Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2407-2414. doi:10.7150/jca.32411 This issue Cite
Review
The Advancement of Long Non-Coding RNAs in Cholangiocarcinoma Development
Department of General Surgery, Peking University Third Hospital, 49 North Garden Rd., Haidian District Beijing, P.R.China, 100191, 86-010-82267332
Received 2018-12-19; Accepted 2019-4-18; Published 2019-5-26
Abstract
Cholangiocarcinoma (CCA) is a malignancy with increasing incidence in recent years. CCA patients are usually diagnosed at advanced stage due to lack of apparent symptoms and specifically diagnostic markers. Nowadays, surgical removal is the only effective method for CCA whereas overall 5-year-survival rate keeps around 10%. Long-noncoding RNA (lncRNA), a subtype of noncoding RNA, is widely studied to be abnormally expressed in multiple cancers including CCA. LncRNA can promote proliferation, migration, invasion and inhibit apoptosis of CCA. Moreover, lncRNA is negatively correlated with the prognosis of CCA. LncRNA may contribute to the development of CCA via modulating gene transcription, sponging microRNA, regulating CCA-related signaling pathways or protein expression. LncRNA is thought to be potential diagnostic markers and therapeutic targets for CCA.
Keywords: cholangiocarcinoma, lncRNA, proliferation, invasion, prognosis
Introduction
Cholangiocarcinoma (CCA) is a malignancy originating from the epithelial cells of biliary tree. It is classified into intrahepatic cholangiocarcinoma (ICC), hilar and distal cholangiocarcinoma according to anatomical location[1]. Both hilar and distal cholangiocarcinoma are known as extrahepatic cholangiocarcinoma (ECC). CCA accounts for about 3% of all alimentary malignancies and is the second most common hepatobiliary malignancy just following hepatocellular carcinoma (HCC). The incidence of CCA is relatively higher in Asian countries compared to western countries. The highest annual morbidity of CCA is in southeast Asia, ranging from 0.1/100,000 to 85/100,000. In contrast, in Europe the morbidity ranges from 0.45/100,000 to 3.36/100,000[2]. Recently, the incidence of CCA tends to show a worldwide rise, especially ICC[3-6]. In US, ICC age-adjusted incidence rate increased from 1.42 to 1.82/100,000 in males and from 0.98 to 1.45 in females between 1999 and 2013[8]. Notably, the increasing ICC incidence is thought to be influenced by the criteria modification of CCA classification in the International Classification of Diseases for Oncology (ICD-O) coding system [3, 9].
Many factors contribute to CCA tumorigenesis. Cholestasis and inflammation are key pathological factors in CCA[10]. Some oncogenes such as TP53, KRAS, FGFR2, IDH1, IDH2 are proved to be associated with CCA[11]. ECC usually leads to bile duct obstructive symptoms including painless jaundice and acute cholangitis while ICC develops as intrahepatic mass with non-specific symptoms such as abdominal pain or cachexy[12, 13]. Lacking of apparent and specific symptoms and sensitive diagnostic markers, most CCA patients are diagnosed at late stage. Currently, surgical removal is the preferred treatment choice for CCA. However, the prognosis keeps very poor with the overall 5-year-survival rate around 10%[14].
RNAs with no protein-coding-function are collectively named non-coding RNA (ncRNA). NcRNA can be classified into microRNA (miRNA), long-noncoding RNA (lncRNA) and circular RNA (circRNA). MiRNA, a ncRNA subtype of 20~22nt, is involved in CCA progress and can be used as circulating prognostic biomarker[15]. LncRNA is ncRNA longer than 200nt in length and lncRNA contributes to a major part of human RNA, according to a transcriptome analysis[16]. However, lncRNA used to be thought as the noise of transcription for many years. Until recently, the function of lncRNA has been reported in multiple pathophysiological conditions. 166 diseases are analyzed to be involved of lncRNA dysfunction, including cancer, neurodegeneration disease and cardiovascular disease[17]. LncRNA has been proved to be associated with the development of different tumor types, including HCC, CCA, prostate cancer and so on[18]. Studies indicate that lncRNA can regulate gene expression in cancer by three levels: the genomic sequence of locus, the act of transcription and the RNA product itself[19, 20]. LncRNA may function by affecting gene transcription, targeting RNA polymerase II, regulating splicing, taking part in epigenetic regulation and interacting with RNA molecules[17]. For example, lncRNA may help forming higher order chromatin structures, such as intra- and inter-chromosomal loops and nuclear compartmentalization. LncRNA transcription can influence functional gene expression [19, 21]. It also can regulate gene expression by interacting with DNA fragment directly, chromatin complexes, DNA/RNA-binding proteins (DBP/RBPs) or modulators of proteins and enzyme cofactors. LncRNA can compete with miRNA as compete endogenous RNA (ceRNA), which contributes to multiple biological processes and tumorigenesis [22-24]. There are lots of studies on lncRNA in various cancers, as well as in CCA. In this review, we would summarize lncRNA's involvement in CCA development and possible mechanisms.
LncRNA in CCA development
LncRNA is abnormally expressed in CCA tissues
Studies from different groups give a whole landscape of upregulated or downregulated lncRNAs in CCA. 1434 lncRNAs were observed to be specifically expressed in bile duct cancer tissues, in which 929 upregulated and 505 down-regulated[25]. Lv, L. and his colleagues reported that 2148 lncRNAs were upregulated while 568 lncRNAs were downregulated in ICC tissues compared with normal tissues[26]. Jianguo Wang's report showed a similar result with 2773 lncRNAs upregulated and 2392 lncRNAs downregulated in ICC tissues [27]. Another study by Zhang, F., et al. identified that 268 lncRNAs were differentially expressed in ECC(78 lncRNAs upregulated, 190 lncRNAs downregulated) [28].
The relationship of some differentially expressed lncRNAs (especially upregulated lncRNAs) with CCA clinicopathological characteristics were also reported. The upregulated lncRNAs are associated with tumor size, tumor stage, lymph node metastasis or postoperative recurrence, etc. For instance, upregulated lncRNA Sox2ot is related with lymph node invasion (P=0.0308), TNM stage (P=0.0072) and postoperative recurrence (P=0.0019) [29]. Besides, both lncRNA UCA1(urothelial carcinoma associated 1) and CCAT1 involve in CCA clinicopathological conditions [30-32]. Interestingly, the upregulation of lncRNA CPS1-IT1 is associated with poorer liver function[33]. To give a better piece of knowledge, we summarized the upregulated lncRNAs and their correlation with CCA clinicopathologic characteristics in Table 1.
LncRNA promotes CCA proliferation, migration and invasion
The deregulated expression of lncRNAs may be related with CCA development and progression. These lncRNAs may promote proliferation, migration and invasion of CCA cells including lncRNA AFAP1- AS1, ASAP1-IT1, CCAT1, CCAT2, H19, HULC, LINC01296, ENST0000058480.1, ENST00000517758.1, MALAT1, NEAT1, PANDAR, SNHG1, Sox2ot, SPRY4-IT1, TP73-AS1 etc. [23, 29, 30, 34-49]. In ICC, LncRNA CPS1-IT1 and TLINC are highly expressed [33, 50]. LncRNA CPS1-IT1 promotes cancer cell proliferation, metastasis and inhibits apoptosis[33]. LncRNA TLINC increases cancer cell migration[50]. In ECC, LncRNA PCAT1 is upregulated, promoting cancer cell proliferation, metastasis and inhibiting apoptosis[51]. The upregulated lncRNAs in CCA and their functions in cancer progression were also summarized in Table 1.
LncRNA promotes EMT in CCA development
Epithelial-mesenchymal transition (EMT) is a biological process, regulating the conversion of cells from epithelial to mesenchymal state. Mesenchymal cells are lack of intercellular junctions and can move throughout the extracellular matrix, in contrast, epithelial cells are less migratory[52]. EMT are associated with embryonic development after fertilization, tissue regeneration, cancer progression and metastasis respectively. Primary tumor epithelial cells convert into invasive and metastatic mesenchymal tumor cells with enhanced mobility via the third type EMT process[52, 53]. It is suggested that lncRNA is involved in CCA tumorigenesis by inducing EMT[31, 37, 39, 54]. In this process, it accompanies with N- cadherin and Vimentin enhancement and E-cadherin downregulation [53]. In contrast, silencing EMT- related-lncRNA reverses the process. For instance, Xia, X. L. et al. silenced a colorectal-cancer-related lncRNA CRNDE and observed E-cadherin was significantly increased, while N-cadherin and Vimentin were lost in ICC cell lines [54]. LncRNA UCA1 also can promote EMT formation[32]. When silencing UCA1, Vimentin and N-cadherin were decreased with enhanced expression of E-cadherin. LncRNA CCAT1 is involved in EMT process in ICC as well. Knockdown of CCAT1 upregulates E-cadherin and downregulates N-cadherin and Vimentin[31]. LncRNA NEAT1 promotes cholangiocarcinoma metastasis through suppressing expression of E-cadherin partly by binding EZH2[44]. There are other lncRNAs which are also reported to induce EMT processes in CCA development (see Table 1). All these studies demonstrate that it may be a common and crucial phenomenon for lncRNA in CCA development via activating EMT. However, the exact mechanisms of lncRNA promoting the EMT need further studies.
Upregulated lncRNAs in CCA
| LncRNA | Cancer | Clinicopathologic Characteristics | Function | Possible Mechanism | Clinical Relevance | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TNM stagea | PORa | OSa | other | Promoting CCA cell proliferation, metastasis and inhibiting apoptosis | Enhancing EMT | Unfavorable prognostic biomarker | Potential therapeutic target | ||||
| AFAP1-AS1 | CCA | √ | - | √ | - | √ | - | 1. Decreasing the expression of c-Myc, Cyclin D1, MMP-2 and MMP-9; 2. Decreasing the AFAP1 expression and promoting cell stress filament integrity | √ | √ | [34, 35] |
| ASAP1-IT1 | CCA | √ | √ | - | - | √ | √ | Interacting with hedgehog signaling pathway | √ | √ | [36] |
| CCAT1 | CCA and ICC | √ | - | √ | Associated with histological differentiation | √ | √ | Sponging to miR-152 as ceRNA | Independent prognostic factor | √ | [30, 31] |
| CCAT2 | CCA | √ | √ | √ | Linked to tumor size | √ | √ | - | √ | √ | [37, 38] |
| CPS1‑IT1 | ICC | √ | √ | - | Associated with poor liver function | √ | √ | Co‑expressed with host gene CPS1 | √ | √ | [33] |
| EPIC1 | CCA | - | - | - | - | √ | - | Directly interacting with Mys | - | - | [69] |
| H19 | CCA | √ | √ | √ | Linked to tumor size | √ | √ | Activing IL-6 by sponging to let-7a/let-7b as ceRNA; | √ | √ | [23, 39] |
| HULC | CCA | - | - | - | - | √ | - | Activing CXCR4 by sponging to miR-372/miR-373 as ceRNA | - | √ | [23] |
| HOTAIR | CCA | √ | √ | - | Linked to tumor size | √ | √ | - | √ | √ | [74] |
| LINC01296 | CCA | √ | - | - | - | √ | - | Modulating MYCN transcription by sponge miR-5095 as ceRNA | - | √ | [40] |
| LncRNAs of ENST00000588480.1 and ENST00000517758.1 | CCA | √ | √ | - | - | √ | - | Affecting signaling pathway including p53 signaling pathway | √ | √ | [41] |
| MALAT1 | CCA and HCCA | √ | - | - | Linked to tumor size | √ | √ | 1.Activing PI3K/Akt pathway; 2. miR-204-dependent CXCR4 regulation as ceRNA | √ | √ | [42, 43] |
| NEAT1 | CCA | - | - | - | - | √ | √ | 1.BAP1 protein dependent; 2. Inhibiting E-cadherin expression by EZH2 | √ | Modulating sensitivity to gemcitabine | [44, 45] |
| PANDAR | CCA | √ | √ | - | - | √ | √ | - | √ | √ | [46] |
| PCAT1 | ECC | - | - | - | - | √ | - | Enhancing Wnt/β-catenin signaling through miR-122 repression and WNT1 expression | - | √ | [51] |
| PVT1 | CCA | - | - | - | - | √ | - | Binding to epigenetic modification complexes, adjusting the expression of ANGPTL4 | - | √ | [66] |
| SNHG1 | CCA | - | - | - | - | √ | - | Modulating cancer-related gene like CDKN1A by co-operating with chromatin-modifying enzymes as EZH2 | √ | √ | [47] |
| Sox2ot | CCA | √ | √ | - | - | √ | - | - | Independent prognostic factor | √ | [29] |
| SPRY4-IT1 | CCA | √ | - | - | - | √ | √ | Recruiting EZH2, LSD1 or DNMT1 via sponging to miR-101-3p | √ | √ | [48] |
| TLINC | ICC | - | - | - | - | Increasing cell migration | - | Infecting cell inflammatory microenvironment induced by TGF-β | - | - | [50] |
| TP73-AS1 | CCA | √ | - | - | Linked to tumor size | √ | - | Suppressing caspase-3 and caspase-9. | - | √ | [49] |
| T-UCRs | CCA | - | - | - | - | √ | - | Downstream of Wnt pathway and sponging to miR-193b | √ | √ | [59] |
| TUG1 | CCA | √ | √ | √ | Linked to tumor size | √ | √ | TUG1/miR-145/Sirt3/GDH regulatory network as ceRNA | √ | √ | [24, 75] |
| UCA1 | CCA | √ | √ | - | - | √ | √ | 1.Facilitating apoptosis via Bcl-2/caspase-3 pathway.2. Activating AKT/GSK-3β/CCND1 axis 3. Upregulating MMP-9 | √ | √ | [32, 76] |
a POR: postoperative recurrence; OS: overall survival; TNM stage:T tumor, N node, M metastasis stage; b√ There exists an association of LncRNA; - Unknown
LncRNA activates oncogenic signaling pathways in CCA development
Cancer-related signaling pathways can be influenced by abnormally expressed lncRNA and may be the reasonable mechanism of CCA development. The phosphatidylinositol 3-kinase(PI3K)/AKT pathway is a key driver in carcinogenesis, modulating cell survival, differentiation, angiogenesis, and cellular migration[55]. LncRNA UCA1 promotes CCA progression by activating AKT/GSK-3β/CCND1 signaling pathway [32]. Knockdown of UCA1 in CCA cell lines, phosphorylated-AKT and phosphorylated- GSK-3β decreased, which further inhibiting the expression of CCND1.
The canonical Wnt/β-catenin pathway is an ancient and evolutionarily conserved signaling pathway that is critical for embryo development and adult tissue homeostasis[56, 57]. Wnt pathway is highly activated in CCA with macrophages activation. Inhibition of Wnt signaling in animal models can markedly reduce cancer cell proliferation and increase apoptosis, resulting in tumor regression[58]. LncRNA PCAT1 is reported to activate Wnt/β-catenin signaling and promote ECC progression[51]. Recent studies further demonstrate that lncRNA may participate in the pathogenesis of CCA in a Wnt pathway dependent manner[51, 59]. For example, transcribed-ultraconserved regions (T-UCRs, a group of lncRNAs that harbor ultraconserved fragments) were proved to be the downstream mediators of the Wnt/β-catenin pathway, which was upregulated considerately in human CCA tissues[59]. After treating with Wnt inhibitors ICG-001 and C-59 in rat CCA models, uc.158- (an intergenic 224nt T-UCR located at chromosome 5) was reduced and cancer cell growth was depressed[59]. Collectively, lncRNA contributes to CCA tumorigenesis and development by regulating crucial oncogenic signaling pathways.
LncRNA influences cellular metabolism in CCA development
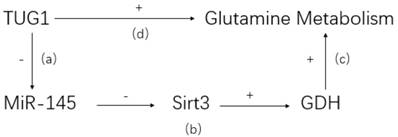
LncRNA may affect pathophysiological metabolism of CCA[60]. It is well known that abnormal glutamine is characteristic in tumor tissues and related to tumorigenesis and development[61]. LncRNA taurine upregulated gene 1 (TUG1), a highly expressed lncRNA in ICC, is demonstrated to promote abnormal glutamine metabolism via TUG1/miR-145/Sirt3/GDH regulatory network (see Figure 1) [24]. The high expression of lncRNA TUG1 is negatively correlated with the expression of miR-145. Co-transfection and rescue studies showed that TUG1 could increase glutamine consumption, α-KG production and ATP levels by counteracting with miR-145. Besides, lncRNA TUG1/miR-145 were able to regulate Sirt3 and GDH expression, which were involved in glutamine metabolism in ICC cells. Therefore, a reasonable explanation is that lncRNA TUG1 promotes CCA proliferation, invasion and migration by affecting glutamine metabolism. It is unclear whether lncRNA also regulates other metabolic processes in CCA development.
TUG1/miR-145/Sirt3/GDH regulatory network. (a) TUG1 can bind to miR-145 directly and inhibit its activity. (b) Sirt3, a positive regulator of GDH, is confirmed to be suppressed by miR-145. (c) GDH could reverse the inhibitive effect of glutamine metabolism caused by miR-145. (d) Knockdown TUG1 in ICC cell would lead to a dramatic decrease of glutamine consumption, indicating a positive glutamine metabolism regulation by TUG1.
The possible acting mechanisms of lncRNA in CCA
LncRNA regulates nearby or host genes transcription
LncRNA may modulate the transcription and expression of nearby mRNA genes[62]. In CCA, the co-expression status of host gene and lncRNA was also studied. CPS1 protein, coded by CPS1 gene, is a crucial enzyme of urea cycle. CPS1 deficiency may lead to central nervous system disorders. The co-expression of CPS1 gene and its lncRNA CPS1 intronic transcript 1 (CPS1‑IT1) was analyzed[33]. CPS1 and CPS1-IT1 were co-upregulated in ICC tissues with CPS1 protein enhanced, which was negatively associated with ICC prognosis. Moreover, CPS1 was depressed by silencing CPS1-IT1, resulting in reduced cell proliferation and enhanced apoptosis. However, the exact regulatory mechanism between CPS1-IT1 and CPS1 in ICC keeps unclear.
Actin filament associated protein 1 antisense RNA 1 (lncRNA AFAP1-AS1) is a type of lncRNA transcribed from the antisense strand of AFAP1 gene locus. The second AFAP1-AS1 exon overlaps with AFAP1 exons 14, 15 and 16. AFAP1-AS1 is involved in multiple human cancers including pancreatic and biliary carcinoma[63, 64]. Knockdown AFAP1-AS1 in CCA cells resulted in elevated expression of AFAP1 mRNA and protein, leading to loss of stress filament integrity. Therefore, AFAP1-AS1 may promote CCA growth, proliferation, and invasion by interfering with AFAP1 expression[35].
LncRNAs act as ceRNAs
| LncRNA | Co-regulatory miRNA | Target of miRNA | Function | Reference |
|---|---|---|---|---|
| T-UCRs | miR-193b | β-catenin | Downstream mediator of the Wnt/β-catenin pathway and specific for cancer development | [59] |
| PCAT1 | miR-122 | WNT1 | Promoting Wnt/β-catenin pathway | [51] |
| H19 | let-7a/let-7b | IL-6 | Activating pivotal inflammation cytokine IL-6 to regulate cancer cell migration and invasion | [23] |
| HULC | miR-372/miR-373 | CXCR4 | Activating Chemokine receptor CXCR4 to regulate cancer cell migration and invasion | [23] |
| MALAT1 | miR-204 | CXCR4 | Modulating HCCA cell growth, migration and invasion through miR-204-dependent CXCR4 | [42] |
| TUG1 | miR-145 | Sirt3 | TUG1/miR-145/Sirt3/GDH network | [24] |
| LINC01296 | miR-5095. | MYCN | LINC01296/miR-5095/MYCN network | [40] |
| SPRY4-IT1 | miR-101-3p. | EZH2 | Enhancing EZH2 expression | [48] |
| CCAT1 | miR-152 | EMT | Leading to EMT activation of ICC cells | [31] |
LncRNA binds EZH2 and mediates epigenetic gene silencing
PRC2 (polycomb repressive complex 2) possesses histone methyltransferase activity and trimethylates histone H3 on lysine 27 (the complex is called H3K27me3), one of transcriptionally silent chromatin markers. Enhancer of zeste homolog 2 (EZH2) is the functional enzymatic component of this complex[65]. Some lncRNAs are reported to directly bind with EZH2 and regulate a spectrum of gene transcription. One study is about the lncRNA SNHG1-EZH2-CDKN1A regulatory network in CCA development. LncRNA SNHG1 is an oncogenic lncRNA in CCA. It directly binds with EZH2, then epigenetically silencing targeting genes by regulating H3K27me3[47]. The main target in this process is CDKN1A. CDKN1A is proved to be negatively involved in CCA tumorigenesis. Silencing SNHG1 leads to high expression of CDKN1A with lower tumor proliferation and higher apoptosis in CCA cell lines. LncRNA SPRY4-IT1 can interact with EZH2, lysine specific demethylase 1 (LSD1) and DNA methyltransferase 1 (DNMT1), serving as a scaffold. The complex suppresses the expression of target genes such as KLF2, LATS2, and further promotes CCA development[48]. Interestingly, SPRY4-IT1 can also positively modulate the expression of EZH2 through sponging miR-101-3p. Moreover, lncRNA PVT1 also can combine with EZH2 to influence the histone methylation of ANGPTL4 promoter, a possible tumor suppressor gene[66]. These findings suggest lncRNA can mediate gene silencing by directly interacting with EZH2 and regulating H3K27me3 in CCA development.
LncRNA sponges miRNA as ceRNA
LncRNA can sponge miRNA directly by miRNA-response elements and regulate miRNA's activity as ceRNA, which is an important lncRNA action mode in CCA[25, 67]. In a study to characterize interactions between survival-related ceRNAs and CCA, seven major ceRNA clusters were identified. Among them, Cluster 23 and 32 were analyzed to be closely related to CCA patients survival by mediating ceRNA regulation of lncRNA[68]. Some lncRNAs have been identified to have specific-binding miRNAs in CCA. Two lncRNAs, H19 and HULC, are upregulated by short-term oxidative stress in CCA cell lines[23]. They promote cell migration and invasion in CCA by activating IL-6 and CXCR4 through sponging let-7a/let-7b and miR-372/miR-373 respectively. There are some other studies of lncRNA as ceRNA in CCA. LINC01296(Long Intergenic Non-protein Coding RNA 1296) contributes to CCA development by sponging miR-5095, further regulating the expression of MYCN proto-oncogene bHLH transcription factor [40]. Oncogenic lncRNA SPRY4-IT1 can enhance the expression of EZH2 by sponging miR-101-3p in CCA[48]. LncRNA PCAT1 (prostate cancer associated transcript 1) is an endogenous competing RNA for miR-122 in ECC, which targets WNT1[51]. Then it upregulates the Wnt/β-catenin signaling pathway and promotes ECC progression. Additionally, TUG1/miR-145/Sirt3/GDH regulatory network is also involved in the binding between TUG1 and miRNA-145[24]. The identified lncRNAs as ceRNAs in CCA have been summarized in Table 2.
LncRNA binds functional proteins
Some lncRNAs are reported to interact with proteins and regulate their localization, activity and functions. For example, lncRNA EPIC1 expression is elevated in CCA. It facilitates CCA progression by directly interacting with MYC protein, then regulating the expression of transcription factor MYC downstream genes [69]. The interaction of lncRNA with protein EZH2 has been discussed in Section 2.2.
Clinical applications of lncRNA in CCA
LncRNA serves as potential diagnostic or prognostic markers
The abnormal upregulated expression of certain lncRNA points to aggressive tumor phenotypes and unfavorable prognosis in CCA. The high expression of lncRNA CCAT1 could be an independent unfavorable prognostic biomarker for CCA[30]. LncRNA UCA1 is also an independent prognostic factor for CCA overall survival (P = 0.014)[32]. The high expression of host gene CPS1 and its associated lncRNA CPS1-IT1 serves as a poor prognostic marker in ICC[33]. Upregulation of AFAP1-AS1 is negatively associated with CCA prognosis[34, 35]. The availability of specimens is an important factor for clinical application. One study demonstrated plasma lncRNA PCAT1, MALAT1, and CPS1-IT1 might serve as potential circulating fingerprints for early diagnosis of hilar cholangiocarcinoma(HCCA)[70]. The possible application of lncRNA as diagnostic or prognostic marker is also summarized in Table 1.
LncRNA may assist chemotherapy in CCA
Knocking down or silencing lncRNA in CCA decreases cell proliferation, migration and invasion and promotes cell apoptosis in vitro. This indicates that lncRNA can be used as potential novel therapeutic targets for CCA patients. Manipulating of lncRNA expression may also influence the sensitivity of CCA cells to chemotherapy. BAP1(BRCA-1 associated protein-1) is a protein coded by BRCA-1, a tumor suppressor gene in many cancers. It is a member of the ubiquitin C-terminal hydrolase superfamily that plays a critical role in chromatin remodeling [45]. Several studies reported late stage ICC was associated with BAP1 loss, suggesting the potential tumor suppressive functions of BAP1 as a chromatin modulator[71-73]. Recently, the negative correlation between BAP1 and lncRNA NEAT1 in CCA is reported [45]. Knockdown of BAP1 induces the upregulation of lncRNA NEAT1 in CCA cell lines. The inverse relationship between lncRNA NEAT1 and BAP1 is also supported by the analysis of the Tumor Cancer Genome Atlas cholangiocarcinoma dataset. Furthermore, BAP1-NEAT1-network can regulate sensitivity to chemotherapy. After depressing BAP1 in CCA cell lines with high NEAT1 expression, it is more sensitive to gemcitabine, TSA, olaparib, b-AP15. Therefore, NEAT1 may be used to assist CCA chemotherapy such as gemcitabine treatment.
Conclusion and Perspective
Related functional analysis has clearly showed the involvement of lncRNA abnormal expression and CCA development. LncRNA can promote CCA progression, EMT, influence signaling pathway and regulate metabolism. It may function via regulating transcription, chromatin status and interacting with miRNA or proteins. Besides, the application of lncRNA as potential diagnostic or prognostic markers is also proposed. The understanding of lncRNA in CCA tumorigenesis is of great value for early detection and novel therapeutic development. What's more, plasma lncRNA as circulating biomarkers may be a meaningful study direction for CCA early diagnosis. More systematic studies are needed to uncover functions and analyze regulatory mechanisms for lncRNA in CCA development.
Abbreviations
CCA: Cholangiocarcinoma; ICC: Intrahepatic Cholangiocarcinoma; ECC: Extrahepatic Cholangiocarcinoma; HCC: Hepatocellular Carcinoma; ICD-O : International Classification of Diseases for Oncology; NcRNA: Non-coding RNA; MiRNA: MicroRNA; LncRNA: Long-noncoding RNA; CircRNA: Circular RNA; DBP/RBPs: DNA/RNA-binding Proteins; CeRNA: Compete Endogenous RNA; LncRNA UCA1: LncRNA urothelial carcinoma associated 1; EMT: Epithelial-mesenchymal Transition; PI3K: Phosphatidylinositol 3-kinase; T-UCRs: Transcribed-ultraconserved Regions; LncRNA TUG1: LncRNA taurine upregulated gene 1; LncRNA CPS1‑IT1: LncRNA CPS1 Intronic Transcript 1; LncRNA AFAP1-AS1: Actin Filament Associated Protein 1 Antisense RNA 1; PRC2: Polycomb repressive complex 2; H3K27me3: Trimethylation of histone H3 on lysine 27; EZH2: Enhancer of zeste homolog 2; LSD1: Lysine specific demethylase 1; DNMT1: DNA methyltransferase 1; LncRNA PCAT1: LncRNA prostate cancer associated transcript 1; HCCA: Hilar Cholangiocarcinoma; BAP1: BRCA-1 associated protein-1; LINC01296: Long Intergenic Non-protein Coding RNA 1296.
Acknowledgements
This work is supported by Beijing Municipal Natural Science Foundation (Grant No.: 7172233)
Competing Interests
The authors have declared that no competing interest exists.
References
1. Blechacz B. Cholangiocarcinoma: Current Knowledge and New Developments. Gut Liver. 2017;11:13-26
2. Banales JM, Cardinale V, Carpino G. et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-80
3. Saha SK, Zhu AX, Fuchs CS. et al. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. Oncologist. 2016;21:594-9
4. Blendis L, Halpern Z. An increasing incidence of cholangiocarcinoma: why? Gastroenterology. 2004;127:1008-9
5. Mukkamalla SKR, Naseri HM, Kim BM. et al. Trends in Incidence and Factors Affecting Survival of Patients With Cholangiocarcinoma in the United States. J Natl Compr Canc Netw. 2018;16:370-6
6. Witjes CD, Karim-Kos HE, Visser O. et al. Intrahepatic cholangiocarcinoma in a low endemic area: rising incidence and improved survival. HPB (Oxford). 2012;14:777-81
7. Tyson GL, Ilyas JA, Duan Z. et al. Secular trends in the incidence of cholangiocarcinoma in the USA and the impact of misclassification. Dig Dis Sci. 2014;59:3103-10
8. Van Dyke AL, Shiels MS, Jones GS. et al. Biliary tract cancer incidence and trends in the United States by demographic group, 1999-2013. Cancer. 2019
9. Khan SA, Emadossadaty S, Ladep NG. et al. Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J Hepatol. 2012;56:848-54
10. Rizvi S, Gores GJ. Molecular pathogenesis of cholangiocarcinoma. Dig Dis. 2014;32:564-9
11. Brivio S, Cadamuro M, Fabris L. et al. Molecular Mechanisms Driving Cholangiocarcinoma Invasiveness: An Overview. Gene Expr. 2018;18:31-50
12. Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308-21
13. Weber SM, Jarnagin WR, Klimstra D. et al. Intrahepatic cholangiocarcinoma: resectability, recurrence pattern, and outcomes. J Am Coll Surg. 2001;193:384-91
14. Everhart JE, Ruhl CE. Burden of digestive diseases in the United States Part III: Liver, biliary tract, and pancreas. Gastroenterology. 2009;136:1134-44
15. Letelier P, Riquelme I, Hernández AH. et al. Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. International Journal of Molecular Sciences. 2016:17
16. Bertone P, Stolc V, Royce TE. et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242-6
17. Chen G, Wang Z, Wang D. et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983-6
18. Iyer MK, Niknafs YS, Malik R. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208
19. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206
20. Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027-38
21. Touat-Todeschini L, Shichino Y, Dangin M. et al. Selective termination of lncRNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 2017;36:2626-41
22. Zhang Y, Xu Y, Feng L. et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7:64148-67
23. Wang WT, Ye H, Wei PP. et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:016-0348
24. Zeng B, Ye H, Chen J. et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8:113650-61
25. Wang X, Hu KB, Zhang YQ. et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs, miRNAs and mRNAs with associated ceRNA network in Cholangiocarcinoma. Cancer Biomark. 2018
26. Lv L, Wei M, Lin P. et al. Integrated mRNA and lncRNA expression profiling for exploring metastatic biomarkers of human intrahepatic cholangiocarcinoma. Am J Cancer Res. 2017;7:688-99
27. Wang J, Xie H, Ling Q. et al. Coding-noncoding gene expression in intrahepatic cholangiocarcinoma. Transl Res. 2016;168:107-21
28. Zhang F, Wan M, Xu Y. et al. Transcriptome analysis reveals dysregulated long non-coding RNAs and mRNAs associated with extrahepatic cholangiocarcinoma progression. Oncol Lett. 2017;14:6079-84
29. Li Z, Li J, Ji D. et al. Overexpressed long noncoding RNA Sox2ot predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene. 2018;645:131-6
30. Jiang XM, Li ZL, Li JL. et al. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21:1242-7
31. Zhang S, Xiao J, Chai Y. et al. LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma Through Suppressing miR-152. Dig Dis Sci. 2017;62:3050-8
32. Xu Y, Yao Y, Leng K. et al. Long non-coding RNA UCA1 indicates an unfavorable prognosis and promotes tumorigenesis via regulating AKT/GSK-3beta signaling pathway in cholangiocarcinoma. Oncotarget. 2017;8:96203-14
33. Ma SL, Li AJ, Hu ZY. et al. Coexpression of the carbamoylphosphate synthase 1 gene and its long noncoding RNA correlates with poor prognosis of patients with intrahepatic cholangiocarcinoma. Mol Med Rep. 2015;12:7915-26
34. Lu X, Zhou C, Li R. et al. Long Noncoding RNA AFAP1-AS1 Promoted Tumor Growth and Invasion in Cholangiocarcinoma. Cell Physiol Biochem. 2017;42:222-30
35. Shi X, Zhang H, Wang M. et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394-404
36. Guo L, Zhou Y, Chen Y. et al. LncRNA ASAP1-IT1 positively modulates the development of cholangiocarcinoma via hedgehog signaling pathway. Biomed Pharmacother. 2018;103:167-73
37. Xu Y, Yao Y, Qin W. et al. Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed Pharmacother. 2018;99:121-7
38. Bai JG, Tang RF, Shang JF. et al. Upregulation of long noncoding RNA CCAT2 indicates a poor prognosis and promotes proliferation and metastasis in intrahepatic cholangiocarcinoma. Mol Med Rep. 2018;17:5328-35
39. Xu Y, Wang Z, Jiang X. et al. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother. 2017;92:17-23
40. Zhang D, Li H, Xie J. et al. Long noncoding RNA LINC01296 promotes tumor growth and progression by sponging miR-5095 in human cholangiocarcinoma. International journal of oncology. 2018;52:1777-86
41. Ge X, Wang Y, Nie J. et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget. 2017;8:69995-70005
42. Tan X, Huang Z, Li X. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J Cell Biochem. 2017;118:3643-53
43. Wang C, Mao ZP, Wang L. et al. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma. 2017;64:725-31
44. Zhang C, Li JY, Tian FZ. et al. Long Noncoding RNA NEAT1 Promotes Growth and Metastasis of Cholangiocarcinoma Cells. Oncol Res. 2018;26:879-88
45. Parasramka M, Yan IK, Wang X. et al. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16:017-0587
46. Xu Y, Jiang X, Cui Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets Ther. 2017;10:2873-83
47. Yu Y, Zhang M, Wang N. et al. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell death & disease. 2018;9:746
48. Xu Y, Yao Y, Jiang X. et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. Journal of experimental & clinical cancer research: CR. 2018;37:81
49. Yao Y, Sun Y, Jiang Y. et al. Enhanced expression of lncRNA TP73-AS1 predicts adverse phenotypes for cholangiocarcinoma and exerts oncogenic properties in vitro and in vivo. Biomed Pharmacother. 2018;106:260-6
50. Merdrignac A, Angenard G, Allain C. et al. A novel transforming growth factor beta-induced long noncoding RNA promotes an inflammatory microenvironment in human intrahepatic cholangiocarcinoma. Hepatol Commun. 2018;2:254-69
51. Zhang F, Wan M, Xu Y. et al. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/beta-catenin-signaling pathway. Biomed Pharmacother. 2017;94:55-62
52. Acloque H, Adams MS, Fishwick K. et al. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438-49
53. Nantajit D, Lin D, Li JJ. The network of epithelial-mesenchymal transition: potential new targets for tumor resistance. J Cancer Res Clin Oncol. 2015;141:1697-713
54. Xia XL, Xue D, Xiang TH. et al. Overexpression of long non-coding RNA CRNDE facilitates epithelial-mesenchymal transition and correlates with poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett. 2018;15:4105-12
55. Robbins HL, Hague A. The PI3K/Akt Pathway in Tumors of Endocrine Tissues. Front Endocrinol (Lausanne). 2015;6:188
56. Saito-Diaz K, Chen TW, Wang X. et al. The way Wnt works: components and mechanism. Growth Factors. 2013;31:1-31
57. Peng Y, Zhang X, Feng X. et al. The crosstalk between microRNAs and the Wnt/beta-catenin signaling pathway in cancer. Oncotarget. 2017;8:14089-106
58. Boulter L, Guest RV, Kendall TJ. et al. WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J Clin Invest. 2015;125:1269-85
59. Carotenuto P, Fassan M, Pandolfo R. et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268-77
60. Sun LY, Li XJ, Sun YM. et al. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol Cancer. 2018;17:127
61. Li T, Le A. Glutamine Metabolism in Cancer. Adv Exp Med Biol. 2018;1063:13-32
62. Yang W, Li Y, Song X. et al. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget. 2017;8:26591-9
63. Liu FT, Xue QZ, Zhu PQ. et al. Long noncoding RNA AFAP1-AS1, a potential novel biomarker to predict the clinical outcome of cancer patients: a meta-analysis. Onco Targets Ther. 2016;9:4247-54
64. Wang Y, Mo Y, Yang X. et al. Long non-coding RNA AFAP1-AS1 is a novel biomarker in various cancers: a systematic review and meta-analysis based on the literature and GEO datasets. Oncotarget. 2017;8:102346-60
65. Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42-8
66. Yu Y, Zhang M, Liu J. et al. Long Non-coding RNA PVT1 Promotes Cell Proliferation and Migration by Silencing ANGPTL4 Expression in Cholangiocarcinoma. Mol Ther Nucleic Acids. 2018;13:503-13
67. Song W, Miao DL, Chen L. Comprehensive analysis of long noncoding RNA-associated competing endogenous RNA network in cholangiocarcinoma. Biochem Biophys Res Commun. 2018;506:1004-12
68. Wan M, Zhang FM, Li ZL. et al. Identifying survival-associated ceRNA clusters in cholangiocarcinoma. Oncol Rep. 2016;36:1542-50
69. Li Y, Cai Q, Li W. et al. Long non-coding RNA EPIC1 promotes cholangiocarcinoma cell growth. Biochem Biophys Res Commun. 2018;504:654-9
70. Shi J. et al. The Plasma LncRNA Acting as Fingerprint in Hilar Cholangiocarcinoma. Cell Physiol Biochem. 2018;49:1694-702
71. Andrici J, Goeppert B, Sioson L. et al. Loss of BAP1 Expression Occurs Frequently in Intrahepatic Cholangiocarcinoma. Medicine (Baltimore). 2016;95:e2491
72. Misumi K, Hayashi A, Shibahara J. et al. Intrahepatic cholangiocarcinoma frequently shows loss of BAP1 and PBRM1 expression, and demonstrates specific clinicopathological and genetic characteristics with BAP1 loss. Histopathology. 2017;70:766-74
73. Mosbeh A, Halfawy K, Abdel-Mageed WS. et al. Nuclear BAP1 loss is common in intrahepatic cholangiocarcinoma and a subtype of hepatocellular carcinoma but rare in pancreatic ductal adenocarcinoma. Cancer Genet. 2018;224-225:21-8
74. Qin W, Kang P, Xu Y. et al. Long non-coding RNA HOTAIR promotes tumorigenesis and forecasts a poor prognosis in cholangiocarcinoma. Sci Rep. 2018;8:12176
75. Xu Y, Leng K, Li Z. et al. The prognostic potential and carcinogenesis of long non-coding RNA TUG1 in human cholangiocarcinoma. Oncotarget. 2017;8:65823-35
76. Tessmann JW, Buss J, Begnini KR. et al. Antitumor potential of 1-thiocarbamoyl-3,5-diaryl-4,5-dihydro-1H-pyrazoles in human bladder cancer cells. Biomed Pharmacother. 2017;94:37-46
Author contact
![]() Corresponding author: Dr. Ling, xiaofenglingedu.cn
Corresponding author: Dr. Ling, xiaofenglingedu.cn

 Global reach, higher impact
Global reach, higher impact