Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2472-2479. doi:10.7150/jca.29426 This issue Cite
Research Paper
MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11
1. Lab for Noncoding RNA & Cancer, School of Life Sciences, Shanghai University, Shanghai 200444, China
2. Experimental Animal Center, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, China
3. Exprimental Center for Life Sciences, School of Life Sciences, Shanghai University, Shanghai 200444, China
*These authors contributed equally to this work
Received 2018-8-23; Accepted 2019-4-12; Published 2019-6-2
Abstract
MicroRNAs (miRNAs) comprise a class of short, non-coding RNAs that directly target 3′UTR of mRNA, causing subsequent degradation or suppression of translation. Here, we verified that miR-199a-5p was significantly down-regulated in mouse NSCLC tissues and human patient samples. To further study the function of miR-199a-5p, lentivirus system was adopted to construct stably over-expressing miR-199a-5p A549, SPC-A1 and H1299 cell lines. Then, miR-199a-5p played a tumor suppression role via directly targeting MAP3K11 gene in non-small cell lung cancer (NSCLC). Elevated miR-199a-5p suppressed cell proliferation and arrested cell cycle in G1 phase. We found that MAP3K11 was negatively correlated with miR-199a-5p in NSCLC patient tissues and mouse xenograft tumors. Our results suggest that miR-199a-5p together with its target gene MAP3K11 is a key factor and constitutes a complicated regulation network in NSCLC.
Keywords: miR-199a-5p, MAP3K11, MAPK pathway, NSCLC, tumor suppression
Introduction
Lung cancer is the leading cause of cancer deaths worldwide [1, 2]. Based on their distinct clinicopathological features, lung cancers are currently divided into two groups: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which accounts for about 80% of all [3-5]. Despite improvements in clinical treatment strategies, the 5-year survival rate after treatment is still very poor; therefore it is crucial to identify better targets for the treatment of human NSCLC [6-9].
MicroRNAs (miRNAs) are short non-coding RNAs of 19-25 nucleotides that can modulate the expression of at least 30% of all human genes, with marked effects on fundamental biological processes such as cell proliferation, apoptosis, differentiation and metabolism [10-13]. MiRNAs can bind to partially complementary sequences of mRNA 3' untranslated region (3'UTR), subsequently causing mRNA degradation or translation inhibition, thus effectively silencing their target genes [14-16]. Hence, not only protein-encoding genes but also non-coding RNAs especially miRNAs, should be considered as key factors in signaling cascades [17-19]. Moreover, the targets of many miRNAs are still unknown or to be verified, even though we can get the putative targets of miRNAs by TargetScan or other bioinformatics tools. Thus, in order to unveil the molecular mechanisms associated with NSCLC progression and development, the target identification of miRNAs is critical, providing new ways for treating lung cancer [20]. Recent findings have highlighted a relevant role for miR-199a-5p in the regulation of tumorigenesis events [21]. MiR-199a-5p directly targets HIF1-α, which is a prominent transcription factor regulating angiogenesis, predominantly via induction of VEGF transcription [22]. Furthermore, hypoxia induces down-regulation of miR-199a-5p, probably through activation of the AKT pathway [23, 24]. On these premises, we investigated the functional role of miR-199-5p in NSCLC.
In the present study, we validated the decrease of miR-199a-5p either in clinical lung adenocarcinoma samples or mouse NSCLC models, and confirmed miR-199a-5p as a tumor suppressor for NSCLC in vitro (human cell lines) and in vivo (xenografted tumor assay). We further dissected the regulatory signaling of miR-199a-5p, and gave the evidence that miR-199a-5p negatively regulates its downstream target MAP3K11, which plays the anti-proliferation roles via MAPK pathway in the lung adenocarcinoma.
Materials and Methods
Cell culture
Human lung cancer cells (A549, SPC-A1, H1299) and Human embryonic kidney cell (HEK-293T) were obtained from the Cell Bank, China Academy of Sciences (Shanghai, China). A549, SPC-A1 and HEK-293T cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) and H1299 in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA), all supplemented with 10% fetal bovine serum (FBS, Hyclone, USA), 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were cultured in a 5% CO2 humidified incubator at 37℃.
Cell proliferation assay
2.0×103 cells per well were seeded in 96-well plates and cell viability was measured at 24, 48, 72 and 96 h. Cell viability was determined by Cell Counting Kit-8 (CCK-8) assay (Dojindo, Japan). At the indicated time, CCK-8 was added and incubated for 2.5 h at 37℃ in 5% CO2 and light absorbance was measured at 450 nm wavelength using a microplate reader, and their growth rates were recorded with three independent experiments.
Cell cycle analysis
Cells (1×106) were digested with a trypsin solution (0.25%) and then fixed in 70% ethanol overnight at -20℃. After washing with cold phosphate-buffered solution (PBS), the fixed cells were re-suspended in PI/RNase Staining Buffer and incubated at 37°C for 30 min in the dark. After staining, samples were analyzed with MoFlo XDP flow cytometry (Beckman Coulter, Inc., Brea, CA, USA). Data from flow cytometry was analyzed using Flow Jo software (Treestar Inc., USA). The flow cytometry analysis was repeated three times.
Colony formation assay
Cells were plated in 6 cm cell plate at 300-500 cells/well and further cultured in complete media for 10-15 d. After removal of the media, the cells were rinsed twice with PBS, and colonies were fixed with methanol for 15 min, stained with 0.1% crystal violet for 10 min and finally photographed using a digital camera (Leica, Germany). Experiments were performed three times.
Quantitative real-time PCR (qRT-PCR) analysis
Total cellular RNA was extracted using TRIzol Regent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. RNAs were reverse-transcribed using M-MLV RTase cDNA Synthesis Kit (TaKaRa, Dalian, China). A cDNA library of miRNAs was constructed by QuantiMir cDNA Kit (TaKaRa, Dalian, China). The level of mRNA or miRNA was quantified by qRT-PCR using SYBR Green PCR master mixture (TaKaRa, Dalian, China). U6 snRNA and 18S RNA was used as endogenous references for miRNAs and mRNAs respectively. Results were expressed using relative quantification (2-ΔΔCt) method. Primer sequences were listed in Supplementary Table 1.
Construction of recombinant expression vectors
The target genes of miR-199a-5p were selected based on the TargetScan (http://www.targetscan.org/) and Starbase (http://starbase.syst.edt.cn/). The 3'UTR of the target genes was sub-cloned downstream of the firefly luciferase reporter gene in the pGL3 vector (Promega, Madison, WI, USA). Mutant 3'UTR of MAP3K11 containing two mutated binding sites of miR-199a-5p in the 3'UTR was constructed. The primer sequences were listed in Supplementary Table 2.
Dual luciferase assay
For dual luciferase assay, HEK-293T cells cultured in 24-well plates were transiently co-transfected with 400 ng luciferase vector pGL3-3′-UTR or mutant pGL3-3′-UTR, miR-199a mimic or miR-NC at a final concentration of 100 nM and 20 ng pRL-SV40 (Promega, Madison, USA) as the control. 48 h after transfection, cells were lysed and reporter gene expression was determined using the Dual-luciferase reporter assay system (Promega, Madison, USA). Experiments were performed three times in triplicates.
Western blot analysis
Total protein was extracted from the cells with RIPA lysis buffer and then quantified using Protein BCA Assay Kit (Bio-Rad, Hercules, California, USA). The protein was separated by 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (PVDF) (Millipore Corporation, Billerica, MA, USA) and then blocked in 5% non-fat powdered milk at room temperature for 1 h. The PVDF membrane was incubated with rabbit anti-MAP3K11 (1:1000, Cell Signaling Technology(CST), Danvers, MA, USA) and rabbit anti-GAPDH (1:1000, Cell Signaling Technology, Danvers, MA, USA) overnight at 4℃ followed by washing and incubating with a goat-anti-rabbit Secondary antibody(1:1000, CST, USA) and the final chemiluminescence ECL (Millipore) detection of band. Protein bands were quantitated by densitometric analysis using Image Lab analysis software and shown as fold of the control after being normalized to GAPDH.
miRNA sequencing
Mouse NSCLC microRNA Solexa sequencing was performed by BGI Tech (Shenzhen, China).
Lentivirus construction and infection
A recombinant lentiviral expression vector (pLenti-miR-199a) with green fluorescence protein gene was constructed. To generate lentiviral particles, the recombinant expression plasmid was co-transfected with a packaging plasmid system (psPAX2 and pMD2G) into HEK-293T cells, and viral particles were collected 48 h after transfection. The A549 and H1299 cells were then infected with the miR-199a lentiviral vector or with a negative control vector without miR-199a (miR-NC) for 24 h. The infection efficiency was preliminary assessed in each experiment under a fluorescence microscope and then measured by sorting the positive cells of green fluorescence via flow cytometry ((Beckman Coulter, USA). The miRNA stably expressed cells were amplified and harvested for further experiments.
Tumor xenograft assay
Six-week-old nude mice were purchased from the Animal Center of the Cancer Institute of Chinese Academy of Medical Science (Shanghai, China) and randomly divided into 2 groups (5 mouse each group). The subcutaneous tumor xenograft model was established as described [25, 26]. Each mouse was subcutaneously injected with 2x106 SPC-A1 cells which stably expressed miR-199a-5p or control. The tumor volume was measured using formula volume = length × width2/2. At the end of the observation, the mouse was euthanized and tumors harvested for analyses. All experimental procedures were carried out under the protocols of school of life science, Shanghai University.
Clinical samples
The specimens were obtained from the Department of Oncology; Shanghai Chest Hospital affiliated to Shanghai Jiao Tong University (Shanghai, China) under ethical assessment. A total of 16 lung cancer tissue samples and 16 normal lung tissues were collected and processed for extraction of RNA.
Statistical analysis
Results were expressed as the mean ± standard error of the mean (SEM) and analyzed using Origin software. Student's t-test between different groups of each experiment and one-way analysis of variance (ANOVA) for three or more group comparisons was used to determine the significance of differences. P < 0.05 was considered statistically significant.
Results
MiR-199a-5p was down-regulated in mouse with lung cancer and human NSCLC tissues
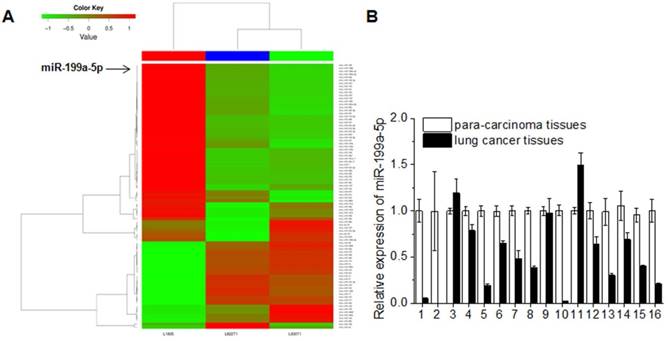
In order to investigate the role of miR-199a-5p in lung carcinogenesis, deep sequencing of mouse NSCLC tissues was performed and the data was processed by cluster analysis. The miRNAs read counts of L1805 (normal mouse lung tissues) less than or equal to 500 was excluded. The fold change limited as up-regulation was equal or greater than 2 fold and down-regulation was less than or equal to 0.5 fold compared to normal mouse lung tissues. miR-199a-5p expression was consistent down-regulation in K-ras+/+ and K-ras+/+/P53-/- lung cancer tissues (Fig. 1A). The expression level of miR-199a-5p in 16 cases NSCLC patients was also detected. The clinical-pathological features of 16 cases NSCLC patients were shown in Supplementary Table 1. Data showed that the miR-199a-5p was down-regulated in 13 cases (81% of total cases) when compared with the corresponding non-tumor lung tissues (Fig. 1B).
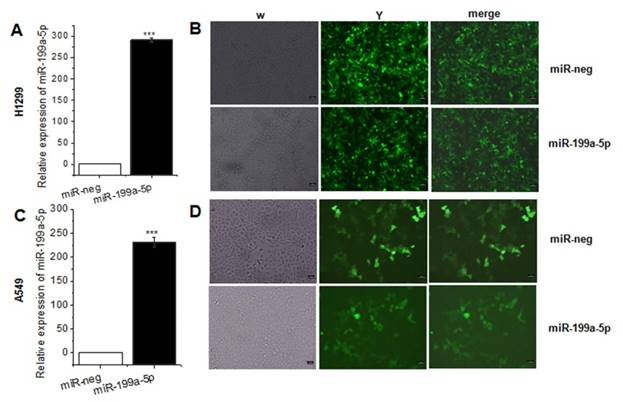
As the previously study presented, miR-199a-5p was downregulated in NSCLC tissues and cell line. For further study the function of miR-199a-5p in NSCLC, it was necessary to constructed miR-199a-5p over-expressed cell line. Lentivirus was packed in HEK-293T cells, and then infected A549, SPC-A1 and H1299 cells. The result of qRT-PCR assay revealed that miR-199a-5p was higher expressed after lentivirus infection (Fig. 2A, 2C and Supplementary Fig. 1) and the positive rates were nearly 99% (Fig. 2B, 2D).
MiR-199a-5p functions as a tumor suppressor in NSCLC
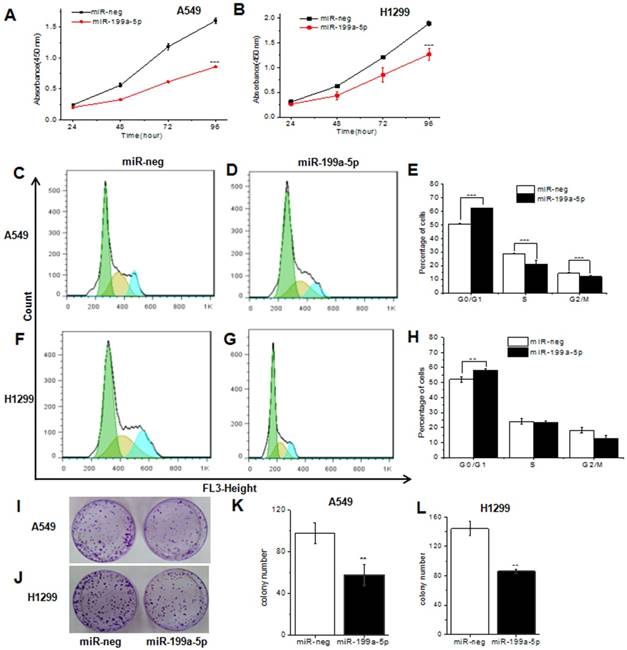
To further study the role of miR-199a-5p on the proliferation inhibition, we used CCK8 for examination. Results showed that the most striking inhibition with miR-199a-5p appeared compared with the miRNA-neg both in A549 and H1299 cells 96 h later (Fig. 3A and 3B).
MiR-199a-5p was down-expression both in mouse lung cancer tissues and human lung cancer tissues. A, deep-sequencing of mouse lung cancer model (L1805, normal lung tissues; L822T1, K-ras+/+ lung cancer tissues; L703T2, K-ras+/+/P53-/-lung cancer tissues) data was analyzed. Read counts of L1805 are equal or greater than 500 were limited. The up-expression of fold change >= 2 and down-expression<= 1/2 were showed. B, Expression of miR-199a-5p was detected by qRT-PCR in 16 lung cancer patient tissues. All experiments were at least repeated in triplicate with similar results. U6 was used as an internal control.
MiR-199a-5p was stable over-expression in lentivirus infected A549 cells and SPC-A1 cells. A, C the expression of miR-199a-5p was detected in A549 and SPC-A1 cells after lentivirus infected 72 hours respectively. U6 was used as an internal control. B, D Infection efficiency was observed in H1299 and A549 cells under fluorescence microscope. ***P<0.001.
In order to determine whether cell cycle arrest resulted in the proliferation inhibition, we next examined the cell cycle by flow-cytometry in miR-199a-5p stably overexpressed A549 and H1299. Results showed that the cell cycle of A549, SPC-A1 and H1299 cells infected with lenti-miR-199a-5p were significantly arrested in the G1 phase when compared to the control (Fig. 3C, 3D, 3F and 3G, Supplementary Fig. 2A and B). There was 62/57% G1 arrest in miR-199a-5p over-expressed A549 and H1299 cells while only 51/52% control cells were G1 arrest respectively. In addition, cell growth was attenuated by miR-199a-5p using monolayer colony formation assays (Fig. 3I and 3J, Supplementary Fig.2C and D). The significant inhibition was observed both in A549 cells (Fig. 3K), H1299 cells (Fig. 3L) and SPC-A1 cells (supplementary Fig. 2C and D) when compared with control. These data strongly suggest that miR-199a-5p contributes to suppression of NSCLC.
MiR-199a-5p negatively regulates MAPK pathway
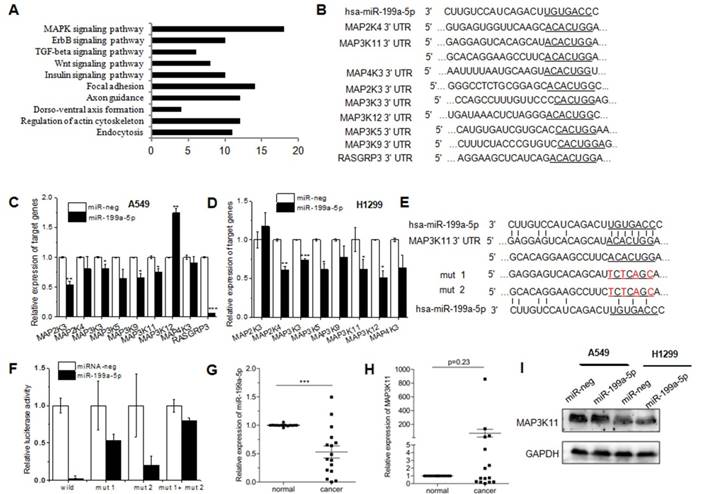
TargetScan and Pictar are two types of software broadly used online to predict the targets of miRNAs. To explore the downstream of miR-199a-5p signaling, TargetScan was used to predict the targets. Then KEGG pathway analysis was performed to analyze all the targets. T0P 10 pathway was showed in Figure 4A, and there were 18 genes clustered into MAPK pathway. Among the top potential targets list, 9 kinds of genes were chosen for further validation (Fig. 4B). qRT-PCR was performed to identify the level of target genes mRNA in miR-199a-5p stably expressed A549 and H1299 cells. Almost all nine genes showed the consistent tendency, except MAP3K12, MAP2K3 and RASGRP3. MAP3K12 demonstrated a significant up-regulation in A549 cells, while the opposite result was observed in H1299 cells. Similarly, MAP2K3 was down-regulated in A549 cells, but there was no difference in H1299 cells (Fig. 4C). The 3′UTR of 9 target genes was sub-cloned into the downstream of the firefly luciferase reporter gene. The results of luciferase reporter assay exhibited the silencing effect by miR-199a-5p only on MAP3K11-3′-UTR wild type. The predicted miR-199a-5p targeting region was further mutated in MAP3K11-3′-UTR as mentioned in Figure 4E. MAP3K11-3′UTR was abolished by mutating the miR-199a-5p seeding region on 3′UTRs, supporting that miR-199a-5p negatively regulates its targets MAP3K11by direct binding on their 3′UTR (Fig. 4F). Then miR-199a-5p and MAP3K11 expression was detected in 16 human patient samples. Negative correlation was observed between miR-199a-5p levels with MAP3K11 (Fig. 4G and H). The protein level of MAP3K11 was also decreased in miR-199a-5p over-expressed human NSCLC cell lines (Fig. 4I). All these data indicated that miR-199a-5p could directly target MAP3K11 by interaction with the 3′UTR of MAP3K11 gene in NSCLC cell line.
MiR-199a-5p could inhibit cell proliferation and cell cycle in NSCLC cell line. A, B The proliferation of A549 and SPC-A1 cell line was determined by CCK-8 assay respectively. C, D, F and G Cell cycle distribution measured by flow cytometry. Cell cycle analysis was performed in A549 and SPC-A1 cell by staining DNA with propidium iodide prior to flow cytometry. E, H The phase ratio (%) of cells was showed. I, J Representative images show the cell growth in A549 cells and SPC-A1 cells. K, L Average colonies in each well for each group were counted from three independent experiments. All experiments were at least repeated in triplicate with similar results. *P<0.05, **P<0.01, ***P<0.001.
MiR-199a-5p targets MAPK pathway in NSCLC cells. KEGG pathway analysis of differentially expressed proteins was shown. Pathway enrichment analysis was performed using the DAVID web application. B, Binding site sequences of MAPK pathway genes mRNA with miR-34a seed sequences were shown. C, D MAPK pathway gene was detected in miR-199a-5p stable over-expressed A549 and SPC-A1 cells. 18S was used as an internal control. E, The luciferase activity was significantly decreased in 293T cells transfected with pGL3-MAP3K11-3′-UTR. F, The luciferase activity was significantly decreased in 293T cells transfected with pGL3-MAP3K11-3′-UTR. G, H The mean expression level of miR-199a-5p and MAP3K11 were shown respectively. I, western-blot was performed to measure the protein level of MAP3K11 in miR-199a-5p over-expressed NSCLC cell lines. All experiments were at least repeated in triplicate with similar results.
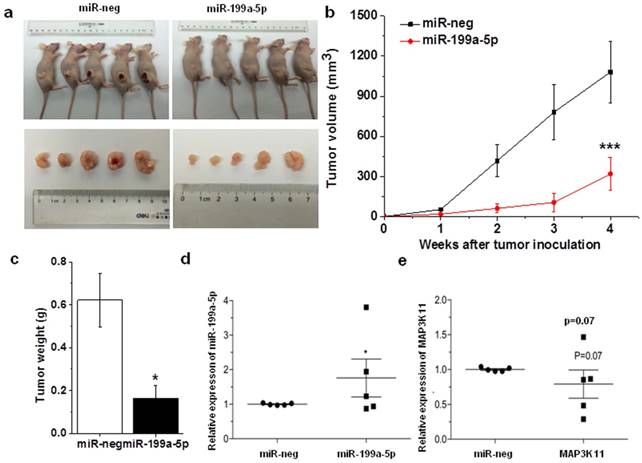
MiR-199a-5p inhibits tumor growth by targeting MAP3K11 in vivo
The antitumor effects of miR-199a-5p were evaluated in a xenograft mouse model using SPC-A1 cells. Stable expressed miR-199a-5p SPC-A1 cells and control (2×106 cells per injection site) were implanted into the flanks of 6-week old, female athymic mouse. Control cells formed tumors 7 d after implantation, while stably expressed miR-199a-5p SPC-A1 cells failed to grow 7 days after injection, and exhibited a marked reduction in tumor size at week 4 post-implantation compared to the control group (Fig. 5A, B and C) and the expression level of miR-199a-5p and MAP3K11 was detected in mouse tumor. Results showed that miR-199a-5p was downregulated, while the expression of MAP3K11 was elevated (Fig. 5D and E). Above results indicate that miR-199a-5p inhibits tumor growth by targeting MAP3K11 in vivo.
Discussion
The last decades were characterized by an extensive delineation of oncogenic pathways in different cancer types, focusing on protein-coding transcripts and more recently on non-coding RNA networks [27]. Typically, miRNAs are considered as tumor suppressors when down-regulated in tumors, or oncogenes when up-regulated in tumors [28, 29]. Recent evidence has revealed that miR-199a-5p plays an important role in the process of disease including tumorigenesis. MiR-199a-5p has been reported as a proliferation inhibition gene in Autosomal Dominant Polycystic Kidney Disease through targeting [30]. The present study showed that miR-199a is frequently down-regulated in HCC tissues and cells and target FZD7 [31]. MiRNA-199a-5p targets hypoxia-inducible factor-1alpha (HIF-1α), which has recently been implicated in the physiology of the hypoxic state and brain injury [22, 32-35].
To the best of our knowledge, the present results revealed for the first time that miR-199a-5p inhibited the proliferation in NSCLC. In the present study, results showed that miR-199a-5p is low expressed in lung cancer tissues compared with lung non-cancer tissues. Elevated level of miR-199a-5p inhibited the proliferation in H1299 and A549 cells. Although the expression level of all 9 MAPK relevant potential target genes was significantly down-regulated in miR-199a-5p stably expressed H1299 and A549 cell line, only MAP3K11 was identified as a target of miR-199a-5p in HEK-293T cells. Two binding regions of miR-199a-5p in MAP3K11 3′-UTR were given by TargetScan software. We testified both two sites can be targeted in truth. And the expression of MAP3K11 was significantly up-regulation in human patient samples, which indicated that the down-regulation miR-199a-5p attributes to the proliferation of NSCLC.
In conclusion, our results suggest that miR-199a-5p inhibits MAPK pathway by targeting MAP3K11 and inhibiting proliferation (Fig. 6). These observations demonstrated that miR-199a might provide a potential therapy for lung cancer patients.
Over-expression miR-199-5p affected tumor growth. A, SPC-A1 cells infected with lentivrius which stable express miR-199a-5p were injected into nude mice. 28 days after the injection, mice were photographed. B, Tumor sizes were measured every four days and growth curves were generated. C, Mouse was sacrificed after 28 days and then tumor weight were measured. D, E the expression of miR-199a-5p and MAP3K11 was detected in mouse xenografted tumor respectively. U6 was used as an internal control for miR-199a-5p. 18S was used as an internal control for MAP3K11. *P<0.05
MiR-199a-5p binds to MAP3K11 involved in the regulation of the MAPK signaling pathway.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by State Key Laboratory of Cell Biology and the National Natural Science Foundation of China (NO. 81601887).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Islami F, Ward EM, Jacobs EJ, Ma J, Goding Sauer A, Lortet-Tieulent J. et al. Potentially preventable premature lung cancer deaths in the USA if overall population rates were reduced to those of educated whites in lower-risk states. Cancer causes & control: CCC. 2015;26:409-18
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
3. Papadakis AI, Sun C, Knijnenburg TA, Xue Y, Grernrum W, Holzel M. et al. SMARCE1 suppresses EGFR expression and controls responses to MET and ALK inhibitors in lung cancer. Cell research. 2015
4. Qiu F, Yang L, Ling X, Yang R, Yang X, Zhang L. et al. Sequence variation in mature microRNA-499 confers unfavorable prognosis of lung cancer patients treated with platinum-based chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015
5. Zhang ZY, Fu SL, Xu SQ, Zhou X, Liu XS, Xu YJ. et al. By downregulating Ku80, hsa-miR-526b suppresses non-small cell lung cancer. Oncotarget. 2014
6. Mataki H, Seki N, Chiyomaru T, Enokida H, Goto Y, Kumamoto T. et al. Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. International journal of oncology. 2015;46:1039-50
7. Ryan BM, Robles AI, McClary AC, Haznadar M, Bowman ED, Pine SR. et al. Identification of a functional SNP in the 3'UTR of CXCR2 that is associated with reduced risk of lung cancer. Cancer research. 2015;75:566-75
8. Jeong HC. Clinical Aspect of MicroRNA in Lung Cancer. Tuberculosis and respiratory diseases. 2014;77:60-4
9. Janssen-Heijnen ML, van Erning FN, De Ruysscher DK, Coebergh JW, Groen HJ. Variation in causes of death in patients with non-small cell lung cancer according to stage and time since diagnosis. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2015
10. Guo Y, Liu J, Elfenbein SJ, Ma Y, Zhong M, Qiu C. et al. Characterization of the mammalian miRNA turnover landscape. Nucleic acids research. 2015
11. Polioudakis D, Abell NS, Iyer VR. miR-503 represses human cell proliferation and directly targets the oncogene DDHD2 by non-canonical target pairing. BMC genomics. 2015;16:40
12. Quan L, Qiu T, Liang J, Li M, Zhang Y, Tao K. Identification of Target Genes Regulated by KSHV miRNAs in KSHV-Infected Lymphoma Cells. Pathology oncology research: POR. 2015
13. Guo R, Gu J, Zhang Z, Wang Y, Gu C. MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB life. 2015
14. Bouamar H, Jiang D, Wang L, Lin AP, Ortega M, Aguiar RC. MicroRNA-155 control of p53 activity is context dependent and mediated by Aicda and Socs1. Molecular and cellular biology. 2015
15. Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F. et al. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene. 2015
16. Xin C, Liu W, Lin Q, Zhang X, Cui P, Li F. et al. Profiling microRNA expression during multi-staged date palm (Phoenix dactylifera L.) fruit development. Genomics. 2015
17. Yang J, Zeng Y. Identification of miRNA-mRNA crosstalk in pancreatic cancer by integrating transcriptome analysis. European review for medical and pharmacological sciences. 2015;19:825-34
18. Cloonan N. Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery. BioEssays: news and reviews in molecular, cellular and developmental biology. 2015;37:379-88
19. Yang B, Liu B, Bi P, Wu T, Wang Q, Zhang J. An integrated analysis of differential miRNA and mRNA expressions in human gallstones. Molecular bioSystems. 2015;11:1004-11
20. Uso M, Jantus-Lewintre E, Sirera R, Bremnes RM, Camps C. miRNA detection methods and clinical implications in lung cancer. Future Oncol. 2014;10:2279-92
21. Kim BK, Yoo HI, Kim I, Park J, Kim Yoon S. FZD6 expression is negatively regulated by miR-199a-5p in human colorectal cancer. BMB reports. 2015
22. He J, Wang M, Jiang Y, Chen Q, Xu S, Xu Q. et al. Chronic arsenic exposure and angiogenesis in human bronchial epithelial cells via the ROS/miR-199a-5p/HIF-1alpha/COX-2 pathway. Environmental health perspectives. 2014;122:255-61
23. Ding G, Huang G, Liu HD, Liang HX, Ni YF, Ding ZH. et al. MiR-199a suppresses the hypoxia-induced proliferation of non-small cell lung cancer cells through targeting HIF1alpha. Molecular and cellular biochemistry. 2013;384:173-80
24. el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K. et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell metabolism. 2013;18:341-54
25. Zhu G, Hong L, Wang S, Junhong C, Yang Z, Yao M. Comparison of two kinds of orthotopic xenograft models for human ovarian cancer. European journal of gynaecological oncology. 2014;35:724-7
26. Pan H, Ma Z, Mao L. [Establishment of nude mouse models of patient-derived lung cancer xenograft]. Zhonghua zhong liu za zhi [Chinese journal of oncology]. 2014;36:571-4
27. Kilic ID, Dodurga Y, Uludag B, Alihanoglu YI, Yildiz BS, Enli Y. et al. microRNA -143 and -223 in obesity. Gene. 2015
28. Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP. et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Medical oncology. 2013;30:477
29. Mito JK, Min HD, Ma Y, Carter JE, Brigman BE, Dodd L. et al. Oncogene-dependent control of miRNA biogenesis and metastatic progression in a model of undifferentiated pleomorphic sarcoma. The Journal of pathology. 2013;229:132-40
30. Sun L, Zhu J, Wu M, Sun H, Zhou C, Fu L. et al. Inhibition of MiR-199a-5p reduced cell proliferation in autosomal dominant polycystic kidney disease through targeting CDKN1C. Medical science monitor: international medical journal of experimental and clinical research. 2015;21:195-200
31. Song J, Gao L, Yang G, Tang S, Xie H, Wang Y. et al. MiR-199a regulates cell proliferation and survival by targeting FZD7. PloS one. 2014;9:e110074
32. Jiang G, Zhou R, He X, Shi Z, Huang M, Yu J. et al. Expression levels of microRNA-199 and hypoxia-inducible factor-1 alpha in brain tissue of patients with intractable epilepsy. The International journal of neuroscience. 2014:1-29
33. Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D. et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget. 2014;5:3039-54
34. Yang X, Lei S, Long J, Liu X, Wu Q. MicroRNA-199a-5p inhibits tumor proliferation in melanoma by mediating HIF-1alpha. Molecular medicine reports. 2016;13:5241-7
35. Jiang G, Zhou R, He X, Shi Z, Huang M, Yu J. et al. Expression levels of microRNA-199 and hypoxia-inducible factor-1 alpha in brain tissue of patients with intractable epilepsy. The International journal of neuroscience. 2016;126:326-34
Author contact
![]() Corresponding authors: Email: zlmaedu.cn &: jhyuhuanshacom
Corresponding authors: Email: zlmaedu.cn &: jhyuhuanshacom

 Global reach, higher impact
Global reach, higher impact