Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(10):2332-2341. doi:10.7150/jca.30789 This issue Cite
Research Paper
Clinicopathological Significance of BRAFV600E Mutation in Colorectal Cancer: An Updated Meta-Analysis
1. Division of Gastrointestinal Surgery, Department of General Surgery, First Affiliated Hospital, Nanjing Medical University, Nanjing 210029, China;
2. Department of General Surgery, Affiliated Hospital of Integrated Chinese and Western Medicine of Nanjing University of Chinese Medicine, Nanjing 210028, China;
3. Lab of cellular and molecular biology, Affiliated Hospital of Integrated Chinese and Western Medicine of Nanjing University of Chinese Medicine, Nanjing 210028, China;
4. Department of Surgical Oncology, Jiangsu Province Hospital of Chinese Medicine, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, China.
*These authors contributed equally to this work.
Received 2018-10-18; Accepted 2019-4-20; Published 2019-5-26
Abstract
Background and Aims: Numerous studies have identified BRAFV600E mutation as a predictive factor of anti-EGFR antibodies in colorectal cancer (CRC). However, the association between BRAFV600E mutation and clinicopathological features remains unclear. Therefore, we aimed to conduct an updated and comprehensive meta-analysis to evaluate the above issues.
Methods: We performed a systematic literature search from PubMed, Web of Science, Embase, and PMC database examining the association between BRAFV600E mutation and clinicopathological features in CRC patients. Odds ratio with 95% confidence interval were used to estimate the effects of BRAFV600E mutation on each clinicopathological parameter with fixed-effect model or random-effect model.
Results: Sixty-one studies published, including 32407 CRC patients from multiple countries, were included in the meta-analysis. The overall BRAFV600E mutation rate was 11.38%, and BRAFV600E mutation was positively related to high disease stage (OR=0.81; 95% CI=0.72-0.92; P=0.001), high T stage (OR=0.51; 95% CI=0.40-0.65; P<0.00001), proximal colon (OR=4.76; 95% CI=3.81-5.96; P<0.00001) or right colon (OR=5.15; 95% CI=4.35-6.10, P<0.00001) tumor location, poor tumor differentiation (OR=0.27; 95% CI=0.21-0.34; P<0.00001), mucinous histology (OR=2.97; 95% CI=2.37-3.72; P<0.00001), K-ras-wild type (OR=0.04; 95% CI=0.02-0.07; P<0.00001), TP53-wild type (OR=0.50; 95% CI=0.31-0.78; P=0.003), deficient DNA mismatch repair (OR=2.93; 95% CI=1.78-4.82; P<0.00001), high microsatellite instability (OR=11.15; 95% CI=8.51-14.61; P<0.00001) and high CpG island methylator phenotype (OR=0.04; 95% CI=0.03-0.08; P<0.00001).
Conclusions: Our updated meta-analysis demonstrated that BRAFV600E mutation was related to poor prognosis of CRC and associated with the distinct molecular phenotypes.
Keywords: colorectal cancer, BRAF mutation, prognosis, meta-analysis
Introduction
Colorectal cancer (CRC), the third most common cancer, causes the fourth most frequent cancer-related deaths worldwide [1]. It has been widely recognized that constitutive activation of the RAS-RAF-MEK- ERK (MAPK) pathway plays a critical roles in CRC development and progression [2]. Gain-of-function mutations of the key protein BRAF in this pathway will constitutively activate this pathway, suggesting the crucial role of BRAF mutation in CRC [3]. The BRAFV600E mutation, inducing the substitution of valine for glutamate at position 600 of the b-raf protein, accounts for approximately 90% of BRAF mutations and has more important significance compared to other BRAF mutation types in CRC, and about 10% of CRC patients harbor the BRAFV600E mutation [3]. Increasing studies have discussed the relationship between BRAFV600E mutation and the effect of anti-EGFR inhibitors in CRC, but the effects of BRAFV600E mutation on the clinicopathological characteristics of CRC remains limited. Therefore, in this article we comprehensively estimate the association between BRAFV600E mutation and clinicopathological characteristics of CRC patients.
Methods
Literature search strategy
We searched PubMed, Web of Science, Embase, and PMC database for relevant publications with the following search terms: (“colorectal cancer” or “rectal cancer” or “colon cancer”) and (“BRAF mutation” or BRAFV600E). Original articles about human studies written in English published before June 18, 2018 were included.
A flow chart of the study selection process.
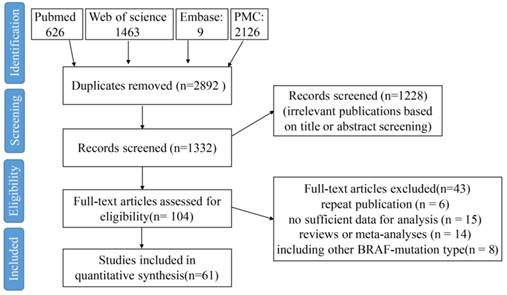
Inclusion criteria
The studies were gone through in accordance to the predetermined selection. The inclusion criteria were: (1) the association between BRAFV600E mutation and clinicopathological characteristics was studied; (2) sufficient published data for calculating an odds ratio (OR) and 95% confidence interval (CI) were reported; (3) the most appropriate article was selected when multiple articles associated with the same patient population were published. The exclusion criteria were: (1) review articles; (2) articles without enough data to analyzed; and (3) single case reports. The quality of each study was assessed using the Newcastle-Ottawa Scale (NOS).
Data extraction
For every appropriate study, the relevant data were extracted, including name of the first author, publication year, country where the study was conducted, follow-up time, number of patients with BRAFV600E mutation, total number of patients, patient demographics (age and gender); clinicopathological characteristics including tumor site, disease stage, T stage, N stage, metastasis status, tumor size, tumor differentiation and mucinous histology; molecular characteristics including KRAS mutation status, CpG island methylator phenotype (CIMP), TP53 mutation status, DNA mismatch repair (MMR) status and microsatellite instability (MSI) status).
Statistical analysis
Meta-analysis was performed using RevMan (Cochrane Collaboration, Oxford, UK). The strength of the association between the BRAFV600E mutation and clinicopathological parameters was assessed by odds ratio (OR) with the corresponding 95% confidence interval (CI). In the course of data pooling, statistical heterogeneity was defined by using chi-square-based Q-test. The I2 value indicates the degree of heterogeneity. A P-value<0.10 and/or I2>50% are considered significant heterogeneity, and then a random-effect model is used. Otherwise, a fixed-effect model is used.
Results
Characteristics of eligible literatures
According to the search terms, a total of 1332 eligible citations were obtained. After screening the abstract, 1228 citations were excluded. Among the remaining 104 citations, 43 citations were excluded because of the reasons shown in Figure 1. Finally, 61 studies published from 2006 to 2018 were included in the meta-analysis (Figure 1). A total of 32407 CRC patients from China, Japan, South Korea, India, French, Sweden, Greece, American, Netherlands, Italy, Germany, Australia, and so on were included, and among these patients, 3688 patients were with BRAFV600E mutation (11.38%). The study sample sizes ranged from 69 to 1980 cases. BRAFV600E mutation rate among all studies ranged from 3.14% to 23.14%, which was consistent with the results in the previous study [4]. All specimens were derived from CRC tissues by either biopsy or surgical resection, and were detected for BRAF mutation status mainly by direct sequencing, pyrosequencing, allele-specific PCR and immunohistochemistry (IHC) method.
The basic characters of the 61 eligible studies were summarized in Supplementary Table 1. Thirty-five studies are with sample size below 500 [5-37, 64], whereas twenty-six studies are with sample size over 500 [38-63]. The earliest study was published in July 2005 [51], and the latest study was published in August 2017 [49]. Most of these studies involved patients with stage I-IV CRC [5, 6, 8, 10, 12, 14, 15, 19, 21, 22, 24, 26, 27, 31, 32, 34, 38, 39, 42, 44-47, 51, 52, 54, 55, 57-60, 64], and six studies only involved patients with stage IV CRC [23, 29, 33, 35, 43, 56]. All the studies have a NOS score of ≥5, and 18 studies have a NOS score of ≥6 (Supplementary Table 1).
Correlation of BRAFV600E mutation with clinicopathological characteristics of CRC patients
Demographic characteristics (Age and Gender)
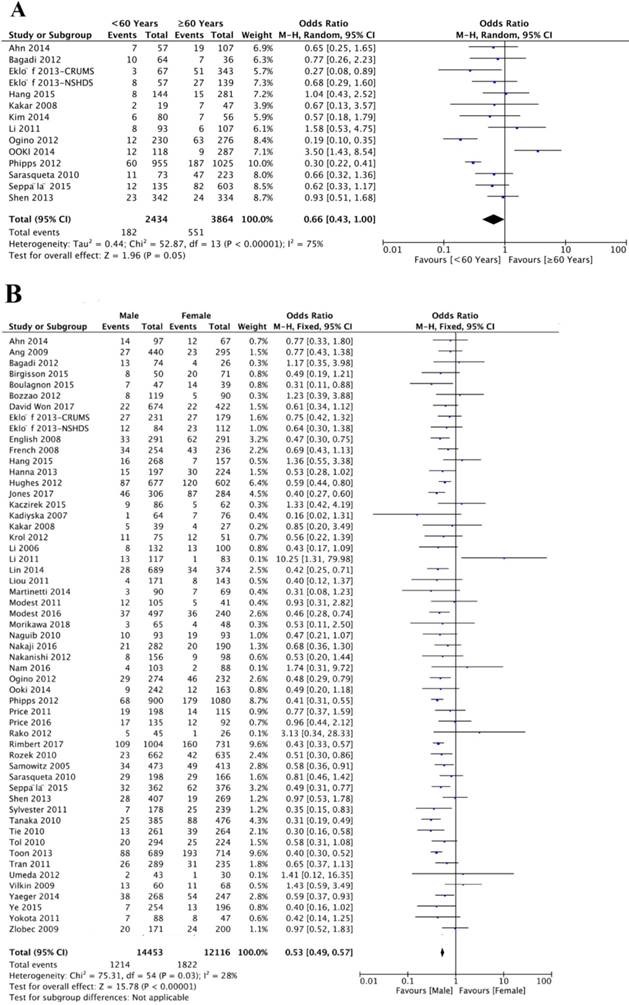
A total of 14 studies investigated the association between BRAFV600E mutation and age. Of 2434 patients younger than 60 years, 182 (7.47%) patients were BRAFV600E mutation positive, and 551 (14.26%) of 3864 patients 60 year or older were BRAFV600E mutation positive. The association between BRAFV600E mutation and age did not reach statistical significance (OR=0.66; 95% CI=0.43-1.00; P=0.05) (Figure 2A, Table 1). Fifty-six studies analyzed the association between BRAFV600E mutation and gender. Of 14453 male patients, 1214 (8.40%) CRC patients were with BRAFV600E mutation, and 1822 (15.04%) of 12048 female patients were with BRAFV600E mutation. There was a significantly negative association between BRAFV600E mutation and male gender (OR=0.53; 95% CI=0.49-0.57; P<0.00001) (Figure 2B, Table 1).
Overall analysis of the association between BRAFV600E mutation and clinicopathological features in CRC patients.
| Clinicopathological features | OR | 95% CI | P value |
|---|---|---|---|
| Demographic characteristics | |||
| age (<60 years) | 0.66 | 0.43-1.00 | 0.05 |
| gender (male) | 0.53 | 0.49-0.57 | <0.00001 |
| Clinical factures | |||
| disease stage (stage I-II) | 0.81 | 0.72-0.92 | 0.001 |
| tumor size (<5cm) | 0.83 | 0.45-1.55 | 0.56 |
| T stage (T1-2) | 0.51 | 0.40-0.65 | <0.00001 |
| N stage (N0) | 0.85 | 0.73-1.00 | 0.05 |
| metastasis (yes) | 1.30 | 0.90-1.88 | 0.16 |
| tumor location (proximal colon) | 4.76 | 3.81-5.96 | <0.00001 |
| tumor location (right colon) | 5.15 | 4.35-6.10 | <0.00001 |
| tumor differentiation (well/moderate) | 0.27 | 0.21-0.34 | <0.00001 |
| mucinous histology (mucinous) | 2.97 | 2.37-3.72 | <0.00001 |
| Molecular features | |||
| K-ras mutation status (mutation) | 0.04 | 0.02-0.07 | <0.00001 |
| TP53 mutation status (mutation) | 0.50 | 0.31-0.78 | 0.003 |
| MMR status (dMMR) | 2.93 | 1.78-4.82 | <0.00001 |
| MSI status (MSI high) | 11.15 | 8.51-14.61 | <0.00001 |
| CIMP phenotype (CIMP low/negative) | 0.04 | 0.03-0.08 | <0.00001 |
Clinical Factures (Disease stage, T stage, N stage, tumor size, metastasis status, tumor site, tumor differentiation, and mucinous histology)
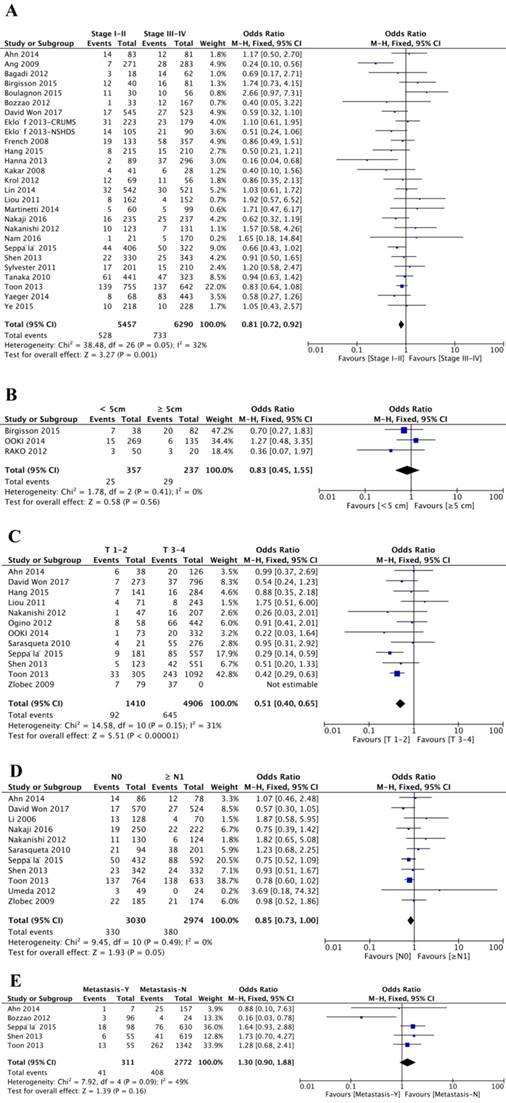
Twenty-six studies analyzed the association between disease stage and BRAFV600E mutation. Of 5457 patients with stage I or II, 528 (9.68%) patients were BRAFV600E mutation positive, and 733 (11.65%) patients was BRAFV600E mutation positive from 6290 patients diagnosed with stage III or IV disease. Stage I or II were negatively related to BRAFV600E mutation (OR=0.81; 95% CI=0.72-0.92; P=0.001), indicating that CRC patients with BRAFV600E mutation trend to have more advanced disease stage (Figure 3A, Table 1). Furthermore, patients with BRAFV600E mutation were also negatively associated with low T stage (OR=0.51; 95% CI=0.40-0.65; P<0.00001) (Figure 3C, Table 1). However, the overall analysis showed BRAFV600E mutation did not statistically significant correlated with tumor size (OR=0.83; 95% CI=0.45-1.55; P=0.56) (Figure 3B, Table 1), N stage (OR=0.85; 95% CI=0.73-1.00; P=0.05) (Figure 3D, Table 1) and metastasis status (OR=1.30; 95% CI=0.90-1.88; P=0.16) (Figure 3E, Table 1).
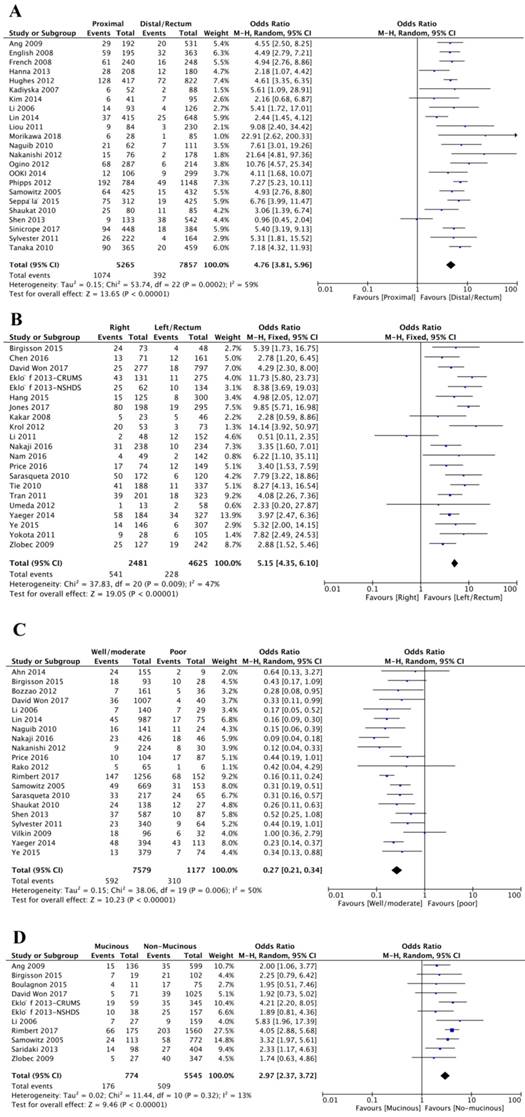
In total, forty-five studies investigated the relationship between BRAFV600E mutation and tumor site. And among these studies, twenty-four studies categorized tumors as proximal colon, distal colon or rectal tumor, and another twenty-one studies classified the tumor as right colon, left colon or rectal tumor. The final results showed that BRAFV600E mutation was significantly associated with proximal colon tumor location (OR=4.76; 95% CI=3.81-5.96; P<0.00001) or right colon tumor location (OR=5.15; 95% CI=4.35-6.10; P<0.00001) (Figure 4A-B, Table 1).
Twenty studies assessed the association between BRAFV600E mutation and tumor differentiation. 592 (7.81%) patients were with BRAFV600E mutation of 7579 patients with well or moderate differentiation, and 310 (26.34%) patients were with BRAFV600E mutation of 1177 patients with poor differentiation. It was obvious that BRAFV600E mutation was negatively associated with well or moderate differentiation, indicating that CRC patients with BRAFV600E mutation trend to have aggressive tumor phenotype (OR=0.27; 95% CI=0.21-0.34; P<0.00001) (Figure 4C, Table 1). Besides, BRAFV600E mutation was also strikingly related to mucinous histology (OR=2.97; 95% CI=2.37-3.72; P<0.00001) (Figure 4D, Table 1).
Molecular Features (K-ras mutation status, TP53 mutation status, MMR capacity, MSI status, and CIMP)
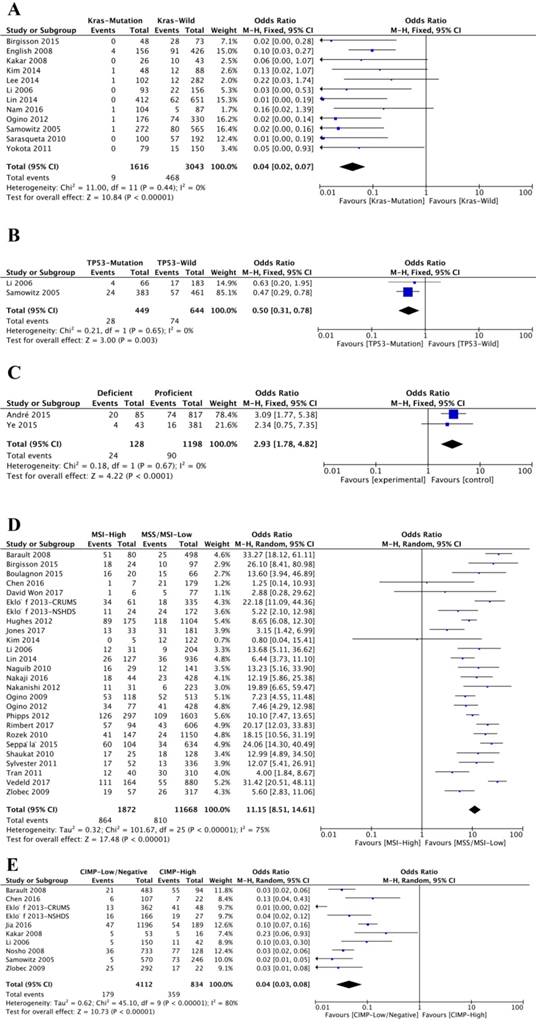
Twelve studies analyzed the association between BRAFV600E mutation and K-ras mutation status. Notably, K-ras mutation and BRAFV600E mutation were negatively related (OR=0.04; 95% CI=0.02-0.07; P<0.00001) (Figure 5A, Table 1). Of 1616 K-ras-mutated patients, only nine (0.56%) ones were BRAFV600E mutated, while 468 (15.38%) patients of 3043 K-ras-wild patients were BRAFV600E mutated. Interestingly, BRAFV600E mutation was also negatively associated with TP53 mutation (OR=0.50; 95% CI=0.31-0.78; P=0.003) (Figure 5B and Table 1).
The association of BRAFV600E mutation with demographics, including age (A) and gender (B).
Meta-analysis of association between BRAFV600E mutation and clinical features, including disease stage (A), tumor size (B), T stage (C), N stage (D) and metastasis status (E).
The association of BRAFV600E mutation with tumor characteristics, including tumor site (A and B), tumor differentiation (C) and mucinous histology (B).
Only two studies investigated the relationship between BRAFV600E mutation and mismatch repair (MMR) capacity. The results showed that 24 (18.75%) patients were BRAFV600E mutation positive from 128 patients with deficient MMR (dMMR) capacity, and 90 (7.51%) patients were BRAFV600E mutation positive from 1198 patients with proficient MMR (pMMR) capacity. BRAFV600E mutation was significantly related to dMMR (OR=2.93; 95% CI=1.78-4.82; P<0.00001) (Figure 5C, Table 1). Twenty-seven studies investigated the BRAFV600E mutation and microsatellite instability (MSI). Of 1872 patients with high microsatellite instability (MSI-High), 864 (46.15%) patients were BRAFV600E mutated, and of 11668 patients with low microsatellite instability (MSI-low) or microsatellite stable (MSS), 810 (6.94%) patients were BRAFV600E mutated. There was a significant association between BRAFV600E mutation and MSI-high (OR=11.15; 95% CI=8.51-14.61; P<0.00001) (Figure 5D, Table 1). Ten studies were analyzed for CpG island methylator phenotype (CIMP) and BRAFV600E mutation. Of 4112 patients with low or negative CIMP, 179 (4.35%) patients were with BRAFV600E mutation, and of 834 patients with high CIMP, 359 (43.05%) patients were with BRAFV600E mutation. According to the result, BRAFV600E mutation was negatively associated with high CMIP (OR=0.04; 95% CI=0.03-0.08; P<0.00001) (Figure 5E and Table 1).
Additional analyses
A funnel plot of effects calculated from individual studies examining the association between BRAFV600E mutation and disease stage was conducted to estimate the presence of publication bias. Because there are small studies with negative results in the literature, no strong indication of publication bias exist among the series of studies included in this meta-analysis.
Discussion
BRAFV600E mutation was an important molecular alternation in CRC patients. In our study, the highest BRAFV600E mutation rate reached to 23.14% and the average BRAFV600E mutation rate was 11.35% among all the involved studies, similar to other reports [4]. Clinicopathological parameters have crucial roles in predicting the prognosis of cancer patients. It is necessary to clarify the relationship between BRAFV600E mutation and clinicopathological parameters in CRC patients [65]. Our meta-analysis indicated that BRAFV600E mutation was significantly associated with female, advanced disease stage, high T stage, proximal or right tumor location, poor tissue differentiation and mucinous phenotype. As high disease stage, high T stage, poor tissue differentiation and mucinous histology were the multiple risk factors of the prognosis in CRC patients, it may be deemed that BRAFV600E mutation was a poor predictive indicator [20, 51, 65]. Our study also showed that BRAFV600E mutation had a crucial association with disease stage, T stage, N stage and tissue differentiation, which demonstrated the important role of BRAFV600E mutation in occurrence and development of CRC.
Intriguingly, tumors located in the proximal colon were 4.76-fold more likely to have BRAFV600E mutation than tumor located in the distal or rectal colon. Moreover, the BRAFV600E mutation was 5.15-fold more frequent in tumors located in the right colon than tumors located in the left or rectal colon. The association between BRAFV600E mutation and tumor sites was very strong and the reason of the association has not been clarified clearly. Previous studies have indicated that colorectal tumors located in different sites have totally different outcomes and specific biomolecular characteristics [66]. Our study also demonstrated that the difference in regard to BRAFV600E mutation in different tumor location. Moreover, the different BRAFV600E mutation rates among different tumor sites might be useful for formulating treatment therapy for CRC located in different tumor sites [67, 68].
Nowadays, in addition to clinicopathologic stage and histological morphology, molecular markers play increasing roles in making therapeutical decision for cancer patients. Melanoma with BRAFV600E mutation is more sensitive to immunotherapy [69]. Deficient MMR status has been demonstrated to predict the response of PD-1 blockade in metastatic CRC [70], and MSI-High also has been recognized as a predictive factor of programmed death ligand-1 inhibitor pembrolizumab in metastatic/refractory CRC [71]. Our results revealed that BRAFV600E mutation was significantly related to K-ras-wild type, TP53-wild type, deficient MMR, high MSI and high CIMP. This association between BRAFV600E mutation and other molecular features cloud be important to understand the molecular distinction between CRC patients with or without BRAFV600E mutation. However, the association between BRAFV600E mutation and the therapeutical response in CRC needs more prospective investigations.
In this article, although we conducted comprehensive and detailed meta-analysis, there are still some limitations. Firstly, with regard to some clinicopathologic characteristics, the number of the involved patients was limited. Small studies are prone to introduce unstable results and related to publication bias. Secondly, most of these studies were retrospective or observational studies (data not shown), which might induce heterogeneity. Thirdly, the mutation detection assays were different among these studies. The most two commonly used methods are direct sequencing and pyrosequencing. Different BRAFV600E mutation assays also affected the accuracy and precision of the pooled estimates.
The association of BRAFV600E mutation with molecular features, including Kras mutation status (A), TP53 mutation status (B), MMR capacity (C), MSI status (D) and CIMP phenotype (E).
In conclusion, our updated and comprehensive meta-analysis based on a large number of clinical data demonstrated that BRAFV600E mutation is a biological predictor for poor prognosis in CRC patients, which helps to elucidate the mechanisms of progression and metastasis of CRC and to develop novel therapeutic strategies for CRC.
Supplementary Material
Supplementary tables.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant No. 81673945 and 81503574 to Cao M); the National Natural Science Foundation of China (Grant No. 81272711 and 81871959 to Shen L), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801 to Shen L), the Key Medical Talents Program of Jiangsu Province (ZDRCA2016014 to Shen L), and the Key R & D Program of Jiangsu Province (Social Development, BE2018758 to Shen L).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fedewa SA. et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-93
2. Safaee AG, Jafarnejad SM, Tan L. et al. The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. PLoS One. 2012;7:e47054
3. Cohen R, Cervera P, Svrcek M. et al. BRAF-Mutated Colorectal Cancer: What Is the Optimal Strategy for Treatment? Curr Treat Options Oncol. 2017;18:9
4. Chen D, Huang JF, Liu K. et al. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e90607
5. Ahn TS, Jeong D, Son MW. et al. The BRAF mutation is associated with the prognosis in colorectal cancer. J Cancer Res Clin Oncol. 2014;140:1863-71
6. Bagadi SB, Sanghvi M, Nair SB. et al. Combined mutational analysis of KRAS, NRAS and BRAF genes in Indian patients with colorectal carcinoma. Int J Biol Markers. 2012;27:27-33
7. Birgisson H, Edlund K, Wallin U. et al. Microsatellite instability and mutations in BRAF and KRAS are significant predictors of disseminated disease in colon cancer. BMC Cancer. 2015;15:125
8. Bozzao C, Varvara D, Piglionica M. et al. Survey of KRAS, BRAF and PIK3CA mutational status in 209 consecutive Italian colorectal cancer patients. Int J Biol Markers. 2012;27:e366-74
9. Chen KH, Lin YL, Liau JY. et al. BRAF mutation may have different prognostic implications in early- and late-stage colorectal cancer. Med Oncol. 2016;33:39
10. Eklof V, Wikberg ML, Edin S. et al. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108:2153-63
11. French AJ, Sargent DJ, Burgart LJ. et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408-15
12. Hang JF, Li AF, Chang SC. et al. Immunohistochemical detection of the BRAF V600E mutant protein in colorectal cancers in Taiwan is highly concordant with the molecular test. Histopathology. 2016;69:54-62
13. Hanna MC, Go C, Roden C. et al. Colorectal cancers from distinct ancestral populations show variations in BRAF mutation frequency. PLoS One. 2013;8:e74950
14. Kadiyska TK, Konstantinova DV, Atanasov VR. et al. Frequency and application of the hot spot BRAF gene mutation (p.V600E) in the diagnostic strategy for Hereditary Nonpolyposis Colorectal Cancer. Cancer Detect Prev. 2007;31:254-6
15. Kakar S, Deng G, Sahai V. et al. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Arch Pathol Lab Med. 2008;132:958-64
16. Kim B, Park SJ, Cheon JH. et al. Clinical meaning of BRAF mutation in Korean patients with advanced colorectal cancer. World J Gastroenterol. 2014;20:4370-76
17. Krol LC, 't Hart NA, Methorst N. et al. Concordance in KRAS and BRAF mutations in endoscopic biopsy samples and resection specimens of colorectal adenocarcinoma. Eur J Cancer. 2012;48:1108-15
18. Lee DW, Kim KJ, Han SW. et al. KRAS mutation is associated with worse prognosis in stage III or high-risk stage II colon cancer patients treated with adjuvant FOLFOX. Ann Surg Oncol. 2015;22:187-94
19. Li HT, Lu YY, An YX. et al. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25:1691-7
20. Li WQ, Kawakami K, Ruszkiewicz A. et al. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2
21. Liou JM, Wu MS, Shun CT. et al. Mutations in BRAF correlate with poor survival of colorectal cancers in Chinese population. Int J Colorectal Dis. 2011;26:1387-95
22. Martinetti D, Costanzo R, Kadare S. et al. KRAS and BRAF mutational status in colon cancer from Albanian patients. Diagn Pathol. 2014;9:187
23. Morikawa T, Inada R, Nagasaka T. et al. BRAF V600E mutation is a predictive indicator of upfront chemotherapy for stage IV colorectal cancer. Oncol Lett. 2018;15:2195-201
24. Naguib A, Mitrou PN, Gay LJ. et al. Dietary, lifestyle and clinicopathological factors associated with BRAF and K-ras mutations arising in distinct subsets of colorectal cancers in the EPIC Norfolk study. BMC Cancer. 2010;10:99
25. Nakaji Y, Oki E, Nakanishi R. et al. Prognostic value of BRAF V600E mutation and microsatellite instability in Japanese patients with sporadic colorectal cancer. J Cancer Res Clin Oncol. 2017;143:151-60
26. Nakanishi R, Harada J, Tuul M. et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18:1042-8
27. Nam SK, Yun S, Koh J. et al. BRAF, PIK3CA, and HER2 oncogenic alterations according to KRAS mutation status in advanced colorectal cancers with distant metastasis. PLoS One. 2016;11:e151865
28. Ooki A, Akagi K, Yatsuoka T. et al. Combined microsatellite instability and BRAF gene status as biomarkers for adjuvant chemotherapy in stage III colorectal cancer. J Surg Oncol. 2014;110:982-8
29. Price TJ, Beeke C, Townsend AR. et al. BRAF mutation testing and metastatic colorectal cancer in the community setting: is there an urgent need for more education? Mol Diagn Ther. 2016;20:75-82
30. Rako I, Jakic-Razumovic J, Katalinic D. et al. Mutation pattern of KRAS and BRAF oncogenes in colorectal cancer patients. Neoplasma. 2012;59:376-83
31. Shaukat A, Arain M, Thaygarajan B. et al. Is BRAF mutation associated with interval colorectal cancers? Dig Dis Sci. 2010;55:2352-6
32. Sylvester BE, Huo D, Khramtsov A. et al. Molecular analysis of colorectal tumors within a diverse patient cohort at a single institution. Clin Cancer Res. 2012;18:350-9
33. Umeda Y, Nagasaka T, Mori Y. et al. Poor prognosis of KRAS or BRAF mutant colorectal liver metastasis without microsatellite instability. J Hepatobiliary Pancreat Sci. 2013;20:223-33
34. Ye JX, Liu Y, Qin Y. et al. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol. 2015;21:1595-605
35. Yokota T, Ura T, Shibata N. et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856-62
36. Zlobec I, Bihl MP, Schwarb H. et al. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367-80
37. Farina-Sarasqueta A, van Lijnschoten G, Moerland E. et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396-402
38. Ang PW, Li WQ, Soong R. et al. BRAF mutation is associated with the CpG island methylator phenotype in colorectal cancer from young patients. Cancer Lett. 2009;273:221-4
39. Barault L, Charon-Barra C, Jooste V. et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541-6
40. English DR, Young JP, Simpson JA. et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev. 2008;17:1774-80
41. Hughes LA, Williamson EJ, van Engeland M. et al. Body size and risk for colorectal cancers showing BRAF mutations or microsatellite instability: a pooled analysis. Int J Epidemiol. 2012;41:1060-72
42. Jia M, Jansen L, Walter V. et al. No association of CpG island methylator phenotype and colorectal cancer survival: population-based study. Br J Cancer. 2016;115:1359-66
43. Jones JC, Renfro LA, Al-Shamsi HO. et al. (Non-V600) BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol. 2017;35:2624-30
44. Lin CC, Lin JK, Lin TC. et al. The prognostic role of microsatellite instability, codon-specific KRAS, and BRAF mutations in colon cancer. J Surg Oncol. 2014;110:451-7
45. Modest DP, Ricard I, Heinemann V. et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016;27:1746-53
46. Nosho K, Irahara N, Shima K. et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698
47. Ogino S, Nosho K, Kirkner GJ. et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90-6
48. Phipps AI, Buchanan DD, Makar KW. et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21:1792-8
49. Rimbert J, Tachon G, Junca A. et al. Association between clinicopathological characteristics and RAS mutation in colorectal cancer. Mod Pathol. 2018;31:517-26
50. Rozek LS, Herron CM, Greenson JK. et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:838-43
51. Samowitz WS, Sweeney C, Herrick J. et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063-9
52. Shen Y, Wang J, Han X. et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628
53. Sinicrope FA, Shi Q, Allegra CJ. et al. Association of DNA mismatch repair and mutations in BRAF and KRAS with survival after recurrence in stage III colon cancers: a secondary analysis of 2 randomized clinical trials. JAMA Oncol. 2017;3:472-80
54. Tanaka N, Huttenhower C, Nosho K. et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. Am J Pathol. 2010;177:2731-40
55. Tie J, Gibbs P, Lipton L. et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer. 2011;128:2075-84
56. Tol J, Dijkstra JR, Klomp M. et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997-2009
57. Toon CW, Walsh MD, Chou A. et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 2013;37:1592-602
58. Tran B, Kopetz S, Tie J. et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer-Am Cancer Soc. 2011;117:4623-32
59. Vedeld HM, Merok M, Jeanmougin M. et al. CpG island methylator phenotype identifies high risk patients among microsatellite stable BRAF mutated colorectal cancers. Int J Cancer. 2017;141:967-76
60. Won DD, Lee JI, Lee IK. et al. The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer. 2017;17:403
61. Yaeger R, Cercek A, Chou JF. et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer-Am Cancer Soc. 2014;120:2316-24
62. Saridaki Z, Tzardi M, Sfakianaki M. et al. BRAFV600E mutation analysis in patients with metastatic colorectal cancer (mCRC) in daily clinical practice: correlations with clinical characteristics, and its impact on patients' outcome. PLoS One. 2013;8:e84604
63. Seppala TT, Bohm JP, Friman M. et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966-75
64. Vilkin A, Niv Y, Nagasaka T. et al. Microsatellite instability, MLH1 promoter methylation, and BRAF mutation analysis in sporadic colorectal cancers of different ethnic groups in Israel. Cancer. 2009;115:760-69
65. Li C, Lee KC, Schneider EB. et al. BRAF V600E mutation and its association with clinicopathological features of papillary thyroid cancer: a meta-analysis. J Clin Endocrinol Metab. 2012;97:4559-70
66. Ulivi P, Scarpi E, Chiadini E. et al. Right- vs. left-sided metastatic colorectal cancer: differences in tumor biology and Bevacizumab efficacy. Int J Mol Sci. 2017;18:pii E1240
67. De Roock W, Claes B, Bernasconi D. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-62
68. van Brummelen E, de Boer A, Beijnen JH. et al. BRAF Mutations as Predictive Biomarker for Response to Anti-EGFR Monoclonal Antibodies. Oncologist. 2017;22:864-72
69. Pires da Silva I, Wang KYX, Wilmott JS. et al. Distinct molecular profiles and immunotherapy treatment outcomes of V600E and V600K BRAF-mutant melanoma. Clin Cancer Res. 2019;25:1272-9
70. He WZ, Hu WM, Kong PF. et al. Systemic neutrophil lymphocyte ratio and mismatch repair status in colorectal cancer patients: correlation and prognostic value. J Cancer. 2018;9:3093-100
71. Marginean EC, Melosky B. Is there a role for programmed death ligand-1 testing and immunotherapy in colorectal cancer with microsatellite instability? part I—colorectal cancer: microsatellite instability, testing, and clinical implications. Arch Pathol Lab Med. 2018;142:17-25
Author contact
![]() Corresponding authors: Prof. Shen L, Division of Gastrointestinal Surgery, Department of General Surgery, First Affiliated Hospital, Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, China; Tel/Fax: +86-25-83724440; E-mail: shenlzedu.cn & shenlzedu.cn. Prof. Cao M, Lab of cellular and molecular biology, Affiliated Hospital of Integrated Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing 210028, China; Tel/Fax: +86-25-52362182; E-mail: mcao1979com.
Corresponding authors: Prof. Shen L, Division of Gastrointestinal Surgery, Department of General Surgery, First Affiliated Hospital, Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, China; Tel/Fax: +86-25-83724440; E-mail: shenlzedu.cn & shenlzedu.cn. Prof. Cao M, Lab of cellular and molecular biology, Affiliated Hospital of Integrated Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing 210028, China; Tel/Fax: +86-25-52362182; E-mail: mcao1979com.

 Global reach, higher impact
Global reach, higher impact