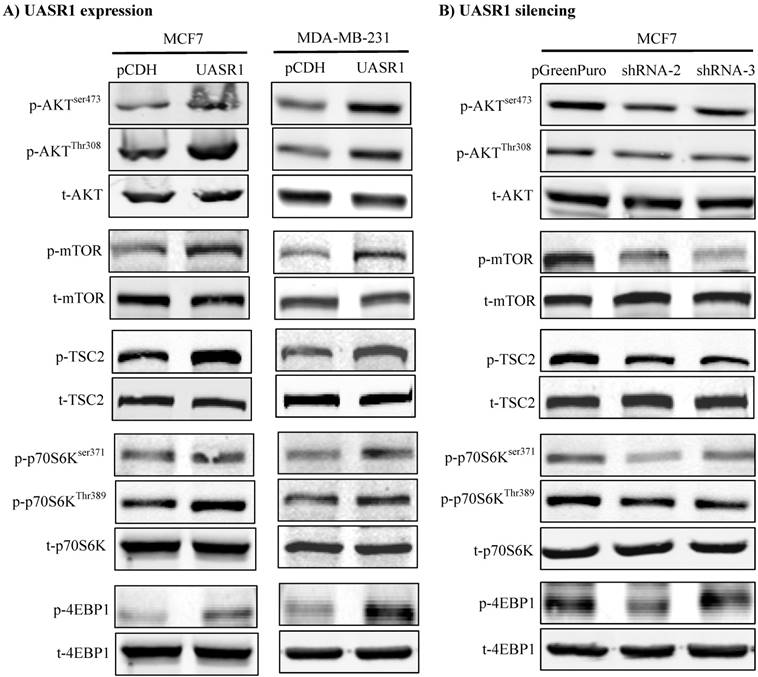
We thus did a broad exploration on the effect of UASR1 on major cell growth signaling pathways, such as AKT, c-Myc and ERK. As a result, UASR1 notably affected the cellular level of pAKT, but not c-Myc and pERK levels. As shown in Fig. 5A, targeted expression of UASR1 increased pAKT (Thr308 and Ser473) in MCF7 and MDA-MB-231 cells, indicating that UASR1 may regulate cell proliferation by oncogenic AKT signaling pathway. We then explored the downstream targets/effectors of AKT, and the results showed that pTSC2 and pmTOR expression was increased by UASR1 (Fig. 5A). The mTOR is an important serine-threonine protein kinase that regulates the proliferation, survival and migration of cells by activating downstream effectors 4EBP1 and p70S6K and protein translation/synthesis (33, 34). Our results showed that p-4EBP1 and p-p70S6K levels were increased by UASR1 (Fig. 5A). In contrast, UASR1 silencing led to a decrease in the pTSC2, pmTOR, p4EBP1 and pp70S6K in MCF7 cells (Fig. 5B) and MDA-MB-231 cells (Supplementary Fig. S2C). Furthermore, rapamycin (20 nM) abolished the induction of cell proliferation by UASR1 (Supplementary Fig. S3). Taken together, these results suggest that UASR1 promotes proliferation of MCF7 and MDA-MB-231 cells through control of the AKT/mTOR pathway.
Activation of the AKT/mTOR pathway by UASR1. Cell lysates were collected with lysis buffer containing phosphatase inhibitors as described in Materials and Methods and then subjected to Western blot analysis of effector proteins pAKT, p-mTOR, p-TSC2, p-p70S6K (Ser371 and Thr389) and p-4EBP1. (A) Activation of the AKT/mTOR pathway by UASR1 expression. (B) Inhibition of the AKT/mTOR pathway by UASR1 silencing.
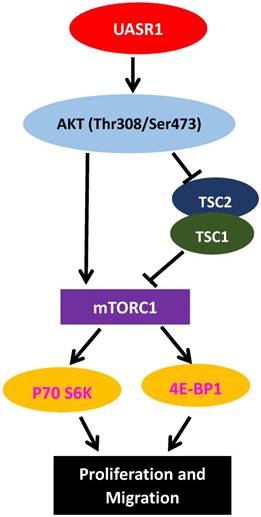
A hypothetic functional model of UASR1. UASR1 promotes cell proliferation and migration through phosphorylation activation of the AKT/mTOR pathway. Expression of UASR1 enhances phosphorylation of AKT. Phosphorylated active AKT in turn phosphorylates and activates mTOR. Activated AKT also phosphorylates and inhibits TSC1/TSC2 complex, further activating mTORC1. Activated mTORC1 then phosphorylates and activates p70S6k and 4E-BP1 to stimulate protein synthesis and cell proliferation.
Discussion
LncRNAs play an important role in tumor development and progression. Abnormal expression of lncRNAs is observed in many types of human cancers, including colorectal cancer, liver cancer, and breast cancer [30, 31]. This study characterized a new lncRNA, UASR1 located in the intron 1 of a tumor suppressor UNC5B gene in the antisense direction. UASR1 was upregulated in breast cancer and promoted growth and migration of breast cancer cells through activation of the AKT/mTOR signaling pathway.
To explore the possible role of UASR1 in breast cancer, we started this study with estimate of the UASR1 expression in tumor tissues. The data showed that UASR1 was upregulated and may play an oncogenic role in breast cancer. Unlimited growth and proliferation and enhanced migration of cancer cells are common features and triggered by oncogenes, leading to development and progression of cancer. Therefore, we investigated the effect of UASR1 expression or silencing on proliferation and migration of breast cancer cells. Our data showed that UASR1 promoted the growth and proliferation of breast cancer cells. UASR1 also enhanced wound healing and migration of breast cancer cells when it was targetedly expressed. In contrast, silencing of UASR1 inhibited the proliferation and migration of breast cancer cells. Furthermore, clonogenic growth of breast cancer cells was also enhanced by UASR1. These results suggest UASR1 as an oncogenic lncRNA in breast cancer.
DNA sequence analyses identified UASR1 as an antisense of the intron 1 of UNC5B gene. Targeted expression or silencing of UASR1 did not affect the expression of UNC5B. Thus UASR1 may not function through UNC5B, and we started to explore the UNC5B-independent mechanisms of action of UASR1 in breast cancer cells. LncRNAs function as a tumorigenic regulator through modulating cellular signaling networks [32]. AKT, also called protein kinase B, is a serine-threonine kinase, regulating a major oncogenic signaling in tumorigenesis [33, 34]. Activated PI3K produces second messenger PIP3 on cell membrane, which serves as a docking site through the PH domain of AKT and translocates AKT in cytoplasm to the cell membrane and in turn, 3-phosphoinositide-dependent protein kinase-1 (PDK1) catalyzes Thr308 phosphorylation of AKT. Furthermore, PDK2 phosphorylates Ser473 of AKT to make full activation of AKT. Activated AKT enters the cytoplasm or nucleus and activates or inhibits downstream effectors, such as mTOR, GSK-3β and FOXO1. Our results showed that UASR1 activated AKT with increased phosphorylation at Thr308 and Ser473, but it is currently unknown how UASR1 activates the phosphorylation of AKT. UASR1 may function as an adaptor to couple AKT with kinases for phosphorylation; UASR1 may also bind to AKT and blocks phosphatases for dephosphorylation. Detailed mechanistic study is warranted.
AKT is a key activator of mTOR signaling. Mammalian target of rapamycin (mTOR) protein is a kinase. AKT directly phosphorylates Ser2448 of mTOR, activating mTOR signaling cascade[35]; Activated AKT also catalyzes Thr1462 phosphorylation of TSC2 and inhibits the formation of TSC1/TSC2 complexes [36]. The TSC1/TSC2 complex is a main negative regulator of mTOR activity, and inhibition of the TSC1/TSC2 complex by Thr1462 phosphorylation would further activate mTOR [36]. Therefore, AKT activates mTOR signaling through both direct phosphorylation activation of mTOR protein and inhibition of the TSC1/TSC2 complex. The activated mTOR then phosphorylates and activates 4E-BP1, and leads to eIF-4E dissociation and activation, enhancing the formation of translation initiation complexes and protein synthesis [37]. In addition, mTOR phosphorylates p70S6K, leading to ribosomal 40S subunit binding to translation complexes and increase of translation efficiency [38]. Activation of the AKT/mTOR signaling pathway is closely related to development of tumors, stimulating cell proliferation and migration [39]. In this study, we found that AKT activated by UASR1 triggered phosphorylation of mTOR that in turn activated phosphorylation of 4EBP1 and P70S6K, stimulating proliferation of breast cancer cells (Fig. 6).
In conclusion, this study characterized a new lncRNA, named UASR1. UASR1 was upregulated in breast cancer tissues and promoted breast cancer cell growth, proliferation, wound healing and migration through activation of the AKT/mTOR signaling pathway. UASR1 plays an oncogenic role in breast cancer and be a potential target for breast cancer management.
Supplementary Material
Supplementary figures and tables.
Abbreviations
ANRIL, antisense non-coding RNA in the INK4 locus; BCAR4, breast cancer anti-estrogen resistance 4; DAPK, death associated protein kinase; HOTTIP, HOXA distal transcript antisense RNA; lincRNA-p21, tumor protein P53 pathway corepressor 1; lncRNA, long non-coding RNA; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PICART1, p53-inducible cancer-associated RNA transcript 1; UASR1, UNC5B antisense RNA1; UNC5B, unc-5 netrin receptor B; XIST, inactive X specific transcripts.
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (81772842 and 81802947).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59
2. Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr.. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646-57
3. Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787-9
4. Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev. 2007;21:11-42
5. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145-66
6. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular cancer. 2011;10:38
7. Mitobe Y, Takayama KI, Horie-Inoue K, Inoue S. Prostate cancer-associated lncRNAs. Cancer letters. 2018;418:159-66
8. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer letters. 2018;419:152-66
9. Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC. et al. BRCA1 is a negative modulator of the PRC2 complex. The EMBO journal. 2013;32:1584-97
10. Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M. et al. Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer letters. 2018;434:1-10
11. Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL. et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer research. 2006;66:5330-7
12. Jia P, Cai H, Liu X, Chen J, Ma J, Wang P. et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer letters. 2016;381:359-69
13. Ohtsuka M, Ling H, Ivan C, Pichler M, Matsushita D, Goblirsch M. et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-beta-Catenin Signaling in Colorectal Cancer. EBioMedicine. 2016;13:113-24
14. Chang YT, Lin TP, Tang JT, Campbell M, Luo YL, Lu SY. et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer letters. 2018;433:43-52
15. Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V. et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864-76
16. Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: there's a long way to go. J Mammary Gland Biol Neoplasia. 2012;17:43-58
17. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
18. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7-30
19. Soudyab M, Iranpour M, Ghafouri-Fard S. The Role of Long Non-Coding RNAs in Breast Cancer. Arch Iran Med. 2016;19:508-17
20. Zhang S, Wang J, Ghoshal T, Wilkins D, Mo YY, Chen Y. et al. lncRNA Gene Signatures for Prediction of Breast Cancer Intrinsic Subtypes and Prognosis. Genes (Basel). 2018:9
21. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Molecular cancer. 2017;16:129
22. Gooding AJ, Zhang B, Jahanbani FK, Gilmore HL, Chang JC, Valadkhan S. et al. The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Scientific reports. 2017;7:12698
23. Shen Y, Ma J, Yan R, Ling H, Li X, Yang W. et al. Impaired self-renewal and increased colitis and dysplastic lesions in colonic mucosa of AKR1B8-deficient mice. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:1466-76
24. Bu Y, Li X, He Y, Huang C, Shen Y, Cao Y. et al. A phosphomimetic mutant of RelA/p65 at Ser536 induces apoptosis and senescence: An implication for tumor-suppressive role of Ser536 phosphorylation. International journal of cancer Journal international du cancer. 2016;138:1186-98
25. Yan R, Zu X, Ma J, Liu Z, Adeyanju M, Cao D. Aldo-keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: Implication for cancer intervention. International journal of cancer Journal international du cancer. 2007;121:2301-6
26. Huang C, Cao Z, Ma J, Shen Y, Bu Y, Khoshaba R. et al. AKR1B10 activates diacylglycerol (DAG) second messenger in breast cancer cells. Molecular carcinogenesis. 2018
27. Huang C, Verhulst S, Shen Y, Bu Y, Cao Y, He Y. et al. AKR1B10 promotes breast cancer metastasis through integrin alpha5/delta-catenin mediated FAK/Src/Rac1 signaling pathway. Oncotarget. 2016;7:43779-91
28. Ma J, Luo DX, Huang C, Shen Y, Bu Y, Markwell S. et al. AKR1B10 overexpression in breast cancer: association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. International journal of cancer Journal international du cancer. 2012;131:E862-71
29. Bhat SA, Gurtoo S, Deolankar SC, Fazili KM, Advani J, Shetty R. et al. A network map of netrin receptor UNC5B-mediated signaling. J Cell Commun Signal. 2018
30. Yang Y, Zhao L, Lei L, Lau WB, Lau B, Yang Q. et al. LncRNAs: the bridge linking RNA and colorectal cancer. Oncotarget. 2017;8:12517-32
31. Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature genetics. 2016;48:500-9
32. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391-407
33. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857-68
34. Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL. et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590-603
35. Sekulic A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM. et al. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer research. 2000;60:3504-13
36. Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. The Biochemical journal. 2008;412:179-90
37. Heesom KJ, Denton RM. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS letters. 1999;457:489-93
38. Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998;273:14484-94
39. Tan X, Zhang Z, Yao H, Shen L. Tim-4 promotes the growth of colorectal cancer by activating angiogenesis and recruiting tumor-associated macrophages via the PI3K/AKT/mTOR signaling pathway. Cancer letters. 2018;436:119-28
Author contact
![]() Corresponding authors: Dr. Deliang Cao, dcaoedu, Dr. Junfei Jin, changliangzijincom or Dr. Xuewen Zhang, xuewen_zhangcom.
Corresponding authors: Dr. Deliang Cao, dcaoedu, Dr. Junfei Jin, changliangzijincom or Dr. Xuewen Zhang, xuewen_zhangcom.

 Global reach, higher impact
Global reach, higher impact